The Need and Opportunity to Update the Inventory of Plant Pathogenic Fungi and Oomycetes in Mexico
Abstract
1. Introduction
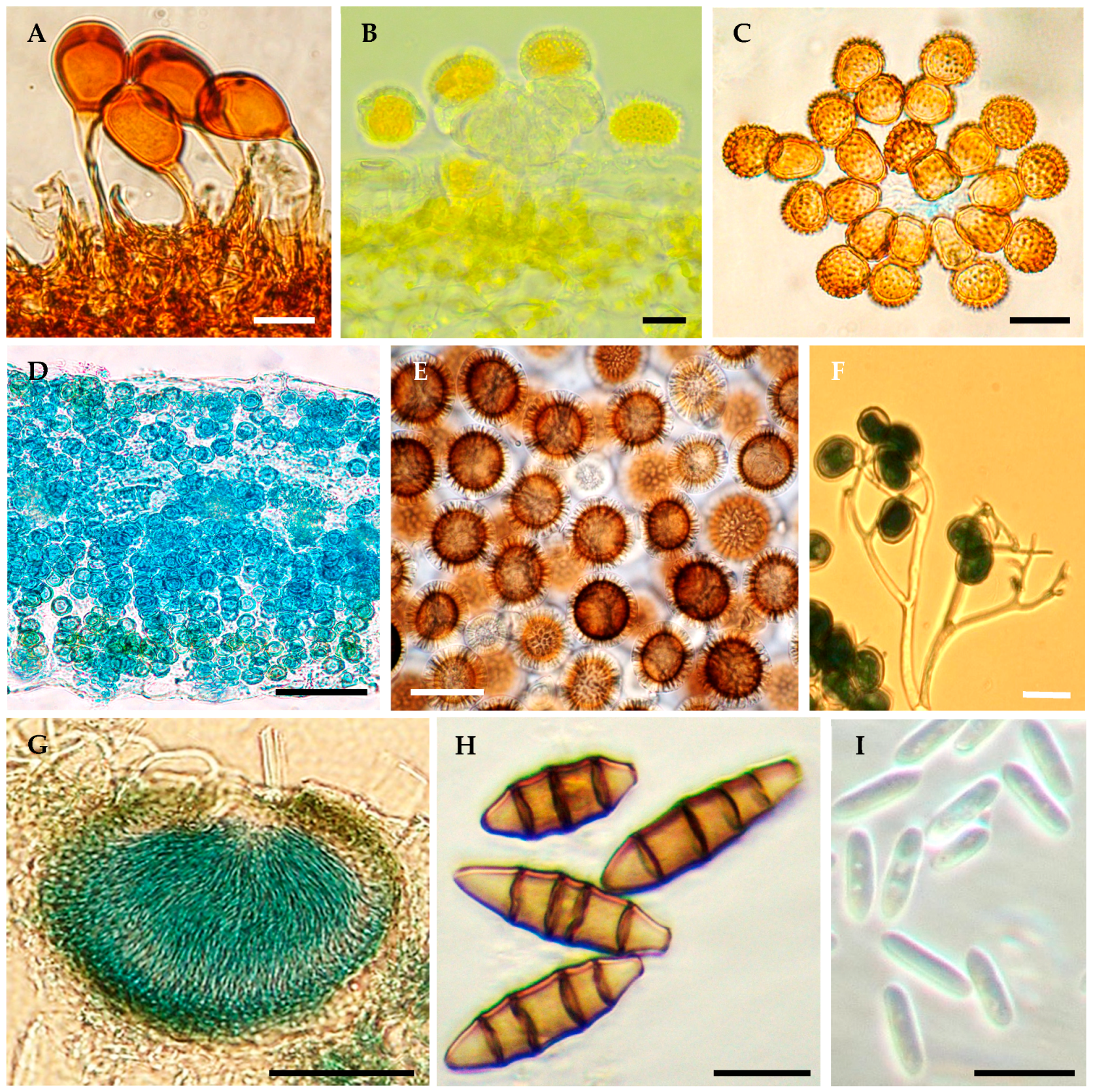
2. Inventory of Plant Pathogenic Fungi and Oomycetes in Mexico
- (a)
- Reports of diseases and phytopathogens based on research that includes morphological characterization, DNA sequence analysis, and verification of pathogenicity through artificial inoculations, except for some obligate parasites. Generally, these studies report the presence of a fungal or oomycete phytopathogen based on the international norms established in ISPM8 [42], so they should be considered valid reports of the presence of the pathogen in a new territory.
