Contrasting Patterns of Fungal and Bacterial Endophytes Inhabiting Temperate Tree Leaves in Response to Thinning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Experimental Design
2.2. Sample Collection and Measurement
2.3. DNA Extraction, PCR Amplification, and Sequencing of Leaf Endophytic Bacterial and Fungal Communities
2.4. Bioinformatics Analysis
2.5. Statistical Analysis
3. Results
3.1. Alpha Diversity of Leaf Endophytic Fungi and Bacteria
3.2. Composition of Leaf Endophytic Fungi and Bacteria
3.3. Co-Occurrence Networks of Leaf Endophytic Fungi and Bacteria
3.4. Functional Prediction of Leaf Endophytic Fungi and Bacteria
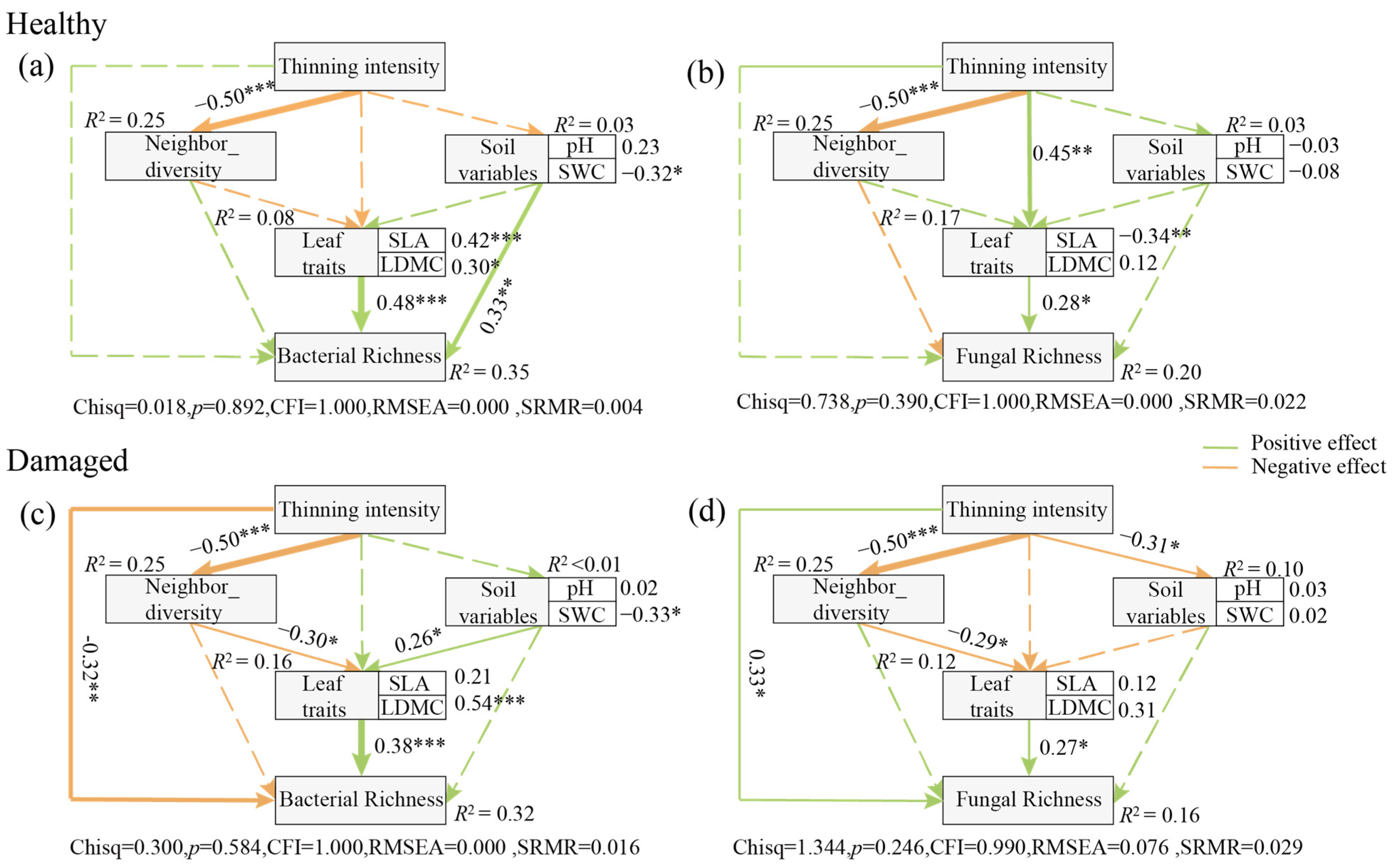
3.5. Drivers Triggering Variations in the Assemblages of Leaf Endophytic Fungi and Bacteria
4. Discussion
4.1. The Contrasting Responses of the Diversity of Leaf Endophytic Fungi and Bacteria to Thinning Intensity
4.2. The Contrasting Trends of the Complexity of Leaf Endophytic Fungal and Bacterial Co-Occurrence Networks
4.3. Leaf Trait-Mediated Thinning Effects on Leaf Fungal and Bacterial Endophytes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhan, C.; Matsumoto, H.; Liu, Y.; Wang, M. Pathways to Engineering the Phyllosphere Microbiome for Sustainable Crop Production. Nat. Food 2022, 3, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.R.; Lundberg, D.S.; del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host Genotype and Age Shape the Leaf and Root Microbiomes of a Wild Perennial Plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef] [PubMed]
- Eschen, R.; Hunt, S.; Mykura, C.; Gange, A.C.; Sutton, B.C. The Foliar Endophytic Fungal Community Composition in Cirsium Arvense Is Affected by Mycorrhizal Colonization and Soil Nutrient Content. Fungal Biol. 2010, 114, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Aydogan, E.L.; Moser, G.; Müller, C.; Kämpfer, P.; Glaeser, S.P. Long-Term Warming Shifts the Composition of Bacterial Communities in the Phyllosphere of Galium Album in a Permanent Grassland Field-Experiment. Front. Microbiol. 2018, 9, 144. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.M.; Porch, R.; Muscettola, I.E.; Vasconcelos, A.L.S.; Sherman, J.K.; Metcalf, C.J.E.; Lindow, S.E.; Koskella, B. Plant Neighborhood Shapes Diversity and Reduces Interspecific Variation of the Phyllosphere Microbiome. ISME J. 2022, 16, 1376–1387. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, P.T.; Whiteman, N.K. Insect Herbivory Reshapes a Native Leaf Microbiome. Nat. Ecol. Evol. 2020, 4, 221–229. [Google Scholar] [CrossRef]
