Abstract
Four new species of jelly fungi were described from northeastern China based on morphological and molecular evidence. These new species were classified into the four genera Sirobasidium (Sirobasidium jilinense), Calocera (Calocera velutina), Dacrymyces (Dacrymyces jauensis), and Dacryopinax (Dacryopinax manghanensis). Maximum likelihood and Bayesian analyses were performed using a combined nuc rDNA internal transcribed spacer region (ITS) and nuc 28S rDNA (nrLSU) dataset for the construction of phylogenetic trees. Morphological descriptions, line illustrations, and the ecological habits of these new species are provided.
1. Introduction
Sirobasidium Lagerh. & Pat. was established in 1892, with the type species being Sirobasidium sanguineum Lagerh. & Pat [1]. Due to the presence of hypobasidia that are divided vertically, the genus was first assigned to Tremellaceae Fr. In 1895, Möller proposed the establishment of Sirobasidiaceae Lindau due to the unique chain-like hypobasidia of this species [2]. Two years later, Sirobasidiaceae was officially published by Lindau [3]. Lowy then discovered that the French record of S. cerasi Bourdot & Galzin did not match the requirements of the genus concerning the size of its basidiomata, the quantity of chain-shaped basidia, and the size of its basidiospores. Lowy also provided a key for the recognized species S. brefeldianum Möller, S. magnum Boedijn, S. albidum, and S. sanguineum [4]. It was not until Bandoni examined the basidia and basidiospores of Sirobasidium that the spindle-shaped epiprobasidia shed by species in this genus were recognized as basidiospores [5]. In 2015, Roedel and Putzmann compiled a key for the Sirobasidium in their study of S. albidum, which included eight species [6]. Unfortunately, molecular information was not provided. At present, the genus contains ten species [3], three of which have been identified in China: S. japonicum, S. magnum, and S. sanguineum [7]. The chain-like hypobasidia and exfoliated epiprobasidia are the two primary characteristics of members of this genus. Phylogenetic analyses have shown the genus to be polyphyletic [8,9,10,11,12,13]. It is currently challenging to confirm the taxonomic status of Sirobasidium, however, as there are currently no molecular sequences available for the type species.
Dacrymycetaceae J. Schröt. (1888) belongs to Basidiomycota [3]. This family includes a variety of small-sized, morphologically diverse, and gelatinous species that are hard to find when dry. The majority of the basidiomata are yellow and contain carotene. The majority of basidiospores are often separated transversely, occasionally they can also be separated longitudinally or obliquely, or not septate when immature, septate by thin or thick walls; germination produces conidia or not; basidia and basidiospores contain oil droplets; hyphidia are present or absent; and with or without clamp connection [14].
In the 1960s and 1970s, McNabb conducted a detailed study on the genera of Dacrymycetaceae based on their morphology and compiled a key [15,16,17]. Since the 21st century, with the development of molecular systematics, researchers have conducted more in-depth research on Dacrymycetaceae species. In 2007, Shirouzu et al. found, in their phylogenetic analysis of Dacrymyces Nees based on 28S, that Dacrymyces was polyphyletic. While some species of Dacrymyces nest in Cerinomycetaceae Jülich or form a separate clade within Dacrymycetes, the majority of Dacrymyces species are found nested in Dacrymycetaceae [18]. In their 2009 study of Dacrymycetes in Japan, Shirouzu et al. described five new species and provided line illustrations [14]. Based on morphological observations, Malysheva (2013) identified the species of Calocera (Fr.) Fr. in Russia, described eight species of the genus, and provided line illustrations [19]. Later, Shirouzu et al. published eight new species of Dacrymycetes from New Zealand [20]. Dacryopinax G.W. Martin species from Brazil and Mexico were studied by Renato et al. and Castro-Santiuste et al., respectively [21,22]. The above studies show that most genera in Dacrymycetaceae are polyphyletic, and species are scattered in various branches of Dacrymycetaceae.
“Flora Fungorum Sinicorum Vol. 2 Tremellales et Dacrymycetales” by Liu was the primary focus of early Chinese research on Dacrymycetaceae species [7]. At the time, only morphological research was included due to the restricted conditions. Chinese researchers have recently studied species such as Calocera and Dacrymyces using multi-gene fragments [23,24,25]. However, most of the specimens used in these studies were collected from southern China, while there are relatively few specimens from northeastern China.
To enrich the species resources of jelly fungi in Northeast China. We have discovered four new species through morphological research and phylogenetic analysis. This study provided a detailed description and discussion of these species.
2. Materials and Methods
2.1. Specimen Collection
The specimens for this study were collected from Jilin Province and the Inner Mongolia Autonomous Region in China and deposited in the herbarium Fungarium of Jilin Agricultural University (FJAU).
2.2. Morphological Studies
Field records of fresh basidiomata, photographs of the habitat, and records after drying were used for macromorphological descriptions. The basidiomata’s color description was derived from Kornerup and Wanscher [26]. Using the technique of freehand slicing, utilizing water and 5% KOH solution as the floating carrier, and dyeing with 1% Phloxine B solution, the structures of basidia, basidiospores, hyphidia, and hyphae were examined under an optical microscope (Olympus CX33, Olympus, Tokyo, Japan). Twenty mature microstructures were chosen at random, examined under a microscope, and measured. “a–b × c–d” indicates the measurement of basidia, basidiospores, and hyphidia; “e–f” indicates the measurement of hyphae. Moreover, we determined the basidiospore’s Q value (Q = length/width). The extreme values of length, width, and diameter are denoted by the letters “a–b”, “c–d”, and “e–f”, respectively. For a given species, “n” denotes the ratio of the total number of measured basidiospores to the total number of measured specimens. All data were collected within a measurement range of 90%. The width was measured by choosing the area that was the widest. The apiculum was not taken into account when measuring the length of basidiospores. Along with the macroscopic morphology and microstructure features, a description and line charts are provided.
2.3. DNA Extraction, PCR and Sequencing
To extract DNA from dry specimens, the plant genome extraction kit from Kangwei Century (CWBIO, Beijing, China) was used. The ITS1F/ITS4 [27] and LR0R/LR7 [28,29] primers were used to amplify and sequence the ITS and nrLSU fragments, respectively. Sangon Bioengineering (Shanghai, China) Co., Ltd. was entrusted with the sequencing work, and the PCR reaction procedure was based on the method of Wang and Bau [30].
2.4. Molecular Phylogenetic Analyses
Sequencher 5.4.5 was used to observe the peak maps, further screen and sort the self-test sequences, and perform blast alignment [31]. ITS and nrLSU sequences were downloaded from GenBank based on the alignment outcomes and morphological similarity [32] (Table 1). The ITS and nrLSU sequences were aligned using the GINS-i algorithm with two iterative cycles only, using the online Mafft tool version 7 [33]. The alignment was manually adjusted and trimmed using MEGA 7.0.
The concatenated alignment of ITS (1–646) and nrLSU (647–1248) of Sirobasidium comprised 64 sequences. The concatenated alignment of ITS (1–682) and nrLSU (683–1517) of Dacrymycetaceae comprised 197 sequences. The best-fit model (edge-unlinked) was selected using the BIC criterion with ModelFinder [34]. The best-fit model of Sirobasidium and Dacrymycetaceae, according to BIC, was both SYM + I + G4. With the use of IQ-TREE [35] and the Shimodaira–Hasegawa-like approximate likelihood ratio test [36], maximum likelihood phylogenies were inferred under the edge–unlinked partition model for 10,000 ultrafast [37] bootstraps. The best-fit partition model (edge-unlinked) was again selected using the BIC criterion with ModelFinder. The best-fit model of Sirobasidium according to BIC was SYM + I + G4 (ITS) and K2P + I + G4 (nrLSU). The best-fit model of Dacrymycetaceae according to BIC was SYM + I + G4 (ITS) and GTR + F + I + G4 (nrLSU). Under the partition model, in which the first 25% of sampled data were discarded as burn-in, Bayesian inference phylogenies were inferred using MrBayes 3.2.6 [38]. Fig Tree v1.4.3 [39] and Photoshop 2021 (Adobe, Sam Jose, CA, USA) were then used to visualize the phylogenetic tree.

Table 1.
Sequence information from phylogenetic trees.
Table 1.
