Applications of the Methylotrophic Yeast Komagataella phaffii in the Context of Modern Biotechnology
Abstract
1. Introduction
2. Applications in the Pharmaceutical Industry
3. Applications in the Production of Renewable Chemicals and Fuels
4. Applications in the Production of Biomaterials
5. Applications in the Food and Feed Industry
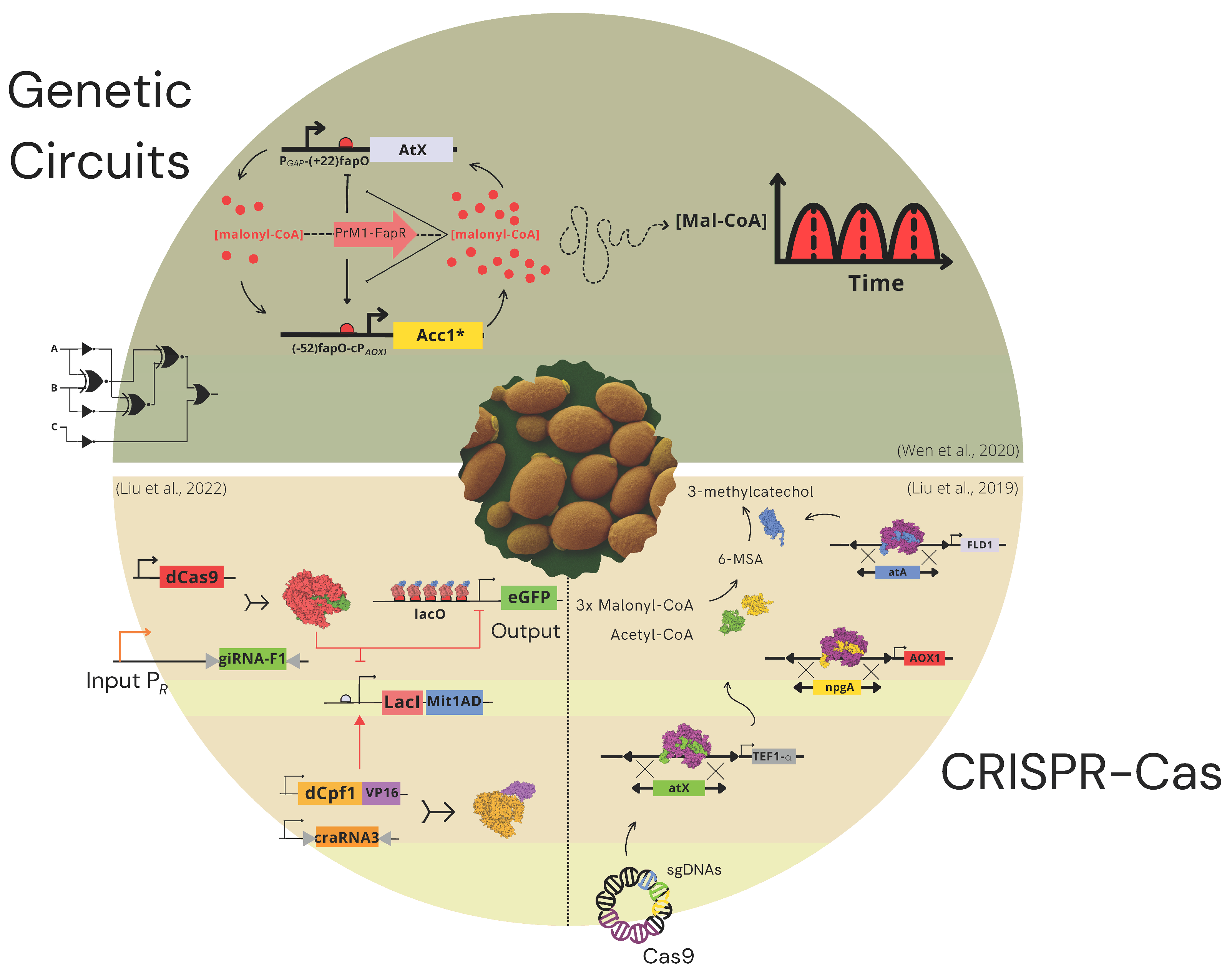
6. Advanced Tools for Synthetic Biology in K. phaffii
7. Synthetic Genetic Circuits
8. CRISPR-Cas Systems as Tools for Gene Editing and Gene Regulation Control
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Marques, W.L.; Raghavendran, V.; Stambuk, B.U.; Gombert, A.K. Sucrose and Saccharomyces cerevisiae: A Relationship Most Sweet. FEMS Yeast Res. 2016, 16, fov107. [Google Scholar] [CrossRef] [PubMed]
- Bernauer, L.; Radkohl, A.; Lehmayer, L.G.K.; Emmerstorfer-Augustin, A. Komagataella phaffii as Emerging Model Organism in Fundamental Research. Front. Microbiol. 2021, 11, 607028. [Google Scholar] [CrossRef] [PubMed]
- Bustos, C.; Quezada, J.; Veas, R.; Altamirano, C.; Braun-Galleani, S.; Fickers, P.; Berrios, J. Advances in Cell Engineering of the Komagataella phaffii Platform for Recombinant Protein Production. Metabolites 2022, 12, 346. [Google Scholar] [CrossRef] [PubMed]
- Paes, B.G.; Steindorff, A.S.; Formighieri, E.F.; Pereira, I.S.; Almeida, J.R.M. Physiological Characterization and Transcriptome Analysis of Pichia pastoris Reveals Its Response to Lignocellulose-Derived Inhibitors. AMB Express 2021, 11, 2. [Google Scholar] [CrossRef] [PubMed]
- Cregg, J.M.; Barringer, K.J.; Hessler, A.Y.; Madden, K.R. Pichia pastoris as a Host System for Transformations. Mol. Cell. Biol. 1985, 5, 3376–3385. [Google Scholar] [CrossRef] [PubMed]
- Darby, R.A.J.; Cartwright, S.P.; Dilworth, M.V.; Bill, R.M. Which Yeast Species Shall I Choose? Saccharomyces cerevisiae versus Pichia pastoris (Review). Methods Mol. Biol. 2012, 866, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia Pastoris: A Highly Successful Expression System for Optimal Synthesis of Heterologous Proteins. J. Cell. Physiol. 2020, 235, 5867–5881. [Google Scholar] [CrossRef] [PubMed]
- Zahrl, R.J.; Peña, D.A.; Mattanovich, D.; Gasser, B. Systems Biotechnology for Protein Production in Pichia pastoris. FEMS Yeast Res. 2017, 17, fox068. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Li, X.; Lü, J.; Mao, P.H.; Jin, X.; Rao, B.; Zheng, P.; Zhou, Y.L.; Liu, S.Y.; Ke, T.; et al. A Visual Method for Direct Selection of High-Producing Pichia pastoris Clones. BMC Biotechnol. 2011, 11, 23. [Google Scholar] [CrossRef]
- GRAS Notices. Available online: https://www.cfsanappsexternal.fda.gov/scripts/fdcc/index.cfm?set=GRASNotices&id=204 (accessed on 18 April 2024).