- (b)
- Lists of diseases and plant pathogens based on literature reviews of studies published in scientific journals and books in the country. León-Gallegos (1978) [43], in his book titled “Enfermedades de cultivos en el Estado de Sinaloa”, conducted a literature review and compiled information regarding the symptoms and control of the main diseases caused by fungi, oomycetes, viruses, and bacteria in various economically important crops. However, it cannot be considered a conclusive report on the presence of such species of plant pathogens, as it lacks the elements described in the previous section. Another example is found in the book “Hongos Fitopatógenos” published by Romero-Cova (1988) [44], which describes the main morphological characteristics used for the identification of various fungi and oomycetes affecting major economically important crops. The work also mentions their distribution, host range, life cycle, and control. However, much of the taxonomic information presented in this work is based solely on a compilation of keys and descriptions from specialized books or journals, so the information provided by this work also does not comply with ISPM 8, and the reports should not be considered valid. However, it is worth noting that the book provides detailed morphological identification of various specimens supported by excellent photomicrographs. More recently, Fernández-Pavía et al. (2015) [45] published a book titled “Enfermedades de Especies Vegetales en México”, which presents an excellent compilation of publications where the presence of plant pathogens in Mexico was recorded or mentioned. However, in this book, some lists and catalogs of fungal and oomycete pathogens were considered, which were not identified through morphological characterization and whose pathogenicity was not verified. Therefore, this source of information should be considered with some reservations.
- (c)
- Lists or catalogs of diseases and plant pathogens without scientific support. These are reports that were not verified by morphology, nor confirmed through DNA sequence analysis, and certainly not subjected to pathogenicity tests. One example is the “Primer catálogo de enfermedades de plantas mexicanas” published by García-Alvarez (1976) [46], which is one of the most extensive lists of plant diseases in Mexico; however, this catalog was generated without scientific evidence or support. Ayala-Escobar et al. (2013), Solano-Báez et al. (2017), Márquez-Licona et al. (2018), and Nieto-López et al. (2018) [47,48,49,50] mentioned that the catalog published by García-Alvarez (1976) [46] lacks morphological characterization data of the causal agents mentioned in the list. Consequently, this list does not comply with ISPM 8 [42], and should not be considered an official report of plant pathogenic species in Mexico.
3. Plant Pathogen Fungal Collections and Plant Pathology Herbaria
4. Reports of Plant Pathogens in Mexico and Their Implications in International Databases
5. Conclusions
- (a)
- Distribution of agricultural important fungi and oomycetes in Mexico.
- (b)
- Detailed information (common name, species, and variety or cultivar of the host plant; scientific name of the pathogen; geographic coordinates, date of collection, collector’s name, identifier’s name, and photographs of symptoms in the field; microphotographs of characteristic structures of the pathogenic fungus, etc.)
- (c)
- Assessment and recording of damage and impacts, by regions or climatic conditions.
- (d)
- Prevalence of the pest in the regional context, and epidemiological studies.
- (e)
- Ease of exchanging specimens of fungi and oomycetes with herbaria or plant pathogens collections from other countries.
- (f)
- Monitoring of fungicide resistance in reference populations of fungi.
- (g)
- Modification or adaptation of biosecurity measures in Mexico.
- (h)
- Evaluation of tolerance or resistance of varieties or cultivars of plant species.
- (i)
- Population structure of fungal and oomycete species.
- (j)
- Studies on comparative genomics, transcriptomics, proteomics, and metabolomics.
6. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bandyopadhyay, R.; Fredericksen, R.A. Contemporary Global Movement of Emerging Plant Diseases. Ann. N. Y. Acad. Sci. 1999, 894, 28–36. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, A.; Pautasso, M.; Jeger, M.J.; Haines-Young, R. Evolution of the international regulation of plant pests and challenges for future plant health. Food Secur. 2010, 2, 49–70. [Google Scholar] [CrossRef]
- Smith, J.J.; Waage, J.; Woodhall, J.W.; Bishop, S.J.; Spence, N.J. The challenge of providing plant pest diagnostic services for Africa. Eur. J. Plant Pathol. 2008, 121, 365–375. [Google Scholar] [CrossRef]
- Gurjar, G.; Kanade, M. Analysis of phytopathogenic fungi isolated from some important crop plants using morpho-molecular tools—Foldscope and ITS region sequencing. Mycol. Prog. 2020, 19, 1475–1493. [Google Scholar] [CrossRef]
- Peng, Y.; Li, S.J.; Yan, J.; Tang, Y.; Cheng, J.P.; Gao, A.J.; Yao, X.; Ruan, J.J.; Xu, B.L. Research Progress on Phytopathogenic Fungi and Their Role as Biocontrol Agents. Front. Microbiol. 2021, 12, 670135. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Akhtar, J.; Kandan, A.; Kumar, S.; Batra, R.; Dubey, S.C. Advance Detection Techniques of Phytopathogenic Fungi: Current Trends and Future Perspectives. In Current Trends in Plant Disease Diagnostics and Management Practices; Kumar, P., Gupta, V.K., Tiwari, A.K., Kamle, M., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 265–298. [Google Scholar]
- McCartney, H.A.; Foster, S.J.; Fraaije, B.A.; Ward, E. Molecular diagnostics for fungal plant pathogens. Pest Manag. Sci. 2003, 59, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Ray, M.; Ray, A.; Dash, S.; Mishra, A.; Achary, K.G.; Nayak, S.; Singh, S. Fungal disease detection in plants: Traditional assays, novel diagnostic techniques and biosensors. Biosens. Bioelectron. 2017, 87, 708–723. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, R.S.; Hyde, K.D.; De Farias, A.R.G.; Bhunjun, C.S.; Ferdinandez, H.S.; Manamgoda, D.S.; Udayanga, D.; Herath, I.S.; Thambugala, K.M.; Manawasinghe, I.S.; et al. What is a species in fungal plant pathogens? Fungal Divers. 2021, 109, 239–266. [Google Scholar] [CrossRef]
- Chethana, K.W.T.; Manawasinghe, I.S.; Hurdeal, V.G.; Bhunjun, C.S.; Appadoo, M.A.; Gentekaki, E.; Raspé, O.; Promputtha, I.; Hyde, K.D. What are fungal species and how to delineate them? Fungal Divers. 2021, 109, 1–25. [Google Scholar] [CrossRef]
- Crous, P.W.; Groenewald, J.Z.; Slippers, B.; Wingfield, M.J. Global food and fibre security threatened by current inefficiencies in fungal identification. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20160024. [Google Scholar] [CrossRef]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.L.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef]
- Taylor, J.W.; Jacobson, D.J.; Kroken, S.; Kasuga, T.; Geiser, D.M.; Hibbett, D.S.; Fisher, M.C. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 2000, 31, 21–32. [Google Scholar] [CrossRef]
- Cai, L.; Udayanga, D.; Manamgoda, D.S.; Maharachchikumbura, S.S.N.; McKenzie, E.H.C.; Guo, L.D.; Liu, X.Z.; Bahkali, A.; Hyde, K.D. The need to carry out re-inventory of plant pathogenic fungi. Trop. Plant Pathol. 2011, 36, 205–213. [Google Scholar] [CrossRef]
- Crous, P.W.; Hawksworth, D.L.; Wingfield, M.J. Identifying and Naming Plant-Pathogenic Fungi: Past, Present, and Future. Annu. Rev. Phytopathol. 2015, 53, 247–267. [Google Scholar] [CrossRef]