Sequence information from phylogenetic trees.
| Species | Locality | Sample No. | GenBank No. | References | |
|---|---|---|---|---|---|
| ITS | nrLSU | ||||
| Bullera alba | USA | CBS 501T | AF444368 | AF075500 | [40] |
| Bullera hannae | USA | CBS 8286T | AF444486 | AF363661 | [40] |
| Bullera penniseticola | USA | CBS 8623T | AF444471 | AF363649 | [40] |
| Bullera unica | USA | CBS 8290T | AF444441 | AF075524 | [40] |
| Calocera bambusicola | China | Wu9910 12 | FJ195751 | - | [23] |
| Calocera cornea | USA | AFTOL ID 438 | AY789083 | AY701526 | [41] |
| Calocera cornea | Sweden | UPS F 940774 | MN595626 | MN595626 | [41] |
| Calocera cornea | Germany | MW 55 | - | AF291302 | [42] |
| Calocera cornea | Canada | CBS 124 84 | AB712437 | AB472738 | [43] |
| Calocera furcata | Russia | H Spirin 10949 | MW191975 | MW159088 | [43] |
| Calocera fusca | New Zealand | PDD 107972 | LC131406 | LC131365 | [20] |
| Calocera glossoides | Ukraine | CWU6247 | MW191968 | MW159084 | [44] |
| Calocera lutea | New Zealand | PDD 107841 | LC131413 | LC131372 | [20] |
| Calocera palmata | Japan | CBS 127 51 | MH856777 | MH868295 | [32] |
| Calocera pedicellata | New Zealand | PDD 107830 | LC131415 | LC131374 | [20] |
| Calocera sinensis | China | FJAU68949 | PP749224 | - | This study |
| Calocera sinensis | China | Dai22645 | OL518939 | OL518948 | [32] |
| Calocera tibetica | China | Dai20171 | MW549777 | MW750403 | |
| Calocera velutina | China | FJAU68950T | PP776476 | PP776564 | This study |
| Calocera velutina | China | FJAU68951 | PP776543 | PP776565 | This study |
| Calocera viscosa | Japan | TUFC12873 | AB712439 | - | [43] |
| Calocera viscosa | China | FJAU68917 | - | PP749235 | This study |
| Calocera viscosa | Japan | 175 | - | AB299048 | [18] |
| Calocera viscosa | Germany | AFTOL ID 1679T | DQ520102 | DQ520102 | [41] |
| Calocera viscosa | Sweden | UPS F 940773 | MN595628 | MN595628 | [41] |
| Cerinomyces cokeri | Canada | TU135089 | MW191983 | MW191983 | [44] |
| Cerinomyces concretus | Colombia | O F 919450T | MW191933 | - | [44] |
| Cerinomyces fugax | USA | HHB 8856T | MW191905 | MW159051 | [44] |
| Cerinomyces hesperidis | USA | NY01782362T | MW191921 | MW159065 | [44] |
| Cerinomyces inermis | New Zealand | PDD87816T | MW191887 | - | [44] |
| Cerinomyces lipoferus | Netherlands | ENZ20001 | MZ147626 | MZ147626 | [44] |
| Cerinomyces nepalensis | Nepal | O F 904088 | MW191896 | - | [44] |
| Cerinomyces neuhoffii | Sweden | UPS F 941020T | MN595625 | MN595625 | [41] |
| Cerinomyces pallidus | USA | ARIZ AN09245 | MZ147624 | - | [44] |
| Cerinomyces paulistanus | Brazil | O Ryvarden 24759T | MW191934 | - | [44] |
| Cerinomyces pinguis | Nepal | O F 904085T | MW191907 | MW159043 | [44] |
| Cerinomyces tortus | Sweden | UPS F 946515 | MW191999 | MW191999 | [44] |
| Cerinomyces volaticus | Russia | LE295748 | MW191901 | MW159047 | [44] |
| Dacrymyces adpressus | Japan | 554 | - | AB472729 | [14] |
| Dacrymyces ancyleus | Japan | MAFF 241177T | AB712448 | AB472713 | [43] |
| Dacrymyces capitatus | China | FJAU68952 | PP749225 | PP749236 | This study |
| Dacrymyces cerebriformis | China | Dai 19832T | OM955202 | OM955197 | [25] |
| Dacrymyces chiangraiensis | Thailand | MFLU 16 0572 | KY498587 | - | [45] |
| Dacrymyces chrysocomus | Japan | CBS 280 84 | AB712451 | AB712427 | [43] |
| Dacrymyces chrysospermus | China | FJAU68956 | PP749226 | PP749237 | This study |
| Dacrymyces citrinus | New Zealand | PDD 107915T | LC131417 | LC131376 | [20] |
| Dacrymyces corticioides | USA | NY02686162 | MW191944 | MW159068 | [44] |
| Dacrymyces cylindricus | New Zealand | PDD 105052T | LC131419 | LC131378 | [20] |
| Dacrymyces cyrtosporus | New Zealand | PDD 107980T | LC131422 | LC131381 | [20] |
| Dacrymyces dictyosporus | Japan | HHB 8618 | AB712454 | AB712429 | [43] |
| Dacrymyces fennicus | Finland | H Miettinen 21174 | MW191957 | MW159071 | [44] |
| Dacrymyces flabelliformis | New Zealand | HHB 18308T | AB712455 | AB712430 | [43] |
| Dacrymyces grandinioides | Kenya | K M 237139T | MW191980 | MW191980 | [44] |
| Dacrymyces intermedius | New Zealand | PDD 107851 | - | LC131384 | [20] |
| Dacrymyces invisibilis | Chile | 14597MDT | MH230100 | MH230102 | [46] |
| Dacrymyces jauensis | China | FJAU68959 | PP776532 | PP776577 | This study |
| Dacrymyces jauensis | China | FJAU68960 | PP776533 | PP776578 | This study |
| Dacrymyces jauensis | China | FJAU68961T | PP776534 | PP776579 | This study |
| Dacrymyces lacrymalis | Japan | TUFC13327 | AB712456 | AB299069 | [43] |
| Dacrymyces longistipitatus | New Zealand | PDD 107997T | LC131426 | LC131387 | [20] |
| Dacrymyces microsporus | China | Wu561 | AB712458 | OL546779 | [32] |
| Dacrymyces minor | Japan | TUFC12836 | AB712458 | - | [43] |
| Dacrymyces minutus | Japan | 648 | AB712459 | AB472733 | [43] |
| Dacrymyces novae-zelandiae | New Zealand | PDD 107953 | LC131428 | LC131391 | [20] |
| Dacrymyces ovisporus | Norway | H Spirin 11145 | MW191960 | MW159073 | [44] |
| Dacrymyces pachysporus | New Zealand | PDD 105004T | LC131429 | LC131392 | [20] |
| Dacrymyces parastenosporus | New Zealand | PDD 104963T | LC131432 | LC131395 | [20] |
| Dacrymyces pezizoides | Japan | TNS F 54909 | LC386890 | LC386894 | [47] |
| Dacrymyces pinacearum | Japan | UPS F 593533 | MN595637 | MN595637 | [41] |
| Dacrymyces puniceus | China | FJAU68955 | PP749227 | PP749238 | This study |
| Dacrymyces san-augustinii | China | Wu544 | OL587813 | OL546781 | [32] |
| Dacrymyces sinostenosporus | China | Dai20003T | MW540888 | MW540890 | [25] |
| Dacrymyces stenosporus | New Zealand | PDD 105018T | LC131433 | LC131396 | [20] |
| Dacrymyces stillatus | Sweden | UPS F 939814 1 | MN595677 | MN595677 | [41] |
| Dacrymyces subalpinus | Japan | 228 | - | AB299060 | [18] |
| Dacrymyces subantarcticensis | New Zealand | PDD 107948 | LC131435 | LC131399 | [20] |
| Dacrymyces subarcticus | Japan | HNo 544 | AB712467 | - | [43] |
| Dacrymyces variisporus | Japan | TUFC14203 | AB712470 | - | [43] |
| Dacrymyces venustus | Ethiopia | O Adane 150 | MW191949 | MW159075 | [44] |
| Dacryomitra pusilla | Sweden | UPS F 176774 | MN595639 | MN595639 | [41] |