- Walsh, G. Biopharmaceutical Benchmarks 2010. Nat. Biotechnol. 2010, 28, 917–924. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. What Are “Biologics” Questions and Answers. 2023. Available online: https://www.fda.gov/about-fda/center-biologics-evaluation-and-research-cber/what-are-biologics-questions-and-answers (accessed on 10 May 2024).
- Drug Approval Package: Kalbitor (Ecallantide) NDA #. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2009/125277s000TOC.cfm (accessed on 9 May 2024).
- European Medicines Agency. Jetrea. 2023. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/jetrea (accessed on 10 May 2024).
- Drug Approval Package: Jetrea (Ocriplasmin) BLA 125422. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2012/125422_jetrea_toc.cfm (accessed on 7 May 2024).
- FDA Commissioner. FDA Approves First Interchangeable Biosimilar Insulin Product for Treatment of Diabetes. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-interchangeable-biosimilar-insulin-product-treatment-diabetes (accessed on 7 May 2024).
- Semglee. European Medicines Agency. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/semglee (accessed on 18 April 2024).
- Biocon’s Insulin Glargine Receives Regulatory Approval in Japan—Biocon. Available online: https://www.biocon.com/biocons-insulin-glargine-receives-regulatory-approval-in-japan/ (accessed on 18 April 2024).
- Ghosh, S.; Bose, S.; Gowda, S.; Mukhopadhyay, P. Biosimilar Insul.—What A Clin. Needs Know? Indian J. Endocrinol. Metab. 2019, 23, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Inpremzia. European Medicines Agency. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/inpremzia (accessed on 18 April 2024).
- Drug Approval Package: MYXREDLIN. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2019/208157Orig1s000TOC.cfm (accessed on 7 May 2024).
- Kirsty (Previously Kixelle). European Medicines Agency. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/kirsty-previously-kixelle#authorisation-details-section (accessed on 18 April 2024).
- ClinicalTrials.gov. Safety Study of Single Intravesical Doses of TTI-1612 in Women with Interstitial Cystitis/Bladder Pain Syndrome. 2023. Available online: https://clinicaltrials.gov/ct2/show/study/NCT01559961 (accessed on 10 May 2024).
- Kamaraj, R.; Manju, M. Biosimilar Current Status in India. Asian J. Pharm. Clin. Res. 2017, 10, 25–28. [Google Scholar]
- Mayer, A.; Francis, R.J.; Sharma, S.K.; Tolner, B.; Springer, C.J.; Martin, J.; Boxer, G.M.; Bell, J.; Green, A.J.; Hartley, J.A.; et al. A Phase I Study of Single Administration of Antibody-Directed Enzyme Prodrug Therapy with the Recombinant Anti–Carcinoembryonic Antigen Antibody-Enzyme Fusion Protein MFECP1 and a Bis-Iodo Phenol Mustard Prodrug. Clin. Cancer Res. 2006, 12, 6509–6516. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency. Vyepti. 2023. Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/vyepti (accessed on 10 May 2024).
- Mease, P.J.; Gottlieb, A.B.; Berman, A.; Drescher, E.; Xing, J.; Wong, R.; Banerjee, S. The Efficacy and Safety of Clazakizumab, an Anti–Interleukin-6 Monoclonal Antibody, in a Phase IIb Study of Adults with Active Psoriatic Arthritis. Arthritis Rheumatol. 2016, 68, 2163–2173. [Google Scholar] [CrossRef] [PubMed]
- Shekhar, C. Pichia Power: India’s Biotech Industry Puts Unconventional Yeast to Work. Chem. Biol. 2008, 15, 201–202. [Google Scholar] [CrossRef] [PubMed]
- Diemert, D.J.; Freire, J.; Valente, V.; Fraga, C.G.; Talles, F.; Grahek, S.; Campbell, D.; Jariwala, A.; Periago, M.V.; Enk, M.; et al. Safety and Immunogenicity of the Na-GST-1 Hookworm Vaccine in Brazilian and American Adults. PLoS Neglected Trop. Dis. 2017, 11, e0005574. [Google Scholar] [CrossRef] [PubMed]
- Keitel, W.A.; Potter, G.E.; Diemert, D.; Bethony, J.; El Sahly, H.M.; Kennedy, J.K.; Patel, S.M.; Plieskatt, J.L.; Jones, W.; Deye, G.; et al. A Phase 1 Study of the Safety, Reactogenicity, and Immunogenicity of a Schistosoma Mansoni Vaccine with or without Glucopyranosyl Lipid A Aqueous Formulation (GLA-AF) in Healthy Adults from a Non-Endemic Area. Vaccine 2019, 37, 6500–6509. [Google Scholar] [CrossRef] [PubMed]
- Sirima, S.B.; Durier, C.; Kara, L.; Houard, S.; Gansane, A.; Loulergue, P.; Bahuaud, M.; Benhamouda, N.; Nebié, I.; Faber, B.; et al. Safety and Immunogenicity of a Recombinant Plasmodium Falciparum AMA1-DiCo Malaria Vaccine Adjuvanted with GLA-SE or Alhydrogel® in European and African Adults: A Phase 1a/1b, Randomized, Double-Blind Multi-Centre Trial. Vaccine 2017, 35, 6218–6227. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, A. Ecallantide (DX-88), a Plasma Kallikrein Inhibitor for the Treatment of Hereditary Angioedema and the Prevention of Blood Loss in on-Pump Cardiothoracic Surgery. Expert Opin. Biol. Ther. 2008, 8, 1187–1199. [Google Scholar] [CrossRef] [PubMed]
- Kalbitor. Withdrawal of the Marketing Authorisation Application. 2023. Available online: https://www.ema.europa.eu/en/medicines/human/withdrawn-applications/kalbitor (accessed on 10 May 2024).
- Stalmans, P.; Benz, M.S.; Gandorfer, A.; Kampik, A.; Girach, A.; Pakola, S.; Haller, J.A. Enzymatic Vitreolysis with Ocriplasmin for Vitreomacular Traction and Macular Holes. N. Engl. J. Med. 2012, 367, 606–615. [Google Scholar] [CrossRef]
- Matli, M.C.; Wilson, A.B.; Rappsilber, L.M.; Sheffield, F.P.; Farlow, M.L.; Johnson, J.L. The First Interchangeable Biosimilar Insulin: Insulin Glargine-Yfgn. J. Diabetes Sci. Technol. 2023, 17, 490–494. [Google Scholar] [CrossRef] [PubMed]
- Gasser, B.; Prielhofer, R.; Marx, H.; Maurer, M.; Nocon, J.; Steiger, M.; Puxbaum, V.; Sauer, M.; Mattanovich, D. Pichia Pastoris: Protein Production Host and Model Organism for Biomedical Research. Future Microbiol. 2013, 8, 191–208. [Google Scholar] [CrossRef] [PubMed]
- de Sá Magalhães, S.; Keshavarz-Moore, E. Pichia Pastoris (Komagataella phaffii) as a Cost-Effective Tool for Vaccine Production for Low- and Middle-Income Countries (LMICs). Bioengineering 2021, 8, 119. [Google Scholar] [CrossRef] [PubMed]
- Mitsubishi Chemical Group. Notice of Termination of the Recombinant Serum Albumin Business. 2023. Available online: https://www.mcgc.com/english/news_release/01328.html (accessed on 10 May 2024).