- Arnold, A.E.; Mejía, L.C.; Kyllo, D.; Rojas, E.I.; Maynard, Z.; Robbins, N.; Herre, E.A. Fungal Endophytes Limit Pathogen Damage in a Tropical Tree. Proc. Natl. Acad. Sci. USA 2003, 100, 15649–15654. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Brettell, L.E.; Singh, B. Linking the Phyllosphere Microbiome to Plant Health. Trends Plant Sci. 2020, 25, 841–844. [Google Scholar] [CrossRef]
- Xu, P.; Fan, X.; Mao, Y.; Cheng, H.; Xu, A.; Lai, W.; Lv, T.; Hu, Y.; Nie, Y.; Zheng, X.; et al. Temporal Metabolite Responsiveness of Microbiota in the Tea Plant Phyllosphere Promotes Continuous Suppression of Fungal Pathogens. J. Adv. Res. 2022, 39, 49–60. [Google Scholar] [CrossRef]
- Liu, H.; Brettell, L.E.; Qiu, Z.; Singh, B.K. Microbiome-Mediated Stress Resistance in Plants. Trends Plant Sci. 2020, 25, 733–743. [Google Scholar] [CrossRef]
- Tian, J.; He, N.; Kong, W.; Deng, Y.; Feng, K.; Green, S.M.; Wang, X.; Zhou, J.; Kuzyakov, Y.; Yu, G. Deforestation Decreases Spatial Turnover and Alters the Network Interactions in Soil Bacterial Communities. Soil Biol. Biochem. 2018, 123, 80–86. [Google Scholar] [CrossRef]
- Carreño-Rocabado, G.; Peña-Claros, M.; Bongers, F.; Alarcón, A.; Licona, J.-C.; Poorter, L. Effects of Disturbance Intensity on Species and Functional Diversity in a Tropical Forest. J. Ecol. 2012, 100, 1453–1463. [Google Scholar] [CrossRef]
- Both, S.; Riutta, T.; Paine, C.E.T.; Elias, D.M.O.; Cruz, R.S.; Jain, A.; Johnson, D.; Kritzler, U.H.; Kuntz, M.; Majalap-Lee, N.; et al. Logging and Soil Nutrients Independently Explain Plant Trait Expression in Tropical Forests. New Phytol. 2019, 221, 1853–1865. [Google Scholar] [CrossRef]
- Xu, R.; Wang, L.; Zhang, J.; Zhou, J.; Cheng, S.; Tigabu, M.; Ma, X.; Wu, P.; Li, M. Growth Rate and Leaf Functional Traits of Four Broad-Leaved Species Underplanted in Chinese Fir Plantations with Different Tree Density Levels. Forests 2022, 13, 308. [Google Scholar] [CrossRef]
- Huang, Y.-L.; Zimmerman, N.; Arnold, A. Observations on the Early Establishment of Foliar Endophytic Fungi in Leaf Discs and Living Leaves of a Model Woody Angiosperm, Populus Trichocarpa (Salicaceae). J. Fungi 2018, 4, 58. [Google Scholar] [CrossRef]
- Murakami, K.; Matsuda, R. Optical and Physiological Properties of a Leaf. In LED Lighting for Urban Agriculture; Kozai, T., Fujiwara, K., Runkle, E.S., Eds.; Springer: Singapore, 2016; pp. 113–123. ISBN 978-981-10-1846-6. [Google Scholar]
- Le Provost, G.; Badenhausser, I.; Le Bagousse-Pinguet, Y.; Clough, Y.; Henckel, L.; Violle, C.; Bretagnolle, V.; Roncoroni, M.; Manning, P.; Gross, N. Land-Use History Impacts Functional Diversity across Multiple Trophic Groups. Proc. Natl. Acad. Sci. USA 2020, 117, 1573–1579. [Google Scholar] [CrossRef]