| Dacryopinax elegans | USA | HHB 18731 | AB712471 | AB712433 | [43] |
| Dacryopinax indacocheae | Venezuela | CRM 72 | AB712472 | AB712434 | [43] |
| Dacryopinax lowyi | Mexico | 27618 | MN733712 | MN733723 | [22] |
| Dacryopinax manghanensis | China | FJAU68940 | PP749230 | PP749241 | This study |
| Dacryopinax manghanensis | China | FJAU68938 | PP749228 | PP749239 | This study |
| Dacryopinax manghanensis | China | FJAU68941 | PP749231 | PP749242 | This study |
| Dacryopinax manghanensis | China | FJAU68943T | PP749233 | PP749244 | This study |
| Dacryopinax manghanensis | China | FJAU68942 | PP749232 | PP749243 | This study |
| Dacryopinax manghanensis | China | FJAU68939 | PP749229 | PP749240 | This study |
| Dacryopinax martinii | Mexico | 682 | MN733715 | MN733726 | [22] |
| Dacryopinax maxidorii | Brazil | RLMA306 | OK257531 | OK257536 | [48] |
| Dacryopinax primogenitus | Costa Rica | MIN 862738 | KT251038 | KT251039 | [49] |
| Dacryopinax sp. | Kenya | H7008759 | MW191959 | MW159091 | [44] |
| Dacryopinax spathularia | China | FJAU68908 | PP749234 | PP749245 | This study |
| Dacryopinax spathularia | USA | H Miettinen 16740 | MW191973 | MW159085 | [44] |
| Dacryopinax spathularia | Indonesia | H Miettinen 20559 | MW191976 | MW159092 | [44] |
| Dacryopinax spathularia | Chinna | Wu 331 | OL614833 | OL616184 | [32] |
| Dacryopinax sphenocarpa | Japan | 534 | - | AB472726 | [14] |
| Dendrodacrys brasiliense | Brazil | INPA 241458T | AB744230 | AB723514 | [50] |
| Dendrodacrys ciprense | Cyprus | UPS F 946590T | OM519385 | OM519385 | [51] |
| Dendrodacrys concrescens | Sweden | UPS F 946602T | OM519390 | OM519390 | [51] |
| Dendrodacrys dendrocalami | Japan | TUFC13914 | AB712453 | AB712428 | [43] |
| Dendrodacrys ellipsosporum | Spain | UPS F 946604T | OM519392 | OM519392 | [51] |
| Dendrodacrys kennedyae | Panama | TAAM192134T | OP529836 | OP529830 | [51] |
| Dendrodacrys laetum | Kenya | H7008757T | OP529841 | OP529841 | [51] |
| Dendrodacrys oblongisporum | Norway | UPS F 946599 | OM519399 | OM519399 | [51] |
| Dendrodacrys rigoratum | Japan | TUFC12845T | AB712447 | - | [14] |
| Ditiola haasii | Czech Republic | PRM 944647 | MW557322 | MW557322 | [52] |
| Ditiola peziziformis | Finland | H Haikonen 2426 | MW191972 | MW159070 | [18] |
| Fellomyces horovitziae | USA | CBS 7515T | - | AF189856 | [53] |
| Femsjonia monospora | Thailand | MFLU 16 0608 | KY498588 | - | [45] |
| Femsjonia uniseptata | Japan | TNS F 54019T | NR_164095 | NG_060435 | [20] |
| Fibulobasidium inconspicuum | USA | CBS 8237T | AF444318 | AF363641 | [40] |
| Fibulobasidium murrhardtense | China | CBS 9109T | GU327540 | AF416648 | [54] |
| Fibulobasidium sirobasidioides | Germany | RJB 12787 | - | AF416644 | [55] |
| Fonsecazyma mujuensis | China | CBS 10308T | KF036595 | DQ333884 | [12] |
| Genolevuria amylolytica | China | CBS 10048T | KF036585 | AY562134 | [12] |
| Genolevuria armeniaca | China | CBS 10050 | KF036587 | AY562140 | [12] |
| Genolevuria bromeliarum | Brazil | BI20 | EU386359 | DQ784566 | [56] |
| Genolevuria tibetensis | China | AS 2 2653T | EF363146 | EF363143 | [57] |
| Guepiniopsis alpina | USA | HAY F 002724 | OR767855 | - | [32] |
| Guepiniopsis buccina | USA | AFTOL ID 888 | DQ206986 | AY745711 | [41] |
| Guepiniopsis estonica | Sweden | UPS F 940137 | MN595632 | MN595632 | [41] |
| Heterotextus miltinus | New Zealand | PDD 107924 | LC131438 | LC131402 | [20] |
| Kockovaella imperatae | USA | CBS 7554T | AB054091 | AF189862 | [53] |
| Pseudotremella allantoinivorans | USA | CBS 9604T | AY315664 | AY315662 | [58] |
| Pseudotremella moriformis | USA | CBS 7810 | AF444331 | AF075493 | [40] |
| Sirobasidium apiculatum | Japan | MY62 1 | LC203425 | LC203426 | [13] |
| Sirobasidium apiculatum | Japan | MY62 4 | LC203428 | LC203429 | [13] |
| Sirobasidium brefeldianum | Spain | AM71 | JN053472 | JN043578 | [9] |
| Sirobasidium brefeldianum | USA | CBS 7805 | AF444330 | AF075492 | [40] |
| Sirobasidium japonicum | Japan | MY111 05 | LC203420 | LC016573 | [13] |
| Sirobasidium japonicum | Japan | MY111 09 | LC203422 | LC203423 | [13] |
| Sirobasidium jilinense | China | FJAU68667 | PP749214 | - | This study |
| Sirobasidium jilinense | China | FJAU68670T | PP749217 | PP749222 | This study |
| Sirobasidium jilinense | China | FJAU68668 | PP749215 | PP749220 | This study |
| Sirobasidium jilinense | China | FJAU68669 | PP749216 | PP749221 | This study |
| Sirobasidium jilinense | China | FJAU68672 | PP749219 | - | This study |
| Sirobasidium jilinense | China | FJAU68671 | PP749218 | PP749223 | This study |
| Sirobasidium magnum | Spain | AM70 | JN053497 | JN043603 | [9] |
| Sirobasidium magnum | Netherlands | CBS 6803 | AF444314 | KY109657 | [40] |
| Sirobasidium magnum | Netherlands | CBS 6964 | KY105421 | KY109658 | [59] |
| Sirobasidium magnum | China | FJAU68996 | PP741731 | PP741733 | This study |
| Sirobasidium magnum | China | FJAU68997 | PP741732 | PP741734 | This study |
| Tremella giraffa | Germany | CCJ 1553 | AF042453 | AF042271 | [60] |
Note: “-” denotes the absence of pertinent genetic data. Sequences newly generated in this study are indicated in bold. The type specimens are those designated with a “T”.
3. Results
3.1. Phylogenetic Analyses
This study generated a total of 46 new sequences, including 24 ITS sequences and 22 nrLSU sequences. These new sequences have been uploaded to GenBank. The multi-locus dataset (ITS + nrLSU) of Sirobasidium had an aligned length of 1248 total characters), including gaps. The multi-locus dataset (ITS + nrLSU) of Dacrymycetaceae had an aligned length of 1517 total characters, including gaps. Only the topological structures of Bayesian inference are displayed, as the topological structures of ML and BI are very similar. Bootstrap support (BS) values ≥ 75% and Bayesian posterior probability (PP) values ≥ 0.75 are indicated on branches (BS/PP) (Figure 1 and Figure 2).
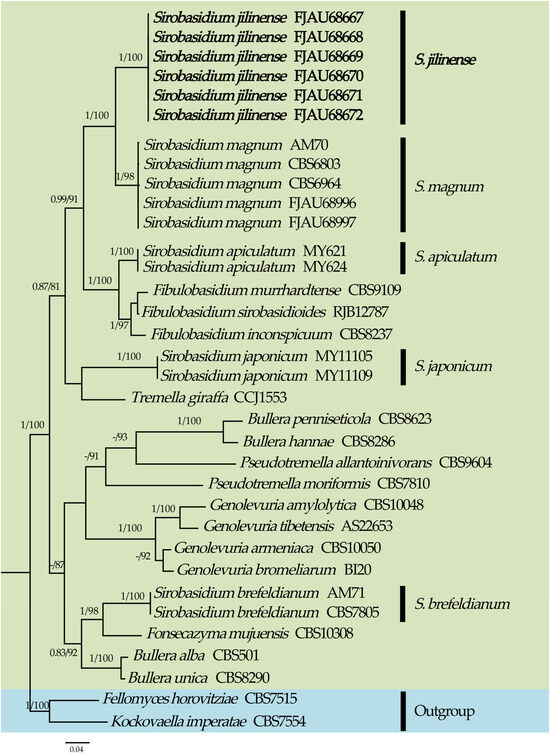
Figure 1.
Phylogeny of Sirobasidium and related species by Bayesian inference based on the ITS + nrLSU dataset. New species are indicated in bold.