- Park, D.-S.; Kim, J.-H.; Lee, S.W.; Jeong, J.-M. Secretory Expression of the α-Subunit of Human Coagulation Factor XIII in the Yeast Pichia pastoris. Biotechnol. Lett. 2002, 24, 97–101. [Google Scholar] [CrossRef]
- Mochizuki, S.; Hamato, N.; Hirose, M.; Miyano, K.; Ohtani, W.; Kameyama, S.; Kuwae, S.; Tokuyama, T.; Ohi, H. Expression and Characterization of Recombinant Human Antithrombin III in Pichia pastoris. Protein Expr. Purif. 2001, 23, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, S.R.; Davidson, R.C.; Sethuraman, N.; Nett, J.H.; Jiang, Y.; Rios, S.; Bobrowicz, P.; Stadheim, T.A.; Li, H.; Choi, B.-K.; et al. Humanization of Yeast to Produce Complex Terminally Sialylated Glycoproteins. Science 2006, 313, 1441–1443. [Google Scholar] [CrossRef]
- Beck, A.; Cochet, O.; Wurch, T. GlycoFi’s Technology to Control the Glycosylation of Recombinant Therapeutic Proteins. Expert Opin. Drug Discov. 2010, 5, 95–111. [Google Scholar] [CrossRef]
- Liu, L.; Gomathinayagam, S.; Hamuro, L.; Prueksaritanont, T.; Wang, W.; Stadheim, A.; Hamilton, S. The Impact of Glycosylation on the Pharmacokinetics of a TNFR2:Fc Fusion Protein Expressed in Glycoengineered Pichia pastoris. Pharm. Res. 2012, 30, 803–812. [Google Scholar] [CrossRef]
- Li, H.; Sethuraman, N.; Stadheim, T.A.; Zha, D.; Prinz, B.; Ballew, N.; Bobrowicz, P.; Choi, B.-K.; Cook, W.J.; Cukan, M.; et al. Optimization of Humanized IgGs in Glycoengineered Pichia pastoris. Nat. Biotechnol. 2006, 24, 210–215. [Google Scholar] [CrossRef]
- Nett, J.H.; Gomathinayagam, S.; Hamilton, S.R.; Gong, B.; Davidson, R.C.; Du, M.; Hopkins, D.; Mitchell, T.; Mallem, M.R.; Nylen, A.; et al. Optimization of Erythropoietin Production with Controlled Glycosylation-PEGylated Erythropoietin Produced in Glycoengineered Pichia pastoris. J. Biotechnol. 2012, 157, 198–206. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, L.; Dan Dumitru, C.; Cummings, N.R.H.; Cukan, M.; Jiang, Y.; Li, Y.; Li, F.; Mitchell, T.; Mallem, M.R.; et al. Glycoengineered Pichia Produced Anti-HER2 Is Comparable to Trastuzumab in Preclinical Study. mAbs 2011, 3, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Science Commentary. Merck Closes Down Glycofi. 2023. Available online: https://www.science.org/content/blog-post/merck-closes-down-glycofi (accessed on 10 May 2024).
- Wu, M.; Shen, Q.; Yang, Y.; Zhang, S.; Qu, W.; Chen, J.; Sun, H.; Chen, S. Disruption of YPS1 and PEP4 Genes Reduces Proteolytic Degradation of Secreted HSA/PTH in Pichia pastoris GS115. J. Ind. Microbiol. Biotechnol. 2013, 40, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Heo, Y.-S. Recent Advances in Antibody Therapeutics. Int. J. Mol. Sci. 2022, 23, 3690. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S. Eptinezumab: First Approval. Drugs 2020, 80, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Schuster, M.; Rigas, J.R.; Orlov, S.V.; Milovanovic, B.; Prabhash, K.; Smith, J.T. ALD518, a Humanized Anti-IL-6 Antibody, Treats Anemia in Patients with Advanced Non-Small Cell Lung Cancer (NSCLC): Results of a Phase II, Randomized, Double-Blind, Placebo-Controlled Trial. J. Clin. Oncol. 2010, 28 (Suppl. S15), 7631. [Google Scholar] [CrossRef]
- BMS Acquires Rights for IL-6 Inhibitor. Nat. Rev. Drug Discov. 2010, 9, 10. [CrossRef]
- Ezzine, A.; M’Hirsi el Adab, S.; Bouhaouala-Zahar, B.; Hmila, I.; Baciou, L.; Marzouki, M.N. Efficient Expression of the Anti-AahI’ Scorpion Toxin Nanobody under a New Functional Form in a Pichia pastoris System. Biotechnol. Appl. Biochem. 2012, 59, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Baghban, R.; Gargari, S.L.M.; Rajabibazl, M.; Nazarian, S.; Bakherad, H. Camelid-Derived Heavy-Chain Nanobody against Clostridium Botulinum Neurotoxin E in Pichia pastoris. Biotechnol. Appl. Biochem. 2016, 63, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Sanofi. NANOBODY Technology Platform. 2023. Available online: https://www.sanofi.com/en/science-and-innovation/research-and-development/technology-platforms/nanobody-technology-platform (accessed on 10 May 2024).