- Ko Ko, T.; Mckenzie, E.; Bahkali, A.; To-Anun, C.; Chukeatirote, E.; Promputtha, I.; Abd-Elsalam, K.A.; Soytong, K.; Wulandari, N.; Sanoamuang, N.; et al. The need for re-inventory of Thai phytopathogens. Chiang Mai J. Sci. 2011, 38, 625–637. [Google Scholar]
- Marin-Felix, Y.; Groenewald, J.Z.; Cai, L.; Chen, Q.; Marincowitz, S.; Barnes, I.; Bensch, K.; Braun, U.; Camporesi, E.; Damm, U.; et al. Genera of phytopathogenic fungi: GOPHY 1. Stud. Mycol. 2017, 86, 99–216. [Google Scholar] [CrossRef]
- Abad, Z.G.; Burgess, T.I.; Redford, A.J.; Bienapfl, J.C.; Srivastava, S.; Mathew, R.; Jennings, K. IDphy: An International Online Resource for Molecular and Morphological Identification of Phytophthora. Plant Dis. 2023, 107, 987–998. [Google Scholar] [CrossRef]
- Dugan, F.M.; Glawe, D.A.; Attanayake, R.N.; Chen, W. The Importance of Reporting New Host-Fungus Records for Ornamental and Regional Crops. Plant Health Prog. 2009, 10, 34. [Google Scholar] [CrossRef]
- Hyde, K.D.; Chomnunti, P.; Crous, P.W.; Groenewald, J.Z.; Damm, U.; Ko, T.W.K.; Shivas, R.G.; Summerell, B.A.; Tan, Y.P. A case for re-inventory of Australia’s plant pathogens. Persoonia Mol. Phylogeny Evol. Fungi 2010, 25, 50–60. [Google Scholar] [CrossRef]
- Drábková, L.Z. DNA Extraction from Herbarium Specimens. In Molecular Plant Taxonomy: Methods and Protocols; Besse, P., Ed.; Humana Press: Totowa, NJ, USA, 2014; pp. 69–84. [Google Scholar]
- Savolainen, V.; Cuénoud, P.; Spichiger, R.; Martinez, M.D.P.; Crèvecoeur, M.; Manen, J.-F. The use of herbarium specimens in DNA phylogenetics: Evaluation and improvement. Plant Syst. Evol. 1995, 197, 87–98. [Google Scholar] [CrossRef]
- Hyde, K.; Udayanga, D.; Manamgoda, D.; Tedersoo, L.; Larsson, E.; Abarenkov, K.; Bertrand, Y.; Oxelman, B.; Hartmann, M.; Kauserud, H.; et al. Incorporating molecular data in fungal systematics: A guide for aspiring researchers. Curr. Res. Environ. Appl. Mycol. 2013, 3, 1–32. [Google Scholar] [CrossRef]
- Hariharan, G.; Prasannath, K. Recent Advances in Molecular Diagnostics of Fungal Plant Pathogens: A Mini Review. Front. Cell. Infect. Microbiol. 2021, 10, 600234. [Google Scholar] [CrossRef]
- Kang, S.; Blair, J.E.; Geiser, D.M.; Khang, C.-H.; Park, S.-Y.; Gahegan, M.; O’Donnell, K.; Luster, D.G.; Kim, S.H.; Ivors, K.L.; et al. Plant Pathogen Culture Collections: It Takes a Village to Preserve These Resources Vital to the Advancement of Agricultural Security and Plant Pathology. Phytopathology 2006, 96, 920–925. [Google Scholar] [CrossRef]
- Abd-Elsalam, K.A.; Yassin, M.A.; Moslem, M.A.; Bahkali, A.H.; De Wit, P.J.G.M.; McKenzie, E.H.C.; Stephenson, S.L.; Cai, L.; Hyde, K.D. Culture collections, the new herbaria for fungal pathogens. Fungal Divers. 2010, 45, 21–32. [Google Scholar] [CrossRef]
- Sette, L.D.; Pagnocca, F.C.; Rodrigues, A. Microbial culture collections as pillars for promoting fungal diversity, conservation and exploitation. Fungal Genet. Biol. 2013, 60, 2–8. [Google Scholar] [CrossRef]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Martinelli, F.; Scalenghe, R.; Davino, S.; Panno, S.; Scuderi, G.; Ruisi, P.; Villa, P.; Stroppiana, D.; Boschetti, M.; Goulart, L.R.; et al. Advanced methods of plant disease detection. A review. Agron. Sustain. Dev. 2015, 35, 1–25. [Google Scholar] [CrossRef]
- Miller, S.A.; Beed, F.D.; Harmon, C.L. Plant Disease Diagnostic Capabilities and Networks. Annu. Rev. Phytopathol. 2009, 47, 15–38. [Google Scholar] [CrossRef]