- Zhang, X.; Guan, D.; Li, W.; Sun, D.; Jin, C.; Yuan, F.; Wang, A.; Wu, J. The Effects of Forest Thinning on Soil Carbon Stocks and Dynamics: A Meta-Analysis. For. Ecol. Manag. 2018, 429, 36–43. [Google Scholar] [CrossRef]
- Baraloto, C.; Hérault, B.; Paine, C.E.T.; Massot, H.; Blanc, L.; Bonal, D.; Molino, J.-F.; Nicolini, E.A.; Sabatier, D. Contrasting Taxonomic and Functional Responses of a Tropical Tree Community to Selective Logging. J. Appl. Ecol. 2012, 49, 861–870. [Google Scholar] [CrossRef]
- Addo-Fordjour, P.; Ofosu-Bamfo, B.; Kwofie, F.; Akyea-Bobi, N.; Rahman, F.A.; Amoah, E. Changes in Liana Community Structure and Functional Traits along a Chronosequence of Selective Logging in a Moist Semi-Deciduous Forest in Ghana. Plant Ecol. Divers. 2020, 13, 75–84. [Google Scholar] [CrossRef]
- Liu, P.; Wang, W.; Bai, Z.; Guo, Z.; Ren, W.; Huang, J.; Xu, Y.; Yao, J.; Ding, Y.; Zang, R. Competition and Facilitation Co-Regulate the Spatial Patterns of Boreal Tree Species in Kanas of Xinjiang, Northwest China. For. Ecol. Manag. 2020, 467, 118167. [Google Scholar] [CrossRef]
- Connell, J.H. Diversity in Tropical Rain Forests and Coral Reefs: High Diversity of Trees and Corals Is Maintained Only in a Nonequilibrium State. Science 1978, 199, 1302–1310. [Google Scholar] [CrossRef] [PubMed]
- Bongers, F.; Poorter, L.; Hawthorne, W.D.; Sheil, D. The Intermediate Disturbance Hypothesis Applies to Tropical Forests, but Disturbance Contributes Little to Tree Diversity. Ecol. Lett. 2009, 12, 798–805. [Google Scholar] [CrossRef]
- Dos Santos, F.A.S.; Johst, K.; Grimm, V. Neutral Communities May Lead to Decreasing Diversity-Disturbance Relationships: Insights from a Generic Simulation Model: Diversity-Disturbance Curve of Neutral Theory. Ecol. Lett. 2011, 14, 653–660. [Google Scholar] [CrossRef]
- Christian, N.; Sedio, B.E.; Florez-Buitrago, X.; Ramírez-Camejo, L.A.; Rojas, E.I.; Mejía, L.C.; Palmedo, S.; Rose, A.; Schroeder, J.W.; Herre, E.A. Host Affinity of Endophytic Fungi and the Potential for Reciprocal Interactions Involving Host Secondary Chemistry. Am. J. Bot. 2020, 107, 219–228. [Google Scholar] [CrossRef]
- Yang, X.; Wang, P.; Xiao, B.; Xu, Q.; Guo, Q.; Li, S.; Guo, L.; Deng, M.; Lu, J.; Liu, L.; et al. Different Assembly Mechanisms of Leaf Epiphytic and Endophytic Bacterial Communities Underlie Their Higher Diversity in More Diverse Forests. J. Ecol. 2023, 111, 970–981. [Google Scholar] [CrossRef]
- Sohrabi, R.; Paasch, B.C.; Liber, J.A.; He, S.Y. Phyllosphere Microbiome. Annu. Rev. Plant Biol. 2023, 74, 539–568. [Google Scholar] [CrossRef]