- Bryan, J.T.; Buckland, B.; Hammond, J.; Jansen, K.U. Prevention of Cervical Cancer: Journey to Develop the First Human Papillomavirus Virus-like Particle Vaccine and the next Generation Vaccine. Curr. Opin. Chem. Biol. 2016, 32, 34–47. [Google Scholar] [CrossRef]
- Wang, M.; Jiang, S.; Wang, Y. Recent Advances in the Production of Recombinant Subunit Vaccines in Pichia pastoris. Bioengineered 2016, 7, 155–165. [Google Scholar] [CrossRef]
- Mani, S.; Tripathi, L.; Raut, R.; Tyagi, P.; Arora, U.; Barman, T.; Sood, R.; Galav, A.; Wahala, W.; de Silva, A.; et al. Pichia pastoris-Expressed Dengue 2 Envelope Forms Virus-Like Particles without Pre-Membrane Protein and Induces High Titer Neutralizing Antibodies. PLoS ONE 2013, 8, e64595. [Google Scholar] [CrossRef] [PubMed]
- Fahimi, H.; Mohammadipour, M.; Haddad Kashani, H.; Parvini, F.; Sadeghizadeh, M. Dengue Viruses and Promising Envelope Protein Domain III-Based Vaccines. Appl. Microbiol. Biotechnol. 2018, 102, 2977–2996. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, V.; Arora, U.; Shukla, R.; Poddar, A.; Shanmugam, R.K.; White, L.J.; Mattocks, M.M.; Raut, R.; Perween, A.; Tyagi, P.; et al. A Tetravalent Virus-like Particle Vaccine Designed to Display Domain III of Dengue Envelope Proteins Induces Multi-Serotype Neutralizing Antibodies in Mice and Macaques Which Confer Protection against Antibody Dependent Enhancement in AG129 Mice. PLoS Neglected Trop. Dis. 2018, 12, e0006191. [Google Scholar] [CrossRef] [PubMed]
- Spice, A.J.; Aw, R.; Bracewell, D.G.; Polizzi, K.M. Synthesis and Assembly of Hepatitis B Virus-Like Particles in a Pichia pastoris Cell-Free System. Front. Bioeng. Biotechnol. 2020, 8, 72. [Google Scholar] [CrossRef] [PubMed]
- Thakur, V.; Pati, N.; Gupta, R.; Sarin, S. Efficacy of Shanvac-B Recombinant DNA Hepatitis B Vaccine in Health Care Workers of Northern India. Hepatobiliary Pancreat. Dis. Int. 2010, 9, 393–397. [Google Scholar] [PubMed]
- Joshi, N.; Kumar, A.; Raghu, M.B.; Bhave, S.; Arulprakash, R.; Bhusari, P.; Rao, R. Immunogenicity and Safety of Hepatitis B Vaccine (Shanvac-B) Using a Novel Pre-Filled Single Use Injection Device Uniject in Indian Subjects. J. Med. Sci. 2004, 58, 472–477. [Google Scholar]
- Leong, H.Y.; Chang, C.-K.; Khoo, K.S.; Chew, K.W.; Chia, S.R.; Lim, J.W.; Chang, J.-S.; Show, P.L. Waste Biorefinery towards a Sustainable Circular Bioeconomy: A Solution to Global Issues. Biotechnol. Biofuels 2021, 14, 87. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Zhang, W.; Wang, Y.; Sheng, J.; Xu, R.; Li, J.; Du, G.; Kang, Z. Biosynthesis of Non-Animal Chondroitin Sulfate from Methanol Using Genetically Engineered Pichia pastoris. Green Chem. 2021, 23, 4365–4374. [Google Scholar] [CrossRef]
- Ata, Ö.; Ergün, B.G.; Fickers, P.; Heistinger, L.; Mattanovich, D.; Rebnegger, C.; Gasser, B. What Makes Komagataella phaffii Non-Conventional? FEMS Yeast Res. 2021, 21, foab059. [Google Scholar] [CrossRef]
- Ergün, B.G.; Laçın, K.; Çaloğlu, B.; Binay, B. Second Generation Pichia pastoris Strain and Bioprocess Designs. Biotechnol. Biofuels Bioprod. 2022, 15, 150. [Google Scholar] [CrossRef]
- Heistinger, L.; Dohm, J.C.; Paes, B.G.; Koizar, D.; Troyer, C.; Ata, Ö.; Steininger-Mairinger, T.; Mattanovich, D. Genotypic and Phenotypic Diversity among Komagataella Species Reveals a Hidden Pathway for Xylose Utilization. Microb. Cell Factories 2022, 21, 70. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Sun, H.; Chen, Z.; Li, Y.; Zhu, T. Construction of Efficient Xylose Utilizing Pichia pastoris for Industrial Enzymeproduction. Microb. Cell Factories 2015, 14, 22. [Google Scholar] [CrossRef] [PubMed]
- Ramos, T.G.S.; Justen, F.; Carneiro, C.V.G.C.; Honorato, V.M.; Franco, P.F.; Vieira, F.S.; Trichez, D.; Rodrigues, C.M.; Almeida, J.R.M. Xylonic Acid Production by Recombinant Komagataella phaffii Strains Engineered with Newly Identified Xylose Dehydrogenases. Bioresour. Technol. Rep. 2021, 16, 100825. [Google Scholar] [CrossRef]
- Gassler, T.; Sauer, M.; Gasser, B.; Egermeier, M.; Troyer, C.; Causon, T.; Hann, S.; Mattanovich, D.; Steiger, M.G. The Industrial Yeast Pichia pastoris Is Converted from a Heterotroph into an Autotroph Capable of Growth on CO2. Nat. Biotechnol. 2020, 38, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Gassler, T.; Baumschabl, M.; Sallaberger, J.; Egermeier, M.; Mattanovich, D. Adaptive Laboratory Evolution and Reverse Engineering Enhances Autotrophic Growth in Pichia pastoris. Metab. Eng. 2022, 69, 112–121. [Google Scholar] [CrossRef] [PubMed]
- de Lima, P.B.A.; Mulder, K.C.L.; Melo, N.T.M.; Carvalho, L.S.; Menino, G.S.; Mulinari, E.; de Castro, V.H.; dos Reis, T.F.; Goldman, G.H.; Magalhães, B.S.; et al. Novel Homologous Lactate Transporter Improves L-Lactic Acid Production from Glycerol in Recombinant Strains of Pichia pastoris. Microb. Cell Factories 2016, 15, 158. [Google Scholar] [CrossRef] [PubMed]
- Louie, T.M.; Louie, K.; DenHartog, S.; Gopishetty, S.; Subramanian, M.; Arnold, M.; Das, S. Production of Bio-Xylitol from d-Xylose by an Engineered Pichia pastoris Expressing a Recombinant Xylose Reductase Did Not Require Any Auxiliary Substrate as Electron Donor. Microb. Cell Factories 2021, 20, 50. [Google Scholar] [CrossRef]
- Liu, Y.; Tu, X.; Xu, Q.; Bai, C.; Kong, C.; Liu, Q.; Yu, J.; Peng, Q.; Zhou, X.; Zhang, Y.; et al. Engineered Monoculture and Co-Culture of Methylotrophic Yeast for de Novo Production of Monacolin J and Lovastatin from Methanol. Metab. Eng. 2018, 45, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Araya-Garay, J.M.; Feijoo-Siota, L.; Rosa-dos-Santos, F.; Veiga-Crespo, P.; Villa, T.G. Construction of New Pichia pastoris X-33 Strains for Production of Lycopene and β-Carotene. Appl. Microbiol. Biotechnol. 2012, 93, 2483–2492. [Google Scholar] [CrossRef]