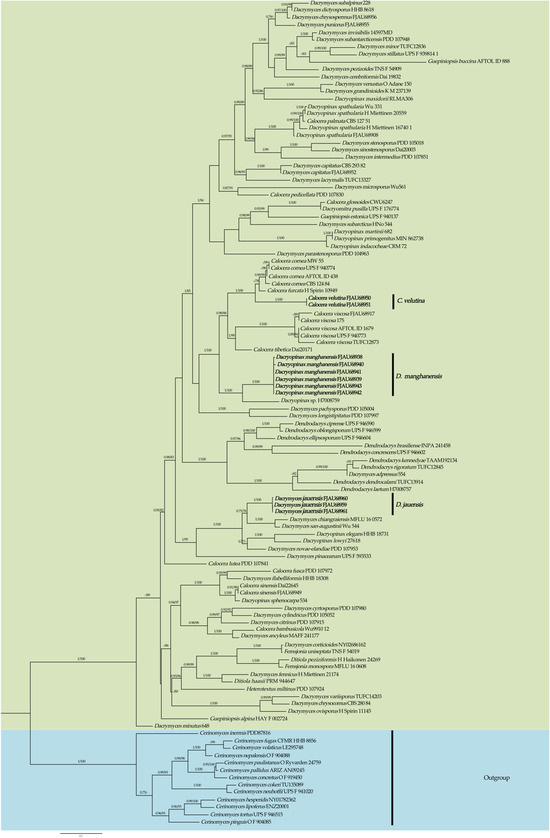
Figure 2.
Phylogeny of Dacrymycetaceae by Bayesian inference based on the ITS + nrLSU dataset. New species are indicated in bold.
From the phylogenetic tree of Sirobasidium and its related species constructed with Fellomyces horovitziae Spaaij, G. Weber & Oberw. and Kockovaella imperatae Nakase, I. Banno & Y. Yamada as outgroups (highlighted in blue), it can be seen that Sirobasidium is polyphyletic (Figure 1). High support rates (PP = 1.00, MLbs = 100%) were attained by the six specimens of the new species Sirobasidium jilinense T. Bau et X. Wang sp. nov., which were grouped in an independent branch. S. jilinense and S. magnum share a sister relationship and received high support (PP = 1.00, MLbs = 100%). The novel species could also be easily distinguished from other species, thanks to its distinct placement in the phylogenetic tree (Figure 1).
From the phylogenetic tree of Dacrymycetaceae constructed with Cerinomycetaceae as an outgroup (highlighted in blue), it can be seen that Dacrymyces, Dacryopinax, and Calocera are polyphyletic. The species belonging to each genus are dispersed across different branches of Dacrymycetaceae (Figure 2).
The two specimens of the recently discovered species Calocera velutina T. Bau et X. Wang sp. nov. were grouped into a separate branch and had a high support rate (PP = 1.00, MLbs = 100%). C. velutina forms a sister clade with C. cornea (Batsch) Fr. and C. furcata (Fr.) Fr., which is highly supported (PP = 1.00, MLbs = 100%). At the same time, the new species can be distinguished from other species in the genus in the phylogenetic tree (Figure 2).
A high support rate (PP = 1.00, MLbs = 100%) was obtained for the three specimens of the new species, Dacrymyces jauensis T. Bau et X. Wang sp. nov. They were grouped into an independent branch. D. jauensis forms a sister relationship with the clade consisting of D. chiangraiensis Ekanayaka, Karun., Q. Zhao & KD Hyde, and D. san-augustinian Kobayasi and gained a lower support rate (PP = 0.75, MLbs = 78%). This suggests that other species among them have not yet been identified. The new species and other species could also be well distinguished in the phylogenetic tree (Figure 2).
The six specimens of the new species Dacryopinax manghanensis T. Bau et X. Wang sp. nov. were clustered into an independent branch and obtained a high support rate (PP = 1.00, MLbs = 100%). It is a sister clade to Dacryopinax sp. (H7008759), with a high support rate (PP = 1.00, MLbs = 100%), and which is well distinguished from other species.
3.2. Taxonomy

Figure 3.
Basidiomata of Sirobasidium jilinense: (a) fresh (FJAU68670); (b) fresh (FJAU68668); (c,d) dry (FJAU68667). Scale bars: (a,b) = 1 cm; (c,d) = 0.5 cm.
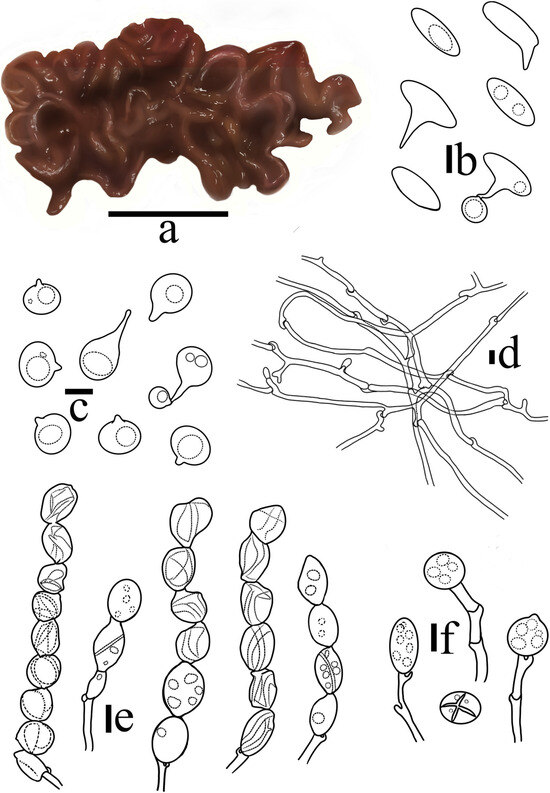
Figure 4.
Sirobasidium jilinense T. Bau et X. Wang, sp. nov. (a) Basidiomata; (b) Epibasidia; (c) Basidiospores; (d) Hyphae; (e) Hypobasidia; (f) Probasidia. (Scale bars: (a) = 1 cm; (b,e,f) = 10 μm; (c,d) = 5 μm).
MycoBank number: 853841
Diagnosis: Sirobasidium jilinense is characterized by cerebriform basidiomata, which are reddish brown to dark reddish brown when fresh and dark brown when dry. The hypobasidia consist of three to nine chains and are divided vertically or obliquely into four cells, whereas the basidiospores are subglobose to broadly ellipsoid, and the hyphae are thick-walled.
Etymology: “jilinense” refers to the discovery of a type specimen in Jilin Province, China.
Type: CHINA. Jilin Province, Jilin, Jiaohe, Shansong Ridge, on rotten wood of Quercus, elev. 577.2 m, 43°75′ N, 127°33′ E, 26 August 2023, T. Bau and Xia Wang, (FJAU68870, holotypus!). Same location, 29 July 2023, Xia Wang, (FJAU68668, paratypus!).
Basidiomata are soft gelatinous when fresh, easily rotting, cerebriform, reddish brown (8E5–E8) to dark reddish brown (8F5–F8), usually remaining coalescing, occasionally separate, 1–3 cm long and 1 cm thick. The hymenium surface is smooth, with full edges, folded, and relatively blunt. Shrinkage when dry, dark brown (8F1–F5), fragile.
Cross section without medulla, composed of the hymenium and intertwined jelly hyphae. Hymenium peripheral, pale yellow, composed of probasidia and hypobasidia. Probasidia 12–20 × 11–16 μm, globose to ellipsoid, light yellow, containing oil droplet-like substances, with clamp connection at the base; hypobasidia, 14–22 × 10–15 μm, ellipsoid to ovoid, the vertical or diagonal partitions are 4-cells, 3–9 chain-like structures, wrinkled at maturity, light yellow, containing oil droplet-like substances, with clamp connection at the base; epibasidia, 13–18 × 5–8 μm, spindle-shaped, detached, containing oil droplet-like substances. Basidiospores 6.7–9.4 × 6.7–7.2 μm, Q = 1.04–1.37 (n = 100/5), subglobose to broadly ellipsoid, apiculate, colorless, smooth, with oil droplets, germinating to produce regenerated spores or germination tubes. Hyphae with rich clamp connection, thick-walled, 2.3–3.6 µm in diameter.
Habitat: In summer and autumn, it grows on fallen trees or branches such as Acer and Quercus in broad-leaved forests.
Distribution: Currently, only known in the Jilin Province, China, Asia.
Additional specimens examined: China. Jilin Province: Jilin, Jiaohe, Shansong Ridge, on fallen branches of Acer, July 24, 2022, Liyang Zhu, FJAU68667 (Z22072428). Same location, Shien Wang, FJAU68669 (E2307209); Jilin, Huandian, Hongshi National Forest Park, on fallen branches of Acer, August 27, 2023, T. Bau & Xia Wang FJAU68671 (W23082706), T. Bau & Mu Liu, FJAU68672 (lm230890).

Figure 5.
Basidiomata of Calocera velutina: (a) fresh (FJAU68950); (b,c) dry (FJAU68951, FJAU68950). Scale bars: (a–c) = 5 mm.