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010, 72, 1–13. [Google Scholar] [CrossRef]
- Aragona, M.; Haegi, A.; Valente, M.T.; Riccioni, L.; Orzali, L.; Vitale, S.; Luongo, L.; Infantino, A. New-Generation Sequencing Technology in Diagnosis of Fungal Plant Pathogens: A Dream Comes True? J. Fungi 2022, 8, 737. [Google Scholar] [CrossRef]
- Baroncelli, R.; Cafà, G. Genomic sequences for fungi. CABI 2021, 231–254. [Google Scholar] [CrossRef]
- Crous, P.W.; Rossman, A.Y.; Aime, M.C.; Allen, W.C.; Burgess, T.; Groenewald, J.Z.; Castlebury, L.A. Names of Phytopathogenic Fungi: A Practical Guide. Phytopathology 2021, 111, 1500–1508. [Google Scholar] [CrossRef]
- Braun, U. The impacts of the discontinuation of dual nomenclature of pleomorphic fungi: The trivial facts, problems, and strategies. IMA Fungus 2012, 3, 81–86. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Crous, P.W.; Redhead, S.A.; Reynolds, D.R.; Samson, R.A.; Seifert, K.A.; Taylor, J.W.; Wingfield, M.J.; Abaci, Ö.; Aime, C.; et al. The Amsterdam Declaration on Fungal Nomenclature. IMA Fungus 2011, 2, 105–111. [Google Scholar] [CrossRef]
- Redhead, S.A.; Demoulin, V.; Hawksworth, D.L.; Seifert, K.A.; Turland, N.J. Fungal Nomenclature at IMC10: Report of the Nomenclature Sessions. IMA Fungus 2014, 5, 449–462. [Google Scholar] [CrossRef]
- Wingfield, M.J.; De Beer, Z.W.; Slippers, B.; Wingfield, B.D.; Groenewald, J.Z.; Lombard, L.; Crous, P.W. One fungus, one name promotes progressive plant pathology. Mol. Plant Pathol. 2012, 13, 604–613. [Google Scholar] [CrossRef]
- Chen, Q.; Bakhshi, M.; Balci, Y.; Broders, K.D.; Cheewangkoon, R.; Chen, S.F.; Fan, X.L.; Gramaje, D.; Halleen, F.; Jung, M.H.; et al. Genera of phytopathogenic fungi: GOPHY 4. Stud. Mycol. 2022, 101, 417–564. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Hernández-Restrepo, M.; Iturrieta-González, I.; García, D.; Gené, J.; Groenewald, J.Z.; Cai, L.; Chen, Q.; Quaedvlieg, W.; Schumacher, R.K.; et al. Genera of phytopathogenic fungi: GOPHY 3. Stud. Mycol. 2019, 94, 1–124. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Hernández-Restrepo, M.; Wingfield, M.J.; Akulov, A.; Carnegie, A.J.; Cheewangkoon, R.; Gramaje, D.; Groenewald, J.Z.; Guarnaccia, V.; Halleen, F.; et al. Genera of phytopathogenic fungi: GOPHY 2. Stud. Mycol. 2019, 92, 47–133. [Google Scholar] [CrossRef] [PubMed]
- FAO. NIMF 8: Determinación de la Situación de Una Plaga en Un Área; Normas Internacionales para Medidas Fitosanitarias; Convención Internacional de Protección Fitosanitaria (CIPF): Rome, Italy, 1996. [Google Scholar]
- León Gallegos, H.M. Enfermedades de Cultivos en el Estado de Sinaloa; Instituto Nacional de Investigaciones Agricolas: Ciudad de Mexico, Mexico, 1978. [Google Scholar]
- Romero-Cova, S. Hongos Fitopatógenos; Universidad Autónoma Chapingo: Texcoco, Mexico, 1988; p. 347. [Google Scholar]
- Fernández-Pavía, S.P.; Gregorio-Cipriano, R.; Rodríguez-Alvarado, G.; Fernández-Pavía, Y.L.; Mondragón-Flores, A.; Gómez-Dorantes, N.; Lozoya-Saldaña, H.; Rodríguez-Fernández, R.; Herrera-Camacho, J. Enfermedades de Especies Vegetales en México; Universidad Michoacana de San Nicolás de Hidalgo: Morelia, Mexico, 2015. [Google Scholar]
- García-Alvarez, M. Primer catálogo de enfermedades de plantas mexicanas. Fitófilo 1976, 71, 35–37. [Google Scholar]