- Jeong, E.; Shim, W.Y.; Kim, J.H. Metabolic Engineering of Pichia Pastoris for Production of Hyaluronic Acid with High Molecular Weight. J. Biotechnol. 2014, 185, 28–36. [Google Scholar] [CrossRef]
- Siripong, W.; Wolf, P.; Kusumoputri, T.P.; Downes, J.J.; Kocharin, K.; Tanapongpipat, S.; Runguphan, W. Metabolic Engineering of Pichia pastoris for Production of Isobutanol and Isobutyl Acetate. Biotechnol. Biofuels 2018, 11, 1. [Google Scholar] [CrossRef]
- De, S.; Mattanovich, D.; Ferrer, P.; Gasser, B. Established Tools and Emerging Trends for the Production of Recombinant Proteins and Metabolites in Pichia pastoris. Essays Biochem. 2021, 65, 293–307. [Google Scholar] [CrossRef] [PubMed]
- Yamada, R.; Ogura, K.; Kimoto, Y.; Ogino, H. Toward the Construction of a Technology Platform for Chemicals Production from Methanol: D-Lactic Acid Production from Methanol by an Engineered Yeast Pichia pastoris. World J. Microbiol. Biotechnol. 2019, 35, 37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Ge, C.; Deng, L.; Tan, T.; Wang, F. C4-Dicarboxylic Acid Production by Overexpressing the Reductive TCA Pathway. FEMS Microbiol. Lett. 2015, 362, fnv052. [Google Scholar] [CrossRef] [PubMed]
- Fina, A.; Brêda, G.C.; Pérez-Trujillo, M.; Freire, D.M.G.; Almeida, R.V.; Albiol, J.; Ferrer, P. Benchmarking Recombinant Pichia Pastoris for 3-Hydroxypropionic Acid Production from Glycerol. Microb. Biotechnol. 2021, 14, 1671–1682. [Google Scholar] [CrossRef] [PubMed]
- Fina, A.; Heux, S.; Albiol, J.; Ferrer, P. Combining Metabolic Engineering and Multiplexed Screening Methods for 3-Hydroxypropionic Acid Production in Pichia pastoris. Front. Bioeng. Biotechnol. 2022, 10, 942304. [Google Scholar]
- Zhang, Q.; Wang, X.; Luo, H.; Wang, Y.; Wang, Y.; Tu, T.; Qin, X.; Su, X.; Huang, H.; Yao, B.; et al. Metabolic Engineering of Pichia pastoris for Myo-Inositol Production by Dynamic Regulation of Central Metabolism. Microb. Cell Factories 2022, 21, 112. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, Z. Production of (2R, 3R)-2,3-Butanediol Using Engineered Pichia pastoris: Strain Construction, Characterization and Fermentation. Biotechnol. Biofuels 2018, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Cai, P.; Wu, X.; Deng, J.; Gao, L.; Shen, Y.; Yao, L.; Zhou, Y.J. Methanol Biotransformation toward High-Level Production of Fatty Acid Derivatives by Engineering the Industrial Yeast Pichia pastoris. Proc. Natl. Acad. Sci. USA 2022, 119, e2201711119. [Google Scholar] [CrossRef]
- Liu, X.-B.; Liu, M.; Tao, X.-Y.; Zhang, Z.-X.; Wang, F.-Q.; Wei, D.-Z. Metabolic Engineering of Pichia pastoris for the Production of Dammarenediol-II. J. Biotechnol. 2015, 216, 47–55. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, S.; Lin, Y.; Li, W.; Wang, D.; Ruan, S.; Yang, Y.; Liang, S. Metabolic Engineering of Pichia pastoris for High-Level Production of Lycopene. ACS Synth. Biol. 2023, 12, 2961–2972. [Google Scholar] [CrossRef] [PubMed]
- Wriessnegger, T.; Augustin, P.; Engleder, M.; Leitner, E.; Müller, M.; Kaluzna, I.; Schürmann, M.; Mink, D.; Zellnig, G.; Schwab, H.; et al. Production of the Sesquiterpenoid (+)-Nootkatone by Metabolic Engineering of Pichia pastoris. Metab. Eng. 2014, 24, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Cai, M.; Shen, W.; Xiao, S.; Zhou, X.; Zhang, Y. Engineered Fungal Polyketide Biosynthesis in Pichia pastoris: A Potential Excellent Host for Polyketide Production. Microb. Cell Factories 2013, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Poirier, Y.; Erard, N.; MacDonald-Comber Petétot, J. Synthesis of Polyhydroxyalkanoate in the Peroxisome of Pichia pastoris. FEMS Microbiol. Lett. 2002, 207, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Zhou, Z.; Wang, P.; Xi, X.; Hu, S.; Xu, R.; Du, G.; Li, J.; Chen, J.; et al. Synthesis of Bioengineered Heparin by Recombinant Yeast Pichia pastoris. Green Chem. 2022, 24, 3180–3192. [Google Scholar] [CrossRef]
- He, J.; Deng, J.; Zheng, Y.; Gu, J. A Synergistic Effect on the Production of S-Adenosyl-l-Methionine in Pichia pastoris by Knocking in of S-Adenosyl-l-Methionine Synthase and Knocking out of Cystathionine-β Synthase. J. Biotechnol. 2006, 126, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Xia, H.; Liu, Y.; Li, J.; Du, G.; Lv, X.; Liu, L. Compartmentalization and Transporter Engineering Strategies for Terpenoid Synthesis. Microb. Cell Factories 2022, 21, 92. [Google Scholar] [CrossRef] [PubMed]
- Peña, D.A.; Gasser, B.; Zanghellini, J.; Steiger, M.G.; Mattanovich, D. Metabolic Engineering of Pichia pastoris. Metab. Eng. 2018, 50, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Rahmati, M.; Pennisi, C.P.; Budd, E.; Mobasheri, A.; Mozafari, M. Biomaterials for Regenerative Medicine: Historical Perspectives and Current Trends. In Cell Biology and Translational Medicine, Volume 4: Stem Cells and Cell Based Strategies in Regeneration; Turksen, K., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–19. [Google Scholar] [CrossRef]
- Brodsky, B.; Ramshaw, J.A.M. Bioengineered Collagens. In Fibrous Proteins: Structures and Mechanisms; Parry, D.A.D., Squire, J.M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 601–629. [Google Scholar] [CrossRef]
- Werten, M.W.T.; Eggink, G.; Cohen Stuart, M.A.; de Wolf, F.A. Production of Protein-Based Polymers in Pichia pastoris. Biotechnol. Adv. 2019, 37, 642–666. [Google Scholar] [CrossRef]
- Aigner, T.B.; DeSimone, E.; Scheibel, T. Biomedical Applications of Recombinant Silk-Based Materials. Adv. Mater. 2018, 30, 1704636. [Google Scholar] [CrossRef]