- Underwood, N.; Inouye, B.D.; Hambäck, P.A. A Conceptual Framework for Associational Effects: When Do Neighbors Matter and How Would We Know? Q. Rev. Biol. 2014, 89, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Rowe, H.I.; Holland, J.D. High Plant Richness in Prairie Reconstructions Support Diverse Leafhopper Communities. Restor. Ecol. 2013, 21, 174–180. [Google Scholar] [CrossRef]
- Atkinson, B.; Bailey, S.; Vaughan, I.P.; Memmott, J. A Comparison of Clearfelling and Gradual Thinning of Plantations for the Restoration of Insect Herbivores and Woodland Plants. J. Appl. Ecol. 2015, 52, 1538–1546. [Google Scholar] [CrossRef]
- Tan, L.-Z.; Fan, C.-Y.; Fan, X.-H. Relationships between Species Diversity or Community Structure and Productivity of Woody-Plants in a Broad-Leaved Korean Pine Forest in Jiaohe, Jilin, China. Chin. J. Plant Ecol. 2017, 11, 1149–1156. [Google Scholar] [CrossRef]
- Yan, Y.; Xin-Na, Z.; Jie, Y.; Chun-Yu, Z.; Xiu-Hai, Z. Composition and Temporal Dynamics of Tree Seedlings at Different Successional Stages of Conifer and Broad-Leaved Mixed Forests in Jiaohe, Jilin Province, China. Chin. J. Plant Ecol. 2016, 40, 127–139. [Google Scholar] [CrossRef]
- Li, M.; Wu, Z.; Qiu, H.; Zhang, C.; Zhao, X. Short-Term Effects of Tending Felling on Ecological Services of Mixed broadleavedKorean Pine Forests at Jiaohe in Jilin Province, Northeastern China. J. Beijing For. Univ. 2019, 41, 40–49. [Google Scholar] [CrossRef]
- Chen, Q. Comparative Study on Photosynthetic Physiological Characteristics of Pinus Koraiensis Seedlings with Different Regeneration Methods in Jiaohe, Jilin, China; Beijing Forestry University: Beijing, China, 2021. [Google Scholar]
- Zhang, Y.; Cao, B.; Pan, Y.; Tao, S.; Zhang, N. Metabolite-Mediated Responses of Phyllosphere Microbiota to Rust Infection in Two Malus Species. Microbiol. Spectr. 2023, 11, e03831-22. [Google Scholar] [CrossRef]
- Li, S.; Deng, Y.; Wang, Z.; Zhang, Z.; Kong, X.; Zhou, W.; Yi, Y.; Qu, Y. Exploring the Accuracy of Amplicon-based Internal Transcribed Spacer Markers for a Fungal Community. Mol. Ecol. Resour. 2020, 20, 170–184. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Wen, T.; Xie, P.; Yang, S.; Niu, G.; Liu, X.; Ding, Z.; Xue, C.; Liu, Y.; Shen, Q.; Yuan, J. ggClusterNet: An R Package for Microbiome Network Analysis and Modularity-based Multiple Network Layouts. iMeta 2022, 1, e32. [Google Scholar] [CrossRef] [PubMed]
- Põlme, S.; Abarenkov, K.; Henrik Nilsson, R.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A User-Friendly Traits Database of Fungi and Fungus-like Stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling Function and Taxonomy in the Global Ocean Microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Lefcheck, J.S. piecewiseSEM: Piecewise Structural Equation Modelling in r for Ecology, Evolution, and Systematics. Methods Ecol. Evol. 2016, 7, 573–579. [Google Scholar] [CrossRef]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; Bodegom, P.M.; Bengtsson-Palme, J.; Anslan, S.; Coelho, L.P.; Harend, H.; et al. Structure and Function of the Global Topsoil Microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef] [PubMed]