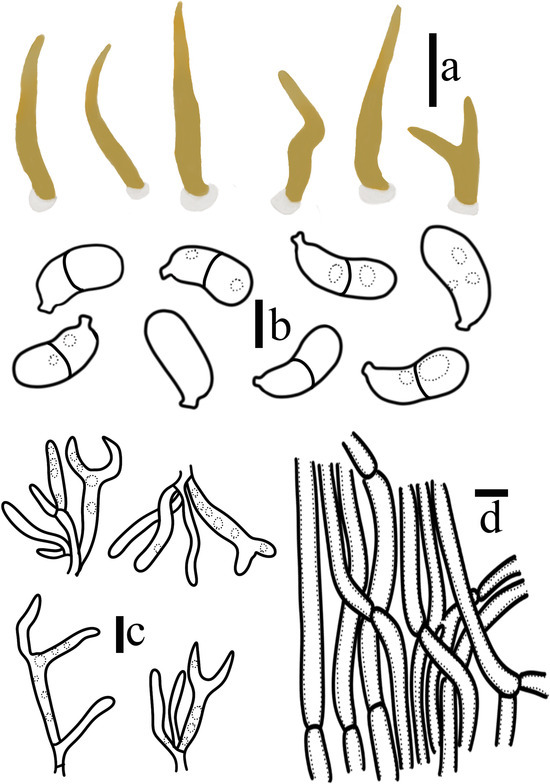
Figure 6.
Calocera velutina T. Bau et X. Wang, sp. nov. (a) Basidiomata; (b) Basidiospores; (c) Basidia and hyphidia; (d) Central hyphae. (Scale bars: (a) = 1 cm; (b) = 5 μm; (c,d) = 10 μm).
MycoBank number: 854036
Diagnosis: Calocera velutina is characterized by stipitate and pileate when young, needle-like when mature, light yellow, with a white disc-shaped villus at the base, simple or dichotomously branched. Hyphae thick-walled, without clamp connection. In summer, they grow on decaying trees in coniferous forests.
Etymology: “velutina” refers to the base of the basidiomata with villus.
Type: CHINA. Jilin Province, Yanbian Korean Autonomous Prefecture, Antu County, Erdaobaihe Town, Forest Industry Cultural Park, on rotten wood of Pinus L., elev. 708 m, 42°43′ N, 128°12′ E, 31 July 2022, T. Bau and Xia Wang, (FJAU68950, holotypus!). Jilin Province, Huadian City, Hongshi National Forest Park, 27 August 2023, T. Bau and Mu Liu, (FJAU68951, paratypus!).
Basidiomata are gelatinous when fresh, stipitate and pileate when young, needle-like when mature, light yellow (4A4–A5), slightly lighter at the tip, with white (4A1) disc-shaped villus at the base, simple or dichotomously branched, up to 15 mm high. The hymenium is located in the upper middle part, and the base is sterile. When dry, it is greyish yellow (4B3–B6, 4C5), and the surface under the body mirror is fine and granular.
The transverse section of the apex of the basidiomata is composed of the hymenium and interwoven hyphae. The transverse section of the basidiomata cylinder presents three annular zones. The central hyphae are densely arranged vertically, surrounded by loosely interlaced hyphae, and the periphery is the hymenium. Hymenium amphigenous is composed of probasidia, basidia, and hyphidia. Probasidia 18–25 × 2.9–4.4 μm, sub-clavate, forked when mature, containing oil droplet-like substances, with septate at the base; basidia 21–33 × 3.5–4.8 μm, clavate, containing oil droplet-like substances, with septate at the base; sterigmata 10–17 × 2.3–3.7 μm, cylindrical, the top gradually becomes pointed and gradually weakens when mature, containing oil droplet-like substances. Hyphidia 19–28 × 1.8–2.7 μm, narrowly cylindrical, simple or slightly curved, with septum at the base. Basidiospores 9.1–10.4 × 4.2–5.3 μm, Q = 1.90–2.27 (n = 40/2), curved cylindrical, narrow top, smooth, thin-walled, mature 1 septate, septate thin-walled, contains oil droplet-like substances. No germination was observed. Central hyphae 4.9–6.8 μm in diameter, septate, slightly rough, thick-walled. Interlaced hyphae, 3.4–5.3 μm in diameter, septate, thick-walled, basal hyphae 2.5–3.8 μm in diameter. All structures without clamp connection.
Habitat: In summer, it grows on decaying trees in coniferous forests.
Distribution: Currently, only known in the Jilin Province, China, Asia.

Figure 7.
Basidiomata of Dacrymyces jauensis: (a) fresh (FJAU68961); (b) dry (FJAU68961). Scale bars: (a) = 1 cm, (b) = 0.5 cm.
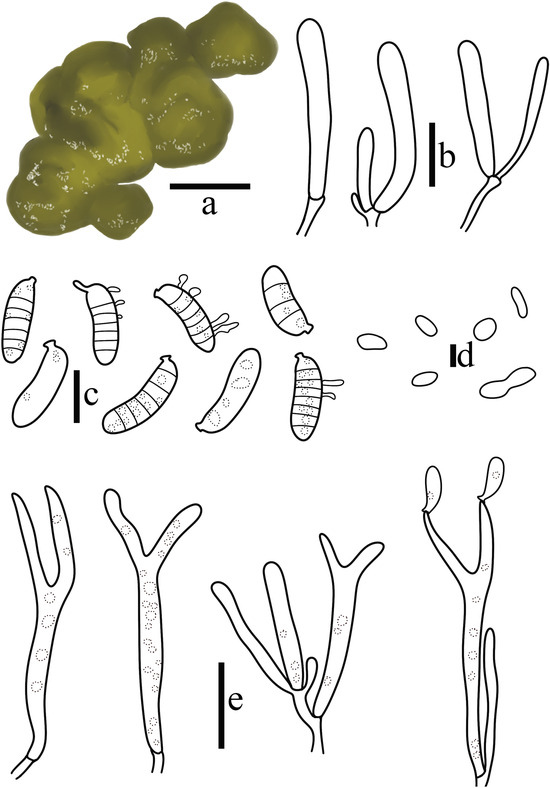
Figure 8.
Dacrymyces jauensis T. Bau et X. Wang, sp. nov. (a) Basidiomata; (b) Probasidia; (c) Basidiospores; (d) Conidia; (e) Basidia. (Scale bars: (a) = 1 cm; (b–d) = 10 μm; (e) = 20 μm).
MycoBank number: 854037
Diagnosis: Dacrymyces jauensis is characterized by basidiomata flat to cushion-shaped when young, cinnamon, golden yellow to brownish yellow; basidiospores sausage-shaped, separated horizontally when mature, usually divided into 7 septate; germination produces rod-shaped conidia; hyphidia absent.
Etymology: “jauensis” refers to the discovery of a type specimen at Jilin Agricultural University.
Type: CHINA. Jilin Province, Changchun city, wild plantation garden of the campus of Jilin Agricultural University, grows on highly decayed wood of broad-leaved trees in broad-leaved forests, 43°81′ N, 125°40′ E, 235 m, 16 July 2022, Xia Wang & Tolgor Bau, (FJAU68961, 22071605W, holotypus!). Same location, 4 July 2022, Xia Wang & Tolgor Bau, (FJAU68959, T22070437W; FJAU68960, T22070438W, paratypus!).
Basidiomata are soft gelatinous when fresh, flat to cushion-shaped when young, cinnamon, golden yellow (4C6), up to 0.5 cm, usually remaining coalescing when mature, brownish yellow (5D6, 5E5), sessile, up to 2.5 cm. Hymenium surface with wrinkles and grooves, edge curved, folded, wavy, blunt. When dry, it shrinks and collapses in a brown (6E6), making it difficult to observe.
The transverse section is composed of hymenium and context hyphae. Hymenium amphigenous. Probasidia 32–47 × 3.7–6.3 μm, cylindrical, subclavate to broadly clavate, thin-walled, transparent, forked when mature, containing oil-dripping substance, basally septate; sterigmata 11–33 × 3.0–4.4 μm, cylindrical, apically acuminate when mature, gradually weakening. Hyphidia absent. Basidiospores 15.9–20.9 × 5.1–6.3 μm, Q = 2.80–3.49 (n = 90/3), sausage-shaped, smooth, thin-walled, transparent, separated horizontally when mature, usually divided into 7 septate, occasionally 0–3 septate, thin-walled, containing oil drop-like substance, and conidia germinate at the septate. Conidia 4.8–12.3 × 1.4–1.8 μm, long cylindrical to rod-shaped. Context hyphae 1.6–3.2 μm in diameter, thin-walled, septate, septate not swollen. All structures without clamp connection.
Habitat: In summer, it grows on highly decayed wood of broad-leaved trees in broad-leaved forests.