- Steffens, D.; Braghirolli, D.I.; Maurmann, N.; Pranke, P. Update on the Main Use of Biomaterials and Techniques Associated with Tissue Engineering. Drug Discov. Today 2018, 23, 1474–1488. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Kwon, I.K.; Park, K. Hydrogels for Delivery of Bioactive Agents: A Historical Perspective. Adv. Drug Deliv. Rev. 2013, 65, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Guo, K.; Zhang, Y.; Zhang, X.; Qin, L.; Wang, X.; Zhu, H.; Guo, Y.; Yang, W.; Li, B.; et al. Adhesive Property and Mechanism of Silkworm Egg Glue Protein. Acta Biomater. 2021, 134, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Garenaux, E.; Maes, E.; Levêque, S.; Brassart, C.; Guerardel, Y. Structural Characterization of Complex O-Linked Glycans from Insect-Derived Material. Carbohydr. Res. 2011, 346, 1093–1104. [Google Scholar] [CrossRef] [PubMed]
- Yukuhiro, K.; Sezutsu, H.; Tsubota, T.; Takasu, Y.; Kameda, T.; Yonemura, N. Insect Silks and Cocoons: Structural and Molecular Aspects. In Extracellular Composite Matrices in Arthropods; Cohen, E., Moussian, B., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 515–555. [Google Scholar] [CrossRef]
- Sampaolesi, S.; Nicotra, F.; Russo, L. Glycans in Nanomedicine, Impact and Perspectives. Future Med. Chem. 2019, 11, 43–60. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Liu, X.; Li, K.; Li, X.; Zhu, J.; Yang, S.; Xu, L.; Yang, M.; Yan, Y.; Yan, J. A Bioinspired Synthetic Fused Protein Adhesive from Barnacle Cement and Spider Dragline for Potential Biomedical Materials. Int. J. Biol. Macromol. 2023, 253, 127125. [Google Scholar] [CrossRef] [PubMed]
- Bolghari, N.; Shahsavarani, H.; Anvari, M.; Habibollahi, H. A Novel Recombinant Chimeric Bio-Adhesive Protein Consisting of Mussel Foot Protein 3, 5, Gas Vesicle Protein A, and CsgA Curli Protein Expressed in Pichia pastoris. AMB Express 2022, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Liang, X.; Yu, S.; Zhou, J. Expression, Characterization, and Application Potentiality Evaluation of Recombinant Human-like Collagen in Pichia pastoris. Bioresour. Bioprocess. 2022, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Sorushanova, A.; Delgado, L.M.; Wu, Z.; Shologu, N.; Kshirsagar, A.; Raghunath, R.; Mullen, A.M.; Bayon, Y.; Pandit, A.; Raghunath, M.; et al. The Collagen Suprafamily: From Biosynthesis to Advanced Biomaterial Development. Adv. Mater. 2019, 31, 1801651. [Google Scholar] [CrossRef]
- Gellermann, P.; Schneider-Barthold, C.; Bolten, S.N.; Overfelt, E.; Scheper, T.; Pepelanova, I. Production of a Recombinant Non-Hydroxylated Gelatin Mimetic in Pichia pastoris for Biomedical Applications. J. Funct. Biomater. 2019, 10, 39. [Google Scholar] [CrossRef]
- Rezvani Ghomi, E.; Nourbakhsh, N.; Akbari Kenari, M.; Zare, M.; Ramakrishna, S. Collagen-Based Biomaterials for Biomedical Applications. J. Biomed. Mater. Res. Part B Appl. Biomater. 2021, 109, 1986–1999. [Google Scholar] [CrossRef]
- Bulleid, N.; John, D.C.A.; Kadler, K. Recombinant Expression Systems for the Production of Collagen. Biochem. Soc. Trans. 2000, 28, 350–353. [Google Scholar] [CrossRef]
- Nokelainen, M.; Tu, H.; Vuorela, A.; Notbohm, H.; Kivirikko, K.I.; Myllyharju, J. High-Level Production of Human Type I Collagen in the Yeast Pichia pastoris. Yeast 2001, 18, 797–806. [Google Scholar] [CrossRef]
- Williams, K.E.; Olsen, D.R. Gelatin Expression from an Engineered Saccharomyces Cerevisiae CUP1 Promoter in Pichia pastoris. Yeast 2021, 38, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Shay, L.K.; Wegner, E.H. High Methionine Content Pichia Pastoris Yeasts 75 Inventors. U.S. Patent 4,439,525, 9 September 1981. [Google Scholar]
- Meng, J.; Liu, S.; Gao, L.; Hong, K.; Liu, S.; Wu, X. Economical Production of Pichia pastoris Single Cell Protein from Methanol at Industrial Pilot Scale. Microb. Cell Factories 2023, 22, 198. [Google Scholar] [CrossRef] [PubMed]
- Rachel, F.; Patrick, O.B.; Jessica, K.; Celeste, H.-S.; Elysia, C. Methods and Compositions for Affecting the Flavor and Aroma Profile of Consumables. U.S. Patent 9,700,067, 12 July 2011. [Google Scholar]
- Jin, Y.; He, X.; Andoh-Kumi, K.; Fraser, R.Z.; Lu, M.; Goodman, R.E. Evaluating Potential Risks of Food Allergy and Toxicity of Soy Leghemoglobin Expressed in Pichia pastoris. Mol. Nutr. Food Res. 2018, 62, 1700297. [Google Scholar] [CrossRef]
- Spohner, S.C.; Müller, H.; Quitmann, H.; Czermak, P. Expression of Enzymes for the Usage in Food and Feed Industry with Pichia pastoris. J. Biotechnol. 2015, 202, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Purkarthofer, T.; Trummer-Gödl, E.; Dib, I.; Weis, R. Pichia pastoris Protein Expression Excellence High-Level Methanol-Free Phytase Production in Pichia pastoris. Available online: https://www.validogen.com (accessed on 10 May 2024).
- Roongsawang, N.; Promdonkoy, P.; Wongwanichpokhin, M.; Sornlake, W.; Puseenam, A.; Eurwilaichitr, L.; Tanapongpipat, S. Coexpression of Fungal Phytase and Xylanase Utilizing the Cis-Acting Hydrolase Element in Pichia pastoris. FEMS Yeast Res. 2010, 10, 909–916. [Google Scholar] [CrossRef]
- Vogl, T.; Glieder, A. Regulation of Pichia Pastoris Promoters and Its Consequences for Protein Production. New Biotechnol. 2013, 30, 385–404. [Google Scholar] [CrossRef]
- Shrivastava, A.; Pal, M.; Sharma, R.K. Pichia as Yeast Cell Factory for Production of Industrially Important Bio-Products: Current Trends, Challenges, and Future Prospects. J. Bioresour. Bioprod. 2023, 8, 108–124. [Google Scholar] [CrossRef]
- Wen, J.; Tian, L.; Xu, M.; Zhou, X.; Zhang, Y.; Cai, M. A Synthetic Malonyl-Coa Metabolic Oscillator in Komagataella phaffii. ACS Synth. Biol. 2020, 9, 1059–1068. [Google Scholar] [CrossRef]
- Liu, Q.; Shi, X.; Song, L.; Liu, H.; Zhou, X.; Wang, Q.; Zhang, Y.; Cai, M. CRISPR-Cas9-Mediated Genomic Multiloci Integration in Pichia pastoris. Microb. Cell Factories 2019, 18, 144. [Google Scholar] [CrossRef]
- Liu, Q.; Song, L.; Peng, Q.; Zhu, Q.; Shi, X.; Xu, M.; Wang, Q.; Zhang, Y.; Cai, M. A Programmable High-Expression Yeast Platform Responsive to User-Defined Signals. Sci. Adv. 2022, 8, eabl5166. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Autin, L.; Olson, A.J. Illustrate: Software for Biomolecular Illustration. Structure 2019, 27, 1716–1720. [Google Scholar] [CrossRef]