- De Vries, F.T.; Griffiths, R.I.; Bailey, M.; Craig, H.; Girlanda, M.; Gweon, H.S.; Hallin, S.; Kaisermann, A.; Keith, A.M.; Kretzschmar, M.; et al. Soil Bacterial Networks Are Less Stable under Drought than Fungal Networks. Nat. Commun. 2018, 9, 3033. [Google Scholar] [CrossRef] [PubMed]
- Mille-Lindblom, C.; Fischer, H.J.; Tranvik, L. Antagonism between Bacteria and Fungi: Substrate Competition and a Possible Tradeoff between Fungal Growth and Tolerance towards Bacteria. Oikos 2006, 113, 233–242. [Google Scholar] [CrossRef]
- Kreutzweiser, D.P.; Hazlett, P.W.; Gunn, J.M. Logging Impacts on the Biogeochemistry of Boreal Forest Soils and Nutrient Export to Aquatic Systems: A Review. Environ. Rev. 2008, 16, 157–179. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Wang, G.; Zhang, Z.; Chen, Y.; Liu, X.; Peng, R. Drivers of Spatial Structure in Thinned Forests. For. Ecosyst. 2024, 11, 100182. [Google Scholar] [CrossRef]
- Waters, C.M.; Gonsalves, L.; Law, B.; Melville, G.; Toole, I.; Brassil, T.; Tap, P. The Effect of Thinning on Structural Attributes of a Low Rainfall Forest in Eastern Australia. For. Ecol. Manag. 2018, 409, 571–583. [Google Scholar] [CrossRef]
- Simmons, L.A.; Anderson, S.H. Effects of Logging Activities on Selected Soil Physical and Hydraulic Properties for a Claypan Landscape. Geoderma 2016, 269, 145–152. [Google Scholar] [CrossRef]
- Yang, H.; Ciais, P.; Frappart, F.; Li, X.; Brandt, M.; Fensholt, R.; Fan, L.; Saatchi, S.; Besnard, S.; Deng, Z.; et al. Global Increase in Biomass Carbon Stock Dominated by Growth of Northern Young Forests over Past Decade. Nat. Geosci. 2023, 16, 886–892. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular Ecological Network Analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef]
- Li, J.; Li, C.; Kou, Y.; Yao, M.; He, Z.; Li, X. Distinct Mechanisms Shape Soil Bacterial and Fungal Co-Occurrence Networks in a Mountain Ecosystem. FEMS Microbiol. Ecol. 2020, 96, fiaa030. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. Microbial Interactions: From Networks to Models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Ma, B.; Wang, H.; Dsouza, M.; Lou, J.; He, Y.; Dai, Z.; Brookes, P.C.; Xu, J.; Gilbert, J.A. Geographic Patterns of Co-Occurrence Network Topological Features for Soil Microbiota at Continental Scale in Eastern China. ISME J. 2016, 10, 1891–1901. [Google Scholar] [CrossRef]
