Candida tropicalis PMT2 Is a Dispensable Gene for Viability but Required for Proper Interaction with the Host
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culturing Conditions
2.2. Complementation of Candida albicans Mutants
| Strain | Organisms | Origin | Genotype | Reference |
|---|---|---|---|---|
| NGY152 | C. albicans | Derived from CAI-4 | ura3Δ-iro1Δ::imm434/ura3Δ-iro1Δ::imm434; RPS1/rps1∆::CIp10 | [46] |
| P2-67 | C. albicans | Derived from CAI-4 | ura3Δ-iro1Δ::imm434/ura3Δ-iro1Δ::imm434; PMT2/pmt2Δ::hisG | [39] |
| CAP2-2341 | C. albicans | Derived from CAI-4 | ura3Δ-iro1Δ::imm434/ura3Δ-iro1Δ::imm434; pmt6Δ::hisG/pmt6Δ::hisG | [45] |
| HMY211 | C. albicans | Derived from P2-67 | As P2-67 but RPS1/rps1∆::CIp10 | This work |
| HMY212 | C