- Xia, P.-F.; Ling, H.; Foo, J.L.; Chang, M.W. Synthetic Genetic Circuits for Programmable Biological Functionalities. Biotechnol. Adv. 2019, 37, 107393. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Monod, J. On the Regulation of Gene Activity. Cold Spring Harb. Symp. Quant. Biol. 1961, 26, 193–211. [Google Scholar] [CrossRef]
- Elowitz, M.B.; Leibler, S. A Synthetic Oscillatory Network of Transcriptional Regulators. Nature 2000, 403, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Gardner, T.S.; Cantor, C.R.; Collins, J.J. Construction of a Genetic Toggle Switch in Escherichia coli. Nature 2000, 403, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Lucks, J.B.; Qi, L.; Whitaker, W.R.; Arkin, A.P. Toward Scalable Parts Families for Predictable Design of Biological Circuits. Curr. Opin. Microbiol. 2008, 11, 567–573. [Google Scholar] [CrossRef]
- Chang, C.H.; Hsiung, H.A.; Hong, K.L.; Huang, C.T. Enhancing the Efficiency of the Pichia Pastoris AOX1 Promoter via the Synthetic Positive Feedback Circuit of Transcription Factor Mxr1. BMC Biotechnol. 2018, 18, 81. [Google Scholar] [CrossRef]
- Gomide, M.d.S.; Leitão, M.d.C.; Coelho, C.M. Biocircuits in Plants and Eukaryotic Algae. Front. Plant Sci. 2022, 13, 982959. [Google Scholar] [CrossRef]
- Sadelain, M.; Papapetrou, E.P.; Bushman, F.D. Safe Harbours for the Integration of New DNA in the Human Genome. Nat. Rev. Cancer 2012, 12, 51–58. [Google Scholar] [CrossRef]
- Piva, L.C.; Marco, J.L.D.; Moraes, L.M.P.D.; Reis, V.C.B.; Torres, F.A.G. Construction and Characterization of Centromeric Plasmids for Komagataella phaffii Using a Color-Based Plasmid Stability Assay. PLoS ONE 2020, 15, e0235532. [Google Scholar] [CrossRef]
- Abramczyk, D.; del Carmen Sanchez Olmos, M.; Rojas, A.A.R.; Schindler, D.; Robertson, D.; McColm, S.; Marston, A.L.; Barlow, P.N. A Supernumerary Synthetic Chromosome in Komagataella phaffii as a Repository for Extraneous Genetic Material. Microb. Cell Factories 2023, 22, 259. [Google Scholar] [CrossRef]
- Weninger, A.; Hatzl, A.M.; Schmid, C.; Vogl, T.; Glieder, A. Combinatorial Optimization of CRISPR/Cas9 Expression Enables Precision Genome Engineering in the Methylotrophic Yeast Pichia pastoris. J. Biotechnol. 2016, 235, 139–149. [Google Scholar] [CrossRef]
- Weninger, A.; Fischer, J.E.; Raschmanová, H.; Kniely, C.; Vogl, T.; Glieder, A. Expanding the CRISPR/Cas9 Toolkit for Pichia pastoris with Efficient Donor Integration and Alternative Resistance Markers. J. Cell. Biochem. 2018, 119, 3183–3198. [Google Scholar] [CrossRef]
- Näätsaari, L.; Mistlberger, B.; Ruth, C.; Hajek, T.; Hartner, F.S.; Glieder, A. Deletion of the Pichia pastoris Ku70 Homologue Facilitates Platform Strain Generation for Gene Expression and Synthetic Biology. PLoS ONE 2012, 7, e0039720. [Google Scholar] [CrossRef]
- Dalvie, N.C.; Leal, J.; Whittaker, C.A.; Yang, Y.; Brady, J.R.; Love, K.R.; Love, J.C. Host-Informed Expression of CRISPR Guide RNA for Genomic Engineering in Komagataella phaffii. ACS Synth. Biol. 2020, 9, 26–35. [Google Scholar] [CrossRef]
- Liao, X.; Li, L.; Jameel, A.; Xing, X.H.; Zhang, C. A Versatile Toolbox for CRISPR-Based Genome Engineering in Pichia pastoris. Appl. Microbiol. Biotechnol. 2021, 105, 9211–9218. [Google Scholar] [CrossRef]
- Gao, J.; Xu, J.; Zuo, Y.; Ye, C.; Jiang, L.; Feng, L.; Huang, L.; Xu, Z.; Lian, J. Synthetic Biology Toolkit for Marker-Less Integration of Multigene Pathways into Pichia pastoris via CRISPR/Cas9. ACS Synth. Biol. 2022, 11, 623–633. [Google Scholar] [CrossRef]
- Zhang, K.; Duan, X.; Cai, P.; Gao, L.; Wu, X.; Yao, L.; Zhou, Y.J. Fusing an Exonuclease with Cas9 Enhances Homologous Recombination in Pichia pastoris. Microb. Cell Factories 2022, 21, 182. [Google Scholar] [CrossRef]
- Zhang, X.; Gu, S.; Zheng, X.; Peng, S.; Li, Y.; Lin, Y.; Liang, S. A Novel and Efficient Genome Editing Tool Assisted by CRISPR-Cas12a/Cpf1 for Pichia pastoris. ACS Synth. Biol. 2021, 10, 2927–2937. [Google Scholar] [CrossRef]
- Deng, M.; Wu, Y.; Lv, X.; Liu, L.; Li, J.; Du, G.; Chen, J.; Liu, Y. Heterologous Single-Strand DNA-Annealing and Binding Protein Enhance CRISPR-Based Genome Editing Efficiency in Komagataella phaffii. ACS Synth. Biol. 2023, 12, 3443–3453. [Google Scholar] [CrossRef]
| Brand Name (If Any) | Manufacturer/Research Institute | Biopharmaceutical | Commercial Approval/Clinical Trials | Reference |
|---|---|---|---|---|
| Kalbitor | Dyax Inc. (Burlington, MA, USA) | Ecallantide | 2009 (USA) | [13] |
| Jetrea | ThromboGenics (Leuven, Belgium) | Ocriplasmin | 2012 (USA) 2013 (Europe) | [14,15] |
| Semglee (USA and Europe) Insugen (India/Japan) | Mylan Pharmaceuticals (Morgantown, WV, USA) Viatris Ltd. (Galway, Ireland) Biocon (Bengaluru, India) | Biosimilar insulin glargine | 2021 (USA) 2018 (Europe) 2016 (Japan) 2004 (India) | [16,17,18,19] |
| Inpremzia (Europe) Myxredlin (USA) | Baxter Holding (Utrecht, The Netherlands) | Biosimilar insulin | 2022 (Europe) 2019 (USA) | [20,21] |
| Kirsty | Mylan Ireland Ltd. (Dublin, Ireland) | Biosimilar insulin aspart | 2021 (Europe) | [22] |
| TTI-1612 | Trillium Pharmaceutics (Brockville, ON, Canada) | Heparin-binding human epidermal growth factor (HB-EGF) | Phase I trials | [23] |
| Shanferon | Shanta Biotech (Hyderabad, India) | Interferon alpha 2b | 2002 (India) | [24] |
| MFECP1 | University College Medical School (London, UK) | Anti-carcinoembryonic antigen antibody–carboxypeptidase G2 | Phase I trials | [25] |
| Vyepti | Lundbeck Pharmaceuticals (Copenhagen, Denmark) | Eptinezumab | 2020 (USA) 2022 (Europe) | [26] |
| Clazakizumab | Bristol-Myers Squibb (New York, NY, USA) | Clazakizumab | Phase II trials | [27] |
| Shanvac-B | Shanta Biotech | Hepatitis B vaccine | 1997 (India) | [28] |
| Recombinant Na-GST-1 | Baylor College of Medicine (Houston, TX, USA) | Hookworm vaccine | Phase I trials | [29] |