Distribution: Currently, only known in the Jilin Province, China, Asia.

Figure 9.
Basidiomata of Dacryopinax manghanensis: (a) fresh (FJAU68943); (b) fresh (FJAU68940); (c) fresh (FJAU68941); (d) dry (FJAU68938). Scale bars: ((a–c) = 0.5 cm; (d) = 0.2 cm).
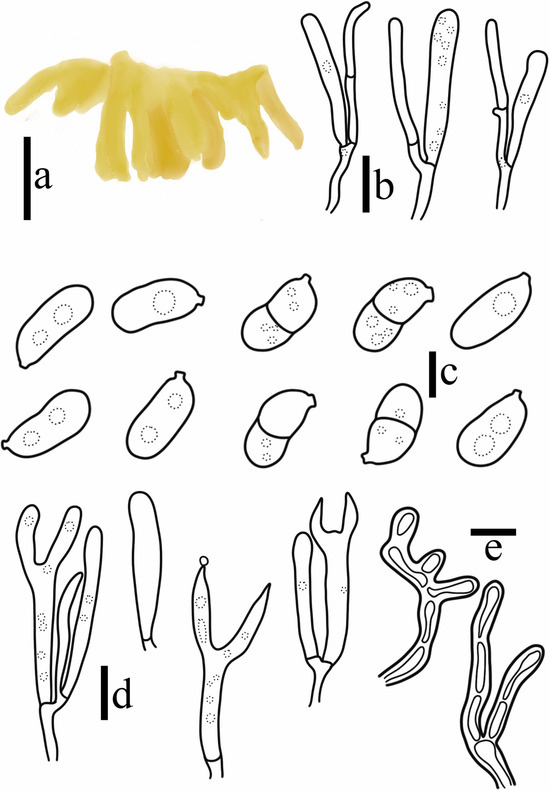
Figure 10.
Dacryopinax manghanensis T. Bau et X. Wang. (a) Basidiomata; (b) Probasidia and hyphidia; (c) Basidiospores; (d) Basidia; (e) Marginal hyphae. ((a) = 0.5 cm; (b) = 5 μm; (c) = 2 μm; (d,e) = 10 μm).
MycoBank number: 853843
Diagnosis: Dacryopinax manghanensis is characterized by small basidiomata, 1–5 mm wide, 1–8 mm high, spathulate and stipitate to substipitate, light yellow, cross section without medulla, the short and fine villi on the sterile surface. Basidiospores curved cylindrical to navicular, with 0–1 septate at maturity, thin-walled septate, and no germination observed; cortical hyphae curved, thick-walled, swollen, and branched.
Etymology: “manghanensis” is a Mongolian word for “sandy land”, which means that type specimens are collected from rotten wood in sandy land.
Type: CHINA. Inner Mongolia Autonomous Region, Tongliao, in the later stage of the left wing of Horqin, Wudantara Forest Farm, fallen branches of broad-leaved trees, 43°01′ N, 122°44′ E, 363 m, 16 July 2022, Weinan Hou & Tolgor Bau, (FJAU68943, H2207159, holotypus!). Same location, 15 July 2023, Hong Cheng & Tolgor Bau, (FJAU68940, C2371524, paratypus!).
When the basidiomata are fresh, soft gelatinous, spathulate, and stipitate to substipitate when young, the stipitate is light yellow (4A4–A5) and slightly hyaline, petaloid when mature, surface with longitudinal ridges, 1–5 mm wide, and 1-8 mm high. Hymenial surface smooth, with entire edges, blunt. The short and fine villi on the sterile surface are visible under a stereomicroscope. When dry, it shrinks, with a white (4A1) base, a dark yellow (4C8) surface on the hymenial, and obvious longitudinal edges on the stipitate.
Cross section without medulla, composed of the hymenium, context hyphae, and cortical hyphae. Hymenium unilateral, occasionally bilateral, composed of basidia and hyphidia. Probasidia 18–27 × 2.5–3.5 μm, cylindrical to clavate, thin-walled, hyaline, forked when mature, containing oil droplet-like substances, with septate at the base; sterigmata 7–13 × 1.8–2.9 μm, the top gradually becomes pointed and gradually weakens when mature. Hyphidia 15–27 × 1.4–2.2 μm, narrow cylindrical, thin-walled, hyaline, simple or slightly curved, with 1 septum at the base. Basidiospores 7.9–9.0 × 3.7–4.9 μm, Q = 1.73–2.07 (n = 80/4), curved cylindrical to navicular, apiculate, smooth, thin-walled, hyaline, mature 0–1 septate, septate thin-walled, contains oil droplet-like substances. No germination was observed. Cortical hyphae 4.5 μm in diameter, cylindrical branching at the end, curved, septum, constriction at intervals, thick-walled. All structures without clamp connection.
Habitat: In summer and autumn, it decays on fallen branches in broad-leaved forests or mixed coniferous and broad-leaved forests.
Distribution: Currently, it is only distributed in China, Inner Mongolia Autonomous Region, and Jilin Province.
Additional specimens examined: CHINA. Jilin Province, Tonghua, Yuhuangshan Park, broad-leaved forest rotting wood, 14 August 2022, Xia Wang, FJAU68941 (2281405W); Changchun, Jilin Agricultural University Campus Back Mountain, 16 September 2022, Xia Wang, FJAU68942 (2291601W); Tonghua, Yushan Park in Ji’an City, 6 July 2023, Zhengqing Chen, FJAU68938 (Q237603); Changchun, Jingyue Pool National Forest Park, 12 July 2023, Xia Wang, FJAU68939 (W23071206).
4. Discussion
Sirobasidium jilinense is easily confused with the type species S. sanguineum in the wild; however, the hypobasidia of the latter are solitary or in chains of 2–4, without a clamp connection at the base, and the clamp connection of hyphae is enlarged (5.5–8 μm). S. jilinense and S. rubrofuscum (Berk.) P. Roberts are similar; the basidiomata are all cerebriform and dark reddish brown, but they can be distinguished by the number of hypobasidia in the chains and the size of the basidiospores. Other species in Sirobasidium are white, off-white, or yellowish and are easily distinguished from S. jilinense (Table 2). Due to its brain-like, reddish-brown basidiomata, S. jilinense is easily confused with individual species of Tremella Pers. and Exidia Fr. in the wild, but it is easily distinguished from the latter under a microscope according to the basidium morphology.

Table 2.
Morphological comparison of Sirobasidium species.
Molecular phylogeny analysis showed that members of Fibulobasidium Bandoni and Sirobasidium jilinense, S. magnum, S. apiculatum, and S. japonicum form a separate branch (PP = 0.99, MLbs = 91%), while S. brefeldianum is located in another branch. This suggests that Sirobasidium is polyphyletic, which is consistent with the results of recent studies [9,10,11,13] (Figure 1). The new species S. jilinense and S. magnum have a sister relationship and obtained a high support rate (PP = 1, MLbs = 100%), which is consistent with the results of morphological studies.
Calocera velutina is characterized by basidiomata that are stipitate and pileate when young, needle-like when mature, yellow with a white disc-shaped villus at the base, and simple or dichotomously branched. The hyphae are thick-walled, without clamp connection. In summer, they grow on decaying trees in coniferous forests. In the wild, C. velutina is easy to be confused with C. cornea (Batsch) Fr., C. sinensis McNabb, and C. tibetica F. Wu, L.F. Fan & Y.C. Dai. They all have fresh yellow basidiomata with sharp, unbranched, or even-branched cylindrical upward-narrowing tops; however, the base of the latter three does not have a white disc-shaped villus [7,23,24]. The basidiomata of C. sinensis are small (1–5 mm), and the hyphae have clamp connections [23]. The basidiospores of C. tibetica are divided into three transverse septates when mature [24]. In the phylogenetic tree, they were also found to have a distant relationship.
Dacrymyces jauensis is characterized by basidiomata flat to cushion-shaped when young, cinnamon, golden yellow to brownish yellow; basidiospores sausage-shaped, separated horizontally when mature, usually divided into seven septate; germination produces rod-shaped conidia; hyphidia absent. Dacrymyces jauensis, Dacrymyces san-augustinii Kobayasi, and Dacrymyces chiangraiensis Ekanayaka, Karun., Q. Zhao & K.D. Hyde present multiple transverse septates when basidiospores mature. In the phylogenetic tree, a close relationship can be observed (Figure 2). However, the basidiomata of Dacrymyces san-augustinii are yellow, and the basidia (38–58 × 5.5–7 µm) and basidiospores (16–27.5 × 6–10 µm) are larger, with hyphidia [14]. The basidiomata of Dacrymyces chiangraiensis are yellow to orange, and the basidia (32–53 × 7–10 µm) and basidiospores (19–23 × 6.5–8 µm) are also large, with hyphidia [45].