- Ayala-Escobar, V.; Gomez-Jaimes, R.; Santiago-Santiago, V.; Madariaga-Navarrete, A.; Castaneda-Vildozola, A.; Nava-Diaz, C. Pseudocercospora cruenta on Vigna unguiculata in Mexico. Australas. Plant Dis. Notes 2013, 8, 115–116. [Google Scholar] [CrossRef]
- Márquez-Licona, G.; Solano-Báez, A.; Pérez-López, M.; Leyva-Mir, S.; Tovar-Pedraza, J. Erysiphe australiana causing powdery mildew on crape myrtle (Lagerstroemia indica) in Mexico. Plant Pathol. Quar. 2018, 8, 48–51. [Google Scholar] [CrossRef]
- Nieto-López, E.H.; Everhart, S.E.; Ayala-Escobar, V.; Camacho-Tapia, M.; Lima, N.B.; Nieto-Angel, R.; Tovar-Pedraza, J.M. First Report of Colletotrichum gloeosporioides Causing Anthracnose of Tejocote (Crataegus gracilior) Fruits in Mexico. Plant Dis. 2018, 102, 1855. [Google Scholar] [CrossRef]
- Solano-Báez, A.; Jiménez-Jiménez, B.; Camacho-Tapia, M.; Leyva-Mir, S.; Nieto-López, E.; Tovar-Pedraza, J. First confirmed report of fig rust caused by Cerotelium fici in Mexico. Plant Pathol. Quar. 2017, 7, 160–163. [Google Scholar] [CrossRef]
- Cotterill, F.P.D. Systematics, biological knowledge and environmental conservation. Biodivers. Conserv. 1995, 4, 183–205. [Google Scholar] [CrossRef]
- De Vero, L.; Boniotti, M.B.; Budroni, M.; Buzzini, P.; Cassanelli, S.; Comunian, R.; Gullo, M.; Logrieco, A.F.; Mannazzu, I.; Musumeci, R.; et al. Preservation, Characterization and Exploitation of Microbial Biodiversity: The Perspective of the Italian Network of Culture Collections. Microorganisms 2019, 7, 685. [Google Scholar] [CrossRef]
- Wen, J.; Ickert-Bond, S.M.; Appelhans, M.S.; Dorr, L.J.; Funk, V.A. Collections-based systematics: Opportunities and outlook for 2050. J. Syst. Evol. 2015, 53, 477–488. [Google Scholar] [CrossRef]
- Card, D.C.; Shapiro, B.; Giribet, G.; Moritz, C.; Edwards, S.V. Museum Genomics. Annu. Rev. Genet. 2021, 55, 633–659. [Google Scholar] [CrossRef]
- Llorente-Bousquets, J.; Koleff-Osorio, P.; Benítez-Díaz, H.; Lara-Morales, L. Síntesis del estado de las colecciones biológicas mexicanas. In Resultados de la Encuesta “Inventario y Diagnóstico de la Actividad Taxonómica en México” 1996–1998; Comisión Nacional para el Conocimiento y Uso de la Biodiversidad: Tlalpan, Mexico, 1999. [Google Scholar]
- Dáttilo, W.; Vásquez-Bolaños, M.; Ahuatzin, D.A.; Antoniazzi, R.; Chávez-González, E.; Corro, E.; Luna, P.; Guevara, R.; Villalobos, F.; Madrigal-Chavero, R.; et al. Mexico ants: Incidence and abundance along the Nearctic–Neotropical interface. Ecology 2020, 101, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Luna-Vega, I.; Espinosa, D.; Rivas, G.; Contreras-Medina, R. Geographical patterns and determinants of species richness in Mexico across selected families of vascular plants: Implications for conservation. Syst. Biodivers. 2013, 11, 237–256. [Google Scholar] [CrossRef]
- CONABIO. La Diversidad Biológica de México: Estudio de País, 1998; Comisión Nacional para el Conocimiento y Uso de la Biodiversidad: Tlalpan, Mexico, 1998; p. 341. [Google Scholar]
- Aguirre-Acosta, E.; Ulloa, M.; Aguilar, S.; Cifuentes, J.; Valenzuela, R. Biodiversidad de hongos en México. Rev. Mex. Biodivers. 2014, 85, 76–81. [Google Scholar] [CrossRef]
- Guzmán, G. Las colecciones de hongos en México y su problemática en la biodiversidad del país. Boletín Soc. Botánica México 1994, 55, 35–37. [Google Scholar] [CrossRef]
- Koleff, P.; Camargo, D.; Hernández-Robles, D.; Lara, M.; Sandoval, O.; Ocegueda, S.; Urquiza-Haas, T. Anexo II: Colecciones Biológicas Institucionales por Entidad Federativa. In Capital Natural de México (Segundo Estudio de País); Comisión Nacional para el Conocimiento y Uso de la Biodiversidad: Tlalpan, Mexico, 2016; Volume IV, pp. 539–564. [Google Scholar]