- Montoya, J.M.; Pimm, S.L.; Solé, R.V. Ecological Networks and Their Fragility. Nature 2006, 442, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Coyle, J.R.; Halliday, F.W.; Lopez, B.E.; Palmquist, K.A.; Wilfahrt, P.A.; Hurlbert, A.H. Using Trait and Phylogenetic Diversity to Evaluate the Generality of the Stress-dominance Hypothesis in Eastern North American Tree Communities. Ecography 2014, 37, 814–826. [Google Scholar] [CrossRef]
- Olesen, J.M.; Bascompte, J.; Dupont, Y.L.; Jordano, P. The Modularity of Pollination Networks. Proc. Natl. Acad. Sci. USA 2007, 104, 19891–19896. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Nuccio, E.E.; Shi, Z.J.; He, Z.; Zhou, J.; Firestone, M.K. The Interconnected Rhizosphere: High Network Complexity Dominates Rhizosphere Assemblages. Ecol. Lett. 2016, 19, 926–936. [Google Scholar] [CrossRef]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.-T.; Weigel, D.; Kemen, E.M. Microbial Hub Taxa Link Host and Abiotic Factors to Plant Microbiome Variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef]
- Carlström, C.I.; Field, C.M.; Bortfeld-Miller, M.; Müller, B.; Sunagawa, S.; Vorholt, J.A. Synthetic Microbiota Reveal Priority Effects and Keystone Strains in the Arabidopsis Phyllosphere. Nat. Ecol. Evol. 2019, 3, 1445–1454. [Google Scholar] [CrossRef] [PubMed]
- Jorge, G.L.; Kisiala, A.; Morrison, E.; Aoki, M.; Nogueira, A.P.O.; Emery, R.J.N. Endosymbiotic Methylobacterium Oryzae Mitigates the Impact of Limited Water Availability in Lentil (Lens Culinaris Medik.) by Increasing Plant Cytokinin Levels. Environ. Exp. Bot. 2019, 162, 525–540. [Google Scholar] [CrossRef]
- Palberg, D.; Kisiała, A.; Jorge, G.L.; Emery, R.J.N. A Survey of Methylobacterium Species and Strains Reveals Widespread Production and Varying Profiles of Cytokinin Phytohormones. BMC Microbiol. 2022, 22, 49. [Google Scholar] [CrossRef]
- Lu, Y.-F.; Chai, C.-Y.; Hui, F.-L. Two New Erythrobasidium Species Inhabiting the Phyllosphere Discovered in the Baotianman Nature Reserve in China. Front. Microbiol. 2024, 15, 1287984. [Google Scholar] [CrossRef] [PubMed]
- Li, A.-H.; Yuan, F.-X.; Groenewald, M.; Bensch, K.; Yurkov, A.M.; Li, K.; Han, P.-J.; Guo, L.-D.; Aime, M.C.; Sampaio, J.P.; et al. Diversity and Phylogeny of Basidiomycetous Yeasts from Plant Leaves and Soil: Proposal of Two New Orders, Three New Families, Eight New Genera and One Hundred and Seven New Species. Stud. Mycol. 2020, 96, 17–140. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Yin, S.; Liu, X.; Zhang, W.; Gu, T.; Shen, Q.; Qiu, H. Fungal Networks in Yield-Invigorating and -Debilitating Soils Induced by Prolonged Potato Monoculture. Soil Biol. Biochem. 2013, 65, 186–194. [Google Scholar] [CrossRef]