There are two kinds of basidiomata of Dacryopinax manghanensis, most of which are clustered and spoon-shaped in the sand of the Inner Mongolia Autonomous Region but petal-shaped in Jilin Province (Figure 9). However, their microstructure is the same, as they are clustered in the same branch in the phylogenetic tree, and the shape of the basidiomata may be related to the environment. There are seven species of Dacryopinax distributed in China. The mature basidiospores of Dacryopinax aurantiaca and Dacryopinax spathularia are 0–1 septate, and they are easily confused with Dacryopinax manghanensis in the wild. However, the basidiospores of Dacryopinax aurantiaca are longer (10.4–13 × 3.5–5 μm) [7]. Basidiospores germinate as conidia or germ tubes and are mainly distributed in South China. Dacryopinax spathularia had obvious villi (visible to the naked eye when dry), with medulla on the transverse section and a bulbous hyphae septum [7]. In the phylogenetic tree, they are not closely related (Figure 2). In addition, the basidiospores of Dacryopinax indacocheae Lowy, Dacryopinax primogenitus, and Dacryopinax sphenocarpa Shirouzu & Tokum. are also divided into 0–1 septate at maturity. However, the basidiomata of Dacryopinax indacocheae are yellowish brown and larger (30 mm wide, 20 mm high). The cortical hyphae of the stipe have chain-like enlarged cells, the basidiospores are longer (8–11.5 × 3–3.5 μm), and the hymenium has abundant conidia [15]. Dacryopinax primogenitus has longer basidia (23–49 × 2.5–4 μm) and longer hyphidia (45.5–48.5 × 2.5–3 μm) and germinates as conidia or germ tubes [49]. The basidiomata of Dacryopinax sphenocarpa are yellow-white or light amber, the hyphae have clamp connections, the base of the probasidia has clamp connections, and the basidiospores are larger (10–16 × 4.5–8.5 μm) [14]. Dacryopinax is polyphyletic, with a total of eight species currently supported by molecular data, located in five distinct clades, consistent with recent findings [6,13,49]. In addition, in the phylogenetic tree constructed in this study, the branch composed of Dacryopinax manghanensis and Dacryopinax sp. (H7008759) was closely related to the branch composed of Calocera species (Figure 2); however, the species of Dacryopinax manghanensis and Calocera were different in morphology, so, it was placed in Dacryopinax.
Regarding jelly fungi, due to their gelatinous texture, researchers tend to focus more on damp environments when collecting specimens. In addition to adding three new species from the humid environment of Jilin Province—S. jilinense, C. velutina, and Dacrymyces jauensis—this study also added a new species from the Horqin Sandy Land—Dacryopinax manghanensis. This shows that jelly fungi have a wide geographical distribution and are well-adapted to different conditions and environments. As such, this type of fungus is worthy of further exploration.
Author Contributions
Conceptualization, T.B. and X.W.; methodology, X.W.; software, X.W.; validation, X.W. and T.B.; formal analysis, X.W.; investigation, X.W. and T.B.; resources, X.W. and T.B.; data curation, X.W. and T.B.; writing—original draft preparation, X.W.; writing—review and editing, X.W. and T.B.; visualization, X.W. and T.B.; supervision, T.B.; project administration, T.B.; funding acquisition, T.B. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Ministry of Education Innovation Team (IRT-15R25).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the sequences have been deposited in GenBank (https://www.ncbi.nlm.nih.gov, accessed on 1 May 2024) and MycoBank (https://www.mycobank.org, accessed on 17 May 2024). The data presented in this study are deposited in the Zenodo repository, accession number doi: 10.5281/zenodo.11206684.
Acknowledgments
We appreciate the help of our team members during the field collection process. We thank the reviewers and responsible editors for their corrections and suggestions.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lagerheim, N.G.; Patouillard, N.T. Sirobasidium nouveau genre d’Hyménomycètes Hétérobasidiés. J. De Bot. 1892, 6, 465–469. [Google Scholar]
- Möller, A. Protobasidiomyceten. In Botanische Mitteilungen aus den Tropen; Gustav Fischer: Jena, Germany, 1895; Volume 8, pp. 1–179. [Google Scholar]
- Index Fungorum. Available online: http://www.indexfungorum.org/Names/Names.asp (accessed on 3 May 2024).
- Lowy, B. A note on Sirobasidium. Mycologia 1956, 48, 324–327. [Google Scholar] [CrossRef]
- Bandoni, R.J. The spores and basidia of Sirobasidium. Mycologia 1957, 49, 250–255. [Google Scholar] [CrossRef]
- Roedel, T.; Putzmann, F. Sirobasidium albidum Lagerh. & Pat. 1892-new for Saxony. Z. Fuer Mykol. 2015, 81, 357–371. [Google Scholar]
- Liu, B. Tremellales & Dacrymycetales. In Flora Fungorum Sinicorum; Science Press: Beijing, China, 1992; Volume 2, pp. 1–93. [Google Scholar]
- Bandoni, R.J.; Sampaio, J.P.; Boekhout, T. Sirobasidium de Lagerheim & Patouillard (1892). Z. Für Mykol. 2011, 81, 357–371. [Google Scholar] [CrossRef]
- Millanes, A.M.; Diederich, P.; Ekman, S.; Wedin, M. Phylogeny and character evolution in the jelly fungi (Tremellomycetes, Basidiomycota, Fungi). Mol. Biol. Evol. 2011, 61, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.; Bauer, R.; Sampaio, J.P.; Oberwinkler, F. 12 Tremellomycetes and related groups. In Systematics and Evolution: Part A; Springer: Berlin/Heidelberg, Germany, 2014; pp. 331–355. [Google Scholar] [CrossRef]
- Liu, X.Z.; Wang, Q.M.; Goker, M.; Groenewald, M.; Kachalkin, A.V.; Lumbsch, H.T.; Millanes, A.M.; Wedin, M.; Yurkov, A.M.; Boekhout, T.; et al. Towards an integrated phylogenetic classification of the Tremellomycetes. Stud. Mycol. 2015, 81, 85–147. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Z.; Wang, Q.M.; Theelen, B.; Groenewald, M.; Bai, F.Y.; Boekhout, T. Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Stud. Mycol. 2015, 81, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Endoh, R.; Masumoto, H.; Yoshihashi, Y.; Ohkuma, M.; Degawa, Y. Taxonomic study of polymorphic basidiomycetous fungi Sirobasidium and Sirotrema: Sirobasidium apiculatum sp. nov., Phaeotremella translucens comb. nov. and rediscovery of Sirobasidium japonicum in Japan. Int. J. Gen. Mol. Microbiol. 2022, 115, 1421–1436. [Google Scholar] [CrossRef]
- Shirouzu, T.; Hirose, D.; Tokumasu, S. Taxonomic study of the Japanese Dacrymycetes. Persoonia 2009, 23, 16–34. [Google Scholar] [CrossRef]
- McNabb, R. Taxonomic studies in the Dacrymycetaceae: III. Dacryopinax Martin. N. Z. J. Bot. 1965, 3, 59–72. [Google Scholar] [CrossRef]
- McNabb, R. Taxonomic studies in the Dacrymycetaceae: II. Calocera (Fries) Fries. N. Z. J. Bot. 1965, 3, 31–58. [Google Scholar] [CrossRef]
- McNabb, R. Taxonomic studies in the Dacrymycetaceae VIII. Dacrymyces Nees ex Fries. N. Z. J. Bot. 1973, 11, 461–524. [Google Scholar] [CrossRef]
- Shirouzu, T.; Hirose, D.; Tokumasu, S. Sequence analyses of the 28S rRNA gene D1/D2 region suggest Dacrymyces (Heterobasidiomycetes, Dacrymycetales) is polyphyletic. Mycoscience 2007, 48, 388–394. [Google Scholar] [CrossRef]
- Malysheva, V. The genus Calocera Dacrymycetales, Basidiomycota in Russia. Mikol. Fitopatol. 2013, 47, 306–315. [Google Scholar]
- Shirouzu, T.; Hosaka, K.; Nam, K.O.; Weir, B.S.; Johnston, P.R.; Hosoya, T. Phylogenetic relationships of eight new Dacrymycetes collected from New Zealand. Persoonia 2017, 38, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, R.L.; Xavier-Santos, S. New records of Dacrymycetes (Fungi: Basidiomycota) from the Cerrado Biome (Brazilian Savanna) and Midwest Region, Brazil. Check List 2017, 13, 335–342. [Google Scholar] [CrossRef]
- Castro-Santiuste, S.; Sierra, S.; Guzman-Davalos, L.; Cifuentes, J.; Evans, T.; Martinez-Gonzalez, C.R.; Alvarado-Sizzo, H.; Luna-Vega, I. Dacryopinax (Fungi: Dacrymycetales) in Mexico. Phytotaxa 2020, 446, 6–22. [Google Scholar] [CrossRef]
- Wu, S.H.; Shih, K.; Yu, S.Y. Calocera bambusicola sp. nov. and C. sinensis newly recorded from Taiwan. Mycotaxon 2011, 115, 163–169. [Google Scholar] [CrossRef]
- Fan, L.F.; Wu, Y.D.; Wu, F.; Dai, Y.C. Calocera tibetica sp. nov.(Dacrymycetaceae, Dacrymycetales) from southwestern China. Phytotaxa 2021, 500, 133–141. [Google Scholar] [CrossRef]
- Lian, Y.P.; Tohtirjap, A.; Wu, F. Two New Species of Dacrymyces (Dacrymycetales, Basidiomycota) from Southwestern China. Diversity 2022, 14, 379. [Google Scholar] [CrossRef]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour; E. Methuen: London, UK, 1978; pp. 1–252. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Hopple Jr, J.S.; Vilgalys, R. Phylogenetic relationships among coprinoid taxa and allies based on data from restriction site mapping of nuclear rDNA. Mycologia 1994, 86, 96–107. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bau, T. A novel exidioides fungi, Myxarium boreale (Auriculariales, Basidiomycota) discovered from Northern China. Phytotaxa 2023, 618, 161–171. [Google Scholar] [CrossRef]
- Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 3 May 2024).