| Sm-TSP-2 | National Institute of Allergy and Infectious Diseases (Bethesda, MD, USA) | Intestinal schistosomiasis vaccine | Phase I trials | [30] |
| PfAMA1-DiCo | Institut National de la Santé et de la Recherche Médicale (Paris, France) | Malaria vaccine | Phase I trials | [31] |
| Product | Substrate | Main genetic Modifications and Strategies | Process | Application | Production (g·L−1) | Yield (g·g−1) | Reference |
|---|---|---|---|---|---|---|---|
| Organic acids | |||||||
| Lactic acid | Glycerol | Integration of heterologous lactate dehydrogenase (LDH) and overexpression of endogenous lactate transporter | Fed-batch | Food, pharmaceutical, textile, and chemical industries | ~28 | 0.7 | [73] |
| Methanol | Multicopy integration of LDH | Batch | 3.48 | 0.22 | [80] | ||
| Malic acid | Glucose and methanol | Combined expression of pc and mdh1 | Batch | Food, pharmaceutical, and chemical industries | 42.28 | - | [81] |
| 3-hydroxypropionic acid | Glycerol | Expression of an engineered mcr; overexpression of ACC1, ACS, ALD6, PDC1; deletion of ArDH | Fed-batch | Building block for production of chemicals, such as acrylic acid and biopolymers | 24.75 to 37.05 | 0.13 to 0.194 | [82,83] |
| Xylonic acid | Xylose | Expression of xylose dehydrogenase (XDH) | Batch | Building block, cleaner agent, and cement additive | 37 | 0.96 | [70] |
| Sugar alcohols | |||||||
| Inositol | Glycerol and glucose | Heterologous protein expression and regulation of carbon flux of glycolysis and pentose phosphate pathway | Fed-batch, high cell density | Pharmaceutical, food and feed industry | 30.71 | - | [84] |
| Xylitol | Xylose | Expression of xylose reductase (XYL1) from Scheffersomyces stipitis and gdh from Bacillus subtilis | Biotransformation | Sweetener with application in food, oral and personal care industries | 320 (mM) | 80% | [74] |
| Biofuels and oleochemicals | |||||||
| 2,3-butanediol | Glucose | Expression of alsS and alsD genes from Bacillus subtilis | Fed-batch | Chemical platform, biofuel | 74.5 | 0.3 | [85] |
| Isobutanol | Glucose | Expression of LlkivD, ScADH7, PpIlv2, PpIlv3, PpIlv5, PpIlv6 | Batch | Chemical platform, biofuel | 2.22 | 0.22 | [78] |
| Free fatty acids (FFA) | Methanol | Deletion of fatty acyl-CoA synthetase genes; expression of MmACL, BbPK—CkPTA; overexpression of DAS2 | Fed-batch | Platform for oleochemical and biofuel production | 23.4 | 0.078 | [86] |
| Terpenoids | |||||||
| Dammarenediol-II | Methanol and glycerol | Expression of PgDDS; overexpressing ERG1; down-regulation of ERG7 | Batch | Bioactive compound for pharmaceutical industry | - | 0.736 (mg.gDCW−1) | [87] |
| Lycopene | Glucose | Expression of heterologous carotenogenic enzymes and regulation of lipid metabolism pathway | Batch | Nutraceutical supplements, food colorants, cosmetic and pharmaceutical industries | 7.24 | 75.48 (mg.gDCW−1) | [88] |
| β-carotene | Glucose | Expression of heterologous carotenogenic enzymes crtE, crtB, crtI and crtL | Batch | - | 339 (μg.gDCW−1) | [76] | |
| (+)-nootkatone | Glucose and methanol | Co-expression of heterologous genes HmHPO, AtCPR, and CnValS; | Fed-batch | Flavor and fragrance compound for the food and cosmetics industries | 0.2 | [89] | |
| Overexpression of endogenous Adh and truncated Hmg1 | |||||||
| Polyketides | |||||||
| 6-Methylsalicylic acid | Methanol | Overexpression of atX and npgA | Fed-batch | Bioactive compounds for pharmaceutical industry | 2.2 | - | [90] |
| Lovastatin and Monacolin J | Methanol | Expression of LovB, LovC, LovG, NpgA, LovA, CPR, LovD, LovF | Fed-batch; co-culture | 0.250 and 0.593 | - | [75] | |
| Biopolymers | |||||||
| Polyhydroxyalkanoates | Oleic acid | Expression and peroxisomal targeting of PaPHA synthase | Batch | Biobased and biodegradable thermoplastic polyesters with broad-range industrial application | 1%.gDCW−1 | - | [91] |
| Hyaluronic acid | Glucose and methanol induction | Overexpression of xhasA2, xhasB from Xenopus laevis and endogenous hasC, hasD, hasE | Fed-batch | Biopolymer used in medical and pharmaceutical industry | 0.8–1.7 | - | [77] |
| Chondroitin sulfate | Methanol | Expression of kfoA, kfoC, tuaD, C4ST, ATPS and APSK | Fed-batch | Medical and nutraceutical industry | 2.1 | - | [65] |
| Heparin | Methanol | Expression of tuaD, kfiC, kfiA, NDST, C5 epi, 2OST,3OST, 6OST | Fed-batch | Anticoagulant drug | 2.08 | - | [92] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moraes, L.M.P.d.; Marques, H.F.; Reis, V.C.B.; Coelho, C.M.; Leitão, M.d.C.; Galdino, A.S.; Porto de Souza, T.P.; Piva, L.C.; Perez, A.L.A.; Trichez, D.; et al. Applications of the Methylotrophic Yeast Komagataella phaffii in the Context of Modern Biotechnology. J. Fungi 2024, 10, 411. https://doi.org/10.3390/jof10060411
Moraes LMPd, Marques HF, Reis VCB, Coelho CM, Leitão MdC, Galdino AS, Porto de Souza TP, Piva LC, Perez ALA, Trichez D, et al. Applications of the Methylotrophic Yeast Komagataella phaffii in the Context of Modern Biotechnology. Journal of Fungi. 2024; 10(6):411. https://doi.org/10.3390/jof10060411
Chicago/Turabian StyleMoraes, Lidia Maria Pepe de, Henrique Fetzner Marques, Viviane Castelo Branco Reis, Cintia Marques Coelho, Matheus de Castro Leitão, Alexsandro Sobreira Galdino, Thais Paiva Porto de Souza, Luiza Cesca Piva, Ana Laura Alfonso Perez, Débora Trichez, and et al. 2024. "Applications of the Methylotrophic Yeast Komagataella phaffii in the Context of Modern Biotechnology" Journal of Fungi 10, no. 6: 411. https://doi.org/10.3390/jof10060411
APA StyleMoraes, L. M. P. d., Marques, H. F., Reis, V. C. B., Coelho, C. M., Leitão, M. d. C., Galdino, A. S., Porto de Souza, T. P., Piva, L. C., Perez, A. L. A., Trichez, D., de Almeida, J. R. M., De Marco, J. L., & Torres, F. A. G. (2024). Applications of the Methylotrophic Yeast Komagataella phaffii in the Context of Modern Biotechnology. Journal of Fungi, 10(6), 411. https://doi.org/10.3390/jof10060411