- CONABIO. Colecciones Biológicas Científicas de México; CONABIO: Tlalpan, Mexico, 2012. [Google Scholar]
- Crous, P.; Verkley, G.; Groenewald, J.; Houbraken, J. Fungal Biodiversity, 2nd ed.; CBS-KNAW Fungal Biodiversity Centre: Utrecht, The Netherlands, 2019; p. 425. [Google Scholar]
- Sharma, S.K.; Saini, S.; Verma, A.; Sharma, P.K.; Lal, R.; Roy, M.; Singh, U.B.; Saxena, A.K.; Sharma, A.K. National Agriculturally Important Microbial Culture Collection in the Global Context of Microbial Culture Collection Centres. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2019, 89, 405–418. [Google Scholar] [CrossRef]
- Shivas, R.G.; Beasley, D.R.; McTaggart, A.R. Online identification guides for Australian smut fungi (Ustilaginomycotina) and rust fungi (Pucciniales). IMA Fungus 2014, 5, 195–202. [Google Scholar] [CrossRef]
- Hyde, K.D.; Nilsson, R.H.; Alias, S.A.; Ariyawansa, H.A.; Blair, J.E.; Cai, L.; De Cock, A.W.A.M.; Dissanayake, A.J.; Glockling, S.L.; Goonasekara, I.D.; et al. One stop shop: Backbones trees for important phytopathogenic genera: I (2014). Fungal Divers. 2014, 67, 21–125. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Hyde, K.D.; Jeewon, R.; Ghobad-Nejhad, M.; Wanasinghe, D.N.; Liu, N.; Phillips, A.J.L.; Oliveira-Filho, J.R.C.; Da Silva, G.A.; Gibertoni, T.B.; et al. One stop shop II: Taxonomic update with molecular phylogeny for important phytopathogenic genera: 26–50 (2019). Fungal Divers. 2019, 94, 41–129. [Google Scholar] [CrossRef]
- Staats, M.; van Baarlen, P.; Schouten, A.; van Kan, J.A.; Bakker, F.T. Positive selection in phytotoxic protein-encoding genes of Botrytis species. Fungal Genet. Biol. 2007, 44, 52–63. [Google Scholar] [CrossRef]
- Staats, M.; van Baarlen, P.; van Kan, J.A. Molecular phylogeny of the plant pathogenic genus Botrytis and the evolution of host specificity. Mol. Biol. Evol. 2005, 22, 333–346. [Google Scholar] [CrossRef]
- Cannon, P.F.; Damm, U.; Johnston, P.R.; Weir, B.S. Colletotrichum—Current status and future directions. Stud. Mycol. 2012, 73, 181–213. [Google Scholar] [CrossRef]
- Braun, U.; Cook, R. Taxonomic Manual of the Erysiphales (Powdery Mildews); CBS-KNAW Fungal Biodiversity Centre: Utrecht, The Netherlands, 2012; p. 707. [Google Scholar]
- Takamatsu, S.; Niinomi, S.; Harada, M.; Havrylenko, M. Molecular phylogenetic analyses reveal a close evolutionary relationship between Podosphaera (Erysiphales: Erysiphaceae) and its rosaceous hosts. Persoonia 2010, 24, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Lombard, L.; Sandoval-Denis, M.; Seifert, K.A.; Schroers, H.J.; Chaverri, P.; Gené, J.; Guarro, J.; Hirooka, Y.; Bensch, K.; et al. Fusarium: More than a node or a foot-shaped basal cell. Stud. Mycol. 2021, 98, 100116. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, A.; Phillips, A.; Li, X.; Hyde, K. Botryosphaeriaceae: Current status of genera and species. Mycosphere 2016, 7, 1001–1073. [Google Scholar] [CrossRef]
- FAO. NIMF 11: Análisis de Riesgo de Plagas para Plagas Cuarentenarias; Normas Internacionales para Medidas Fitosanitarias; Convención Internacional de Protección Fitosanitaria (CIPF): Rome, Italy, 2013. [Google Scholar]
- Burgman, M.; Roberts, B.; Sansford, C.; Griffin, R.; Mengersen, K. The Role of Pest Risk Analysis in Plant Biosecurity; Springer: Dordrecht, The Netherlands, 2014; pp. 235–267. [Google Scholar]