- Matsumoto, H.; Fan, X.; Wang, Y.; Kusstatscher, P.; Duan, J.; Wu, S.; Chen, S.; Qiao, K.; Wang, Y.; Ma, B.; et al. Bacterial Seed Endophyte Shapes Disease Resistance in Rice. Nat. Plants 2021, 7, 60–72. [Google Scholar] [CrossRef] [PubMed]
- González-Teuber, M.; Palma-Onetto, V.; Aguilera-Sammaritano, J.; Mithöfer, A. Roles of Leaf Functional Traits in Fungal Endophyte Colonization: Potential Implications for Host–Pathogen Interactions. J. Ecol. 2021, 109, 3972–3987. [Google Scholar] [CrossRef]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The Soil Microbiome Influences Grapevine-Associated Microbiota. mBio 2015, 6, e02527-14. [Google Scholar] [CrossRef] [PubMed]
- Wright, I.J.; Reich, P.B.; Westoby, M.; Ackerly, D.D.; Baruch, Z.; Bongers, F.; Cavender-Bares, J.; Chapin, T.; Cornelissen, J.H.C.; Diemer, M.; et al. The Worldwide Leaf Economics Spectrum. Nature 2004, 428, 821–827. [Google Scholar] [CrossRef]
- Reich, P.B. The World-wide ‘Fast–Slow’ Plant Economics Spectrum: A Traits Manifesto. J. Ecol. 2014, 102, 275–301. [Google Scholar] [CrossRef]
- Biere, A.; Bennett, A.E. Three-way Interactions between Plants, Microbes and Insects. Funct. Ecol. 2013, 27, 567–573. [Google Scholar] [CrossRef]
- Schuldt, A.; Hönig, L.; Li, Y.; Fichtner, A.; Härdtle, W.; Von Oheimb, G.; Welk, E.; Bruelheide, H. Herbivore and Pathogen Effects on Tree Growth Are Additive, but Mediated by Tree Diversity and Plant Traits. Ecol. Evol. 2017, 7, 7462–7474. [Google Scholar] [CrossRef]
- García-Guzmán, G.; Dirzo, R. Patterns of Leaf-pathogen Infection in the Understory of a Mexican Rain Forest: Incidence, Spatiotemporal Variation, and Mechanisms of Infection. Am. J. Bot. 2001, 88, 634–645. [Google Scholar] [CrossRef] [PubMed]

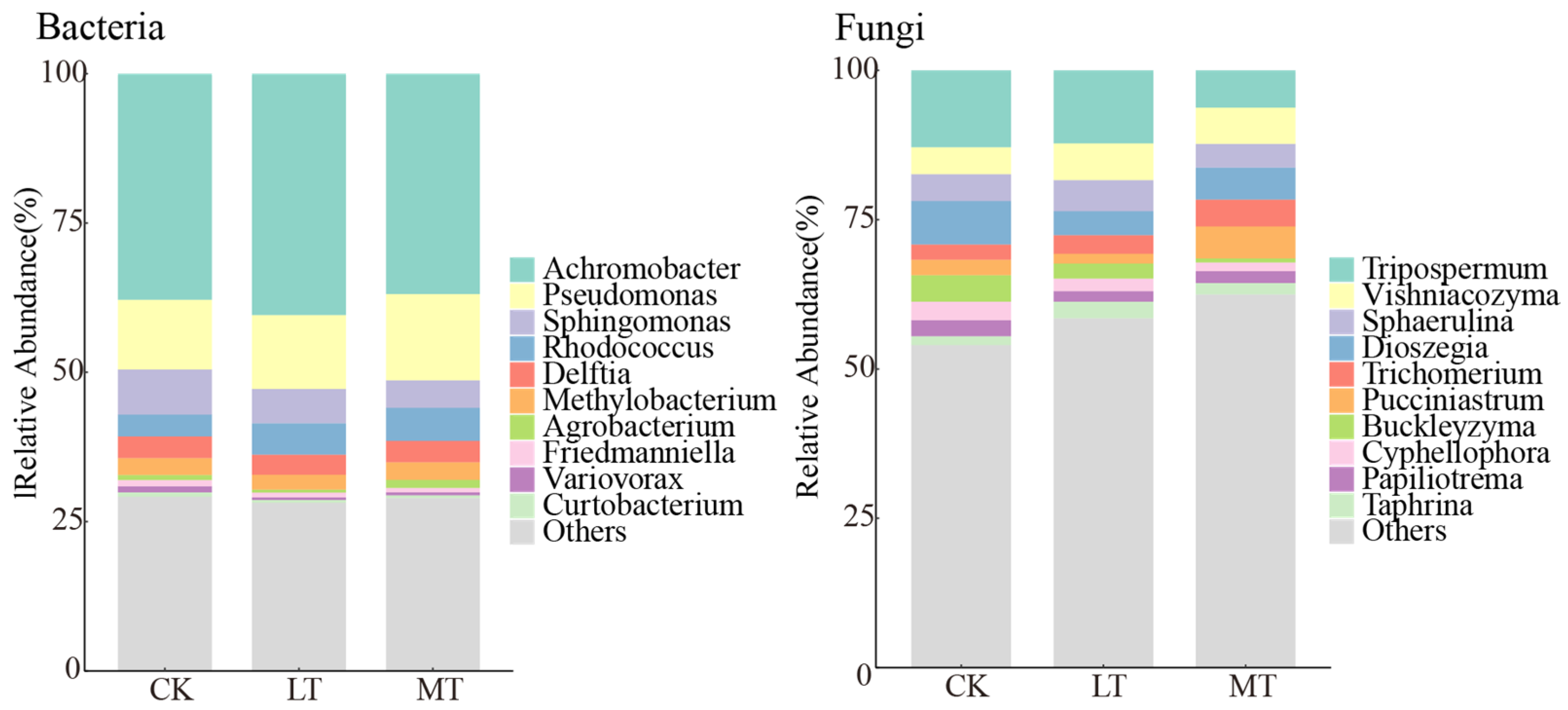



| Factor | Bacteria | Fungi | ||||||
|---|---|---|---|---|---|---|---|---|
| Richness | Shannon Index | Richness | Shannon Index | |||||
| Chisq | p | Chisq | p | Chisq | p | Chisq | p | |
| Thinning intensity (T) | 4.40 | 0.111 | 3.11 | 0.21 | 14.8 | <0.001 | 10.2 | 0.006 |
| Species (S) | 80.2 | <0.001 | 26.3 | <0.001 | 14.7 | 0.005 | 14.3 | 0.006 |
| Damaged (D) | 19.1 | <0.001 | 21.5 | <0.001 | 1.05 | 0.305 | 1.14 | 0.286 |
| T × S | 6.49 | 0.592 | 3.28 | 0.915 | 13.0 | 0.112 | 13.6 | 0.092. |
| T × D | 0.21 | 0.899 | 0.32 | 0.852 | 0.69 | 0.71 | 0.27 | 0.875 |
| S × D | 20.8 | <0.001 | 13.9 | <0.001 | 5.13 | 0.274 | 3.80 | 0.434 |
| T × S × D | 4.96 | 0.762 | 6.26 | 0.617 | 2.75 | 0.949 | 7.89 | 0.444 |