- Nucleotide. Available online: https://www.ncbi.nlm.nih.gov/nucleotide (accessed on 25 April 2024).
- MAFFT version 7. Available online: https://mafft.cbrc.jp/alignment/server/index.html (accessed on 3 May 2024).
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree; v1. 4.3 2006–2016; Tree Figure Drawing Tool; Online publication; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2019. [Google Scholar]
- Scorzetti, G.; Fell, J.W.; Fonseca, A.; Statzell-Tallman, A. Systematics of basidiomycetous yeasts: A comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2002, 2, 495–517. [Google Scholar] [CrossRef] [PubMed]
- Zamora, J.C.; Ekman, S. Phylogeny and character evolution in the Dacrymycetes, and systematics of Unilacrymaceae and Dacryonaemataceae fam. nov. Persoonia 2020, 44, 161–205. [Google Scholar] [CrossRef]
- Wei, M.; Oberwinkler, F. Phylogenetic relationships in Auriculariales and related groups–hypotheses derived from nuclear ribosomal DNA sequences. Mycol. Res. 2001, 105, 403–415. [Google Scholar] [CrossRef]
- Shirouzu, T.; Hirose, D.; Oberwinkler, F.; Shimomura, N.; Maekawa, N.; Tokumasu, S. Combined molecular and morphological data for improving phylogenetic hypothesis in Dacrymycetes. Mycologia 2013, 105, 1110–1125. [Google Scholar] [CrossRef]
- Savchenko, A.; Zamora, J.C.; Shirouzu, T.; Spirin, V.; Malysheva, V.; Koljalg, U.; Miettinen, O. Revision of Cerinomyces (Dacrymycetes, Basidiomycota) with notes on morphologically and historically related taxa. Stud. Mycol. 2021, 99, 100117. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; Jeewon, R.; Maharachchikumbura, S.S.; Liu, J.-K.; Bhat, D.J.; Jones, E.G.; McKenzie, E.H.; Camporesi, E.; Bulgakov, T.S. Fungal diversity notes 491–602: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2017, 83, 1–261. [Google Scholar] [CrossRef]
- Phookamsak, R.; Hyde, K.D.; Jeewon, R.; Bhat, D.J.; Jones, E.G.; Maharachchikumbura, S.S.; Raspé, O.; Karunarathna, S.C.; Wanasinghe, D.N.; Hongsanan, S. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019, 95, 1–273. [Google Scholar] [CrossRef]
- Shirouzu, T.; Sanjo, K. Rediscovery of Dacrymyces pezizoides (Dacrymycetes, Basidiomycota) 80 years after its original description. Phytotaxa 2018, 371, 293–300. [Google Scholar] [CrossRef]
- Mendes-Alvarenga, R.L.; Gibertoni, T.B. Dacrymyces flavobrunneus sp. nov. and two new combinations in Dacrymyces Nees based on morphological and phylogenetic data. Mycol. Prog. 2022, 21, 96. [Google Scholar] [CrossRef]
- McLaughlin, D.J.; Healy, R.A.; Kumar, T.K.; McLaughlin, E.G.; Shirouzu, T.; Binder, M. Cultural and cytological characterization of Dacryopinax primogenitus, a new species in the Dacrymycetes with a fully sequenced genome. Mycologia 2016, 108, 457–468. [Google Scholar] [CrossRef]
- Shirouzu, T.; Ishikawa, N.K.; Hirose, D.; Maekawa, N. A new Amazonian species of Calocera with dendroid and multi-headed basidiocarp. Mycoscience 2013, 54, 252–256. [Google Scholar] [CrossRef]
- Zamora, J.; Savchenko, A.; González-Cruz, Á.; Prieto-García, F.; Olariaga, I.; Ekman, S. Dendrodacrys: A new genus for species with branched hyphidia in Dacrymyces s. l., with the description of four new species. Fungal Syst. Evol. 2022, 9, 27–42. [Google Scholar] [CrossRef]
- Zamora, J.C.; Holec, J. Ditiola haasii (Basidiomycota, Dacrymycetaceae)—Taxonomy and ecology of a rare species from Central Europe. Phytotaxa 2021, 522, 121–130. [Google Scholar] [CrossRef]
- Fell, J.W.; Boekhout, T.; Fonseca, A.; Scorzetti, G.; Statzell-Tallman, A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 2000, 50, 1351–1371. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.M.; Bai, F.Y.; Fungsin, B.; Boekhout, T.; Nakase, T. Proposal of Mingxiaea gen. nov. for the anamorphic basidiomycetous yeast species in the Bulleribasidium clade (Tremellales) based on molecular phylogenetic analysis, with six new combinations and four novel species. Int. J. Syst. Evol. 2011, 61, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, J.P.; Weiss, M.; Gadanho, M.; Bauer, R. New taxa in the Tremellales: Bulleribasidium oberjochense gen. et sp. nov., Papiliotrema bandonii gen. et sp. nov. and Fibulobasidium murrhardtense sp. nov. Mycologia 2002, 94, 873–887. [Google Scholar] [CrossRef]
- Landell, M.F.; Inacio, J.; Fonseca, A.; Vainstein, M.H.; Valente, P. Cryptococcus bromeliarum sp. nov., an orange-coloured basidiomycetous yeast isolated from bromeliads in Brazil. Int. J. Syst. Evol. Microbiol. 2009, 59, 910–913. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.M.; Wang, S.A.; Jia, J.H.; Bai, F.Y. Cryptococcus tibetensis sp. nov., a novel basidiomycetous anamorphic yeast species isolated from plant leaves. J. Gen. Appl. Microbiol. 2007, 53, 281–285. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Middelhoven, W.J. Cryptococcus allantoinivorans sp. nov., an anamorphic basidiomycetous yeast (Tremellales) physiologically resembling other species of the Cryptococcus laurentii complex that degrade polysaccharides and C2 compounds. Antonie Van Leeuwenhoek 2005, 87, 101–108. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; Szöke, S.; Cardinali, G.; Eberhardt, U.; Stielow, B.; De Vries, M.; Verkleij, G.; Crous, P.; Boekhout, T. DNA barcoding analysis of more than 9 000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 2016, 85, 91–105. [Google Scholar] [CrossRef]
- Chen, C.J. Morphological and Molecular Studies in the Genus Tremella; Cramer, J., Ed.; Cornell University: Ithaca, NY, USA, 1998; pp. 1–225. [Google Scholar]
- Kisimova-Horovitz, L.; Oberwinkler, F.; Gómez, L.D. Basidiomicetos resupinados de Costa Rica. Especies nuevas o raras de Atractiellales (Auriculariales sl), Exidiaceae, Sirobasidiaceae y Tremellaceae. Rev. Biol. Trop. 2000, 48, 539–554. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.; de Meijer, A. Macromycetes from the state of Parana, Brazil. 6. Sirobasiciaceae & Tremellaceae. Mycotaxon 1997, 64, 261–283. [Google Scholar]
- Gilbertson, R.; Adaskaveg, J. Studies on wood-rotting basidiomycetes of Hawaii. Mycotaxon 1993, 49, 369–397. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).