- Soliman, T.; Mourits, M.C.M.; Oude Lansink, A.G.J.M.; van der Werf, W. Economic impact assessment in pest risk analysis. Crop Prot. 2010, 29, 517–524. [Google Scholar] [CrossRef]

| Pathogen | Marker for Genus-Level Identification | Marker for Species-Level Identification | Reference |
|---|---|---|---|
| Alternaria | LSU, SSU | ITS, GAPDH, RPB2, TEF1 | [67] |
| Bipolaris | LSU | ITS, GAPDH, TEF1 | [17,66,67] |
| Botrytis | ITS | GAPDH, RPB2, HSP60, NEP1, NEP2 | [66,68,69] |
| Cladosporium | LSU | ACT, TEF1, TUB2 | [17] |
| Colletotrichum | ITS | ACT, APMAT, APN2, CAL, CHS-1, GAPDH, GS, HIS3, SOD2, TUB2 | [17,66,70] |
| Curvularia | LSU, ITS | ITS, GAPDH, TEF | [17,66,67] |
| Diaporthe | ITS | CAL, HIS3, TEF1, TUB2 | [41,66] |
| Elsinoe | ITS | RPB2, TEF1 | [67] |
| Erysiphacea | ITS | ITS, LSU | [71,72] |
| Entyloma | ITS | ITS | [67] |
| Fusarium | ITS | TEF1, RPB1, RPB2, TUB2, cmdA | [73] http://isolate.fusariumdb.org/ (accessed on 25 May 2024) |
| Gaeumannomyces | LSU | ITS, TEF1, RPB1 | [41] |
| Lasiodiplodia | ITS | TEF1, TUB2 | [66,74] |
| Monilinia | ITS | TEF1 | [17] |
| Neofusicoccum | SSU, LSU | ITS, TEF1, TUB2, RPB2 | [17,66,67,74] |
| Phyllosticta | LSU | ITS, ACT, GAPDH, TEF1 | [41,66,67] |
| Phytophthora | ITS | LSU, TUB2, COX2, NAD9, RPS10 | [66] www.phytophthoradb.org (accessed on 25 May 2024) |
| Puccinia | ITS | ITS, LSU | [17,66] |
| Pyricularia | LSU | ITS, ACT, CAL, RPB1 | [41] |
| Pyrenophora | LSU | ITS, GAPDH | [66] |
| Pythium | LSU, SSU | ITS, COX1, COX2 | [66] |
| Tilletia | LSU | ITS | [67] |
| Ustilago | LSU | ITS | [66] |
| Venturia | LSU, SSU | ITS, TEF1, TUB2 | [17,67] |
| Verticillium | ITS | ACT | [66] |
| Wilsonomyces | LSU | ITS, TEF | [17] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tovar-Pedraza, J.M.; Solano-Báez, A.R.; Leyva-Mir, S.G.; Tlapal-Bolaños, B.; Camacho-Tapia, M.; García-León, E.; Ayala-Escobar, V.; Nava-Díaz, C.; Quezada-Salinas, A.; Santiago-Santiago, V.; et al. The Need and Opportunity to Update the Inventory of Plant Pathogenic Fungi and Oomycetes in Mexico. J. Fungi 2024, 10, 395. https://doi.org/10.3390/jof10060395
Tovar-Pedraza JM, Solano-Báez AR, Leyva-Mir SG, Tlapal-Bolaños B, Camacho-Tapia M, García-León E, Ayala-Escobar V, Nava-Díaz C, Quezada-Salinas A, Santiago-Santiago V, et al. The Need and Opportunity to Update the Inventory of Plant Pathogenic Fungi and Oomycetes in Mexico. Journal of Fungi. 2024; 10(6):395. https://doi.org/10.3390/jof10060395
Chicago/Turabian StyleTovar-Pedraza, Juan Manuel, Alma Rosa Solano-Báez, Santos Gerardo Leyva-Mir, Bertha Tlapal-Bolaños, Moisés Camacho-Tapia, Elizabeth García-León, Victoria Ayala-Escobar, Cristian Nava-Díaz, Andrés Quezada-Salinas, Víctor Santiago-Santiago, and et al. 2024. "The Need and Opportunity to Update the Inventory of Plant Pathogenic Fungi and Oomycetes in Mexico" Journal of Fungi 10, no. 6: 395. https://doi.org/10.3390/jof10060395
APA StyleTovar-Pedraza, J. M., Solano-Báez, A. R., Leyva-Mir, S. G., Tlapal-Bolaños, B., Camacho-Tapia, M., García-León, E., Ayala-Escobar, V., Nava-Díaz, C., Quezada-Salinas, A., Santiago-Santiago, V., Beltrán-Peña, H., Hernandez-Hernandez, M. A., Juárez-Cruz, K. J., & Márquez-Licona, G. (2024). The Need and Opportunity to Update the Inventory of Plant Pathogenic Fungi and Oomycetes in Mexico. Journal of Fungi, 10(6), 395. https://doi.org/10.3390/jof10060395