| Thinning Intensity | Edges | Nodes | Edge Density | Average Degree | Relative Modularity | |
|---|---|---|---|---|---|---|
| Bacteria | CK | 3286 | 470 | 0.030 | 13.983 | 1.761 |
| LT | 1439 | 388 | 0.019 | 7.418 | 1.232 | |
| MT | 2574 | 448 | 0.026 | 11.491 | 1.48 | |
| Fungi | CK | 17,922 | 1134 | 0.028 | 31.609 | 6.761 |
| LT | 21,922 | 1264 | 0.027 | 34.687 | 7.198 | |
| MT | 23,597 | 1271 | 0.029 | 37.131 | 7.823 |
| Factor | Thinning Intensity | ||
|---|---|---|---|
| CK | LT | MT | |
| Ca (g/kg) | 2.33 ± 0.19 a | 2.14 ± 0.18 a | 2.26 ± 0.19 a |
| Cu (mg/kg) | 6.49 ± 0.32 a | 6.39 ± 0.42 a | 6.06 ± 0.39 a |
| K (g/kg) | 9.84 ± 0.63 a | 9.69 ± 0.55 a | 9.03 ± 0.54 a |
| Mg (g/kg) | 2.31 ± 0.15 a | 2.21 ± 0.12 a | 2.31 ± 0.14 a |
| Zn (mg/kg) | 37.30 ± 2.43 a | 33.07 ± 3.12 ab | 24.84 ± 2.35 b |
| P (g/kg) | 2.3 ± 0.08 a | 2.06 ± 0.06 b | 2.09 ± 0.07 ab |
| C (g/kg) | 461.13 ± 2.99 a | 461 ± 4.78 a | 462.07 ± 5.17 a |
| N (g/kg) | 24.90 ± 1.11 a | 22.14 ± 0.8 a | 22.56 ± 0.88 a |
| C/N | 20.48 ± 1.21 a | 22.41 ± 1.19 a | 22.28 ± 1.24 a |
| SLA (cm2/g) | 204.27 ± 16.88 a | 155.06 ± 13.64 b | 160.02 ± 11.88 ab |
| LA (cm2) | 27.54 ± 2.53 a | 23.17 ± 2.03 a | 25.49 ± 2.04 a |
| LDMC (g/g) | 0.32 ± 0.02 ab | 0.31 ± 0.01 b | 0.35 ± 0.01 a |
| LL (cm) | 10.11 ± 0.37 a | 9.67 ± 0.29 a | 9.76 ± 0.33 a |
| LW (cm) | 4.80 ± 0.41 a | 4.44 ± 0.39 a | 4.74 ± 0.41 a |
| LT (mm) | 0.17 ± 0.01 a | 0.17 ± 0.01 a | 0.16 ± 0.01 a |
| Neighbor_diversity | 0.99 ± 0.08 a | 0.65 ± 0.06 b | 0.35 ± 0.08 c |
| pH | 4.40 ± 0.07 a | 4.29 ± 0.08 a | 3.99 ± 0.08 b |
| SWC (%) | 47.05 ± 0.88 a | 45.22 ± 1.25 a | 45.57 ± 1.62 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Li, C.; Zhao, X.; Zhang, C.; He, X.; Qu, L.; Zhang, N. Contrasting Patterns of Fungal and Bacterial Endophytes Inhabiting Temperate Tree Leaves in Response to Thinning. J. Fungi 2024, 10, 470. https://doi.org/10.3390/jof10070470
Liu B, Li C, Zhao X, Zhang C, He X, Qu L, Zhang N. Contrasting Patterns of Fungal and Bacterial Endophytes Inhabiting Temperate Tree Leaves in Response to Thinning. Journal of Fungi. 2024; 10(7):470. https://doi.org/10.3390/jof10070470
Chicago/Turabian StyleLiu, Beiping, Chunhuan Li, Xiuhai Zhao, Chunyu Zhang, Xinyi He, Laiye Qu, and Naili Zhang. 2024. "Contrasting Patterns of Fungal and Bacterial Endophytes Inhabiting Temperate Tree Leaves in Response to Thinning" Journal of Fungi 10, no. 7: 470. https://doi.org/10.3390/jof10070470
APA StyleLiu, B., Li, C., Zhao, X., Zhang, C., He, X., Qu, L., & Zhang, N. (2024). Contrasting Patterns of Fungal and Bacterial Endophytes Inhabiting Temperate Tree Leaves in Response to Thinning. Journal of Fungi, 10(7), 470. https://doi.org/10.3390/jof10070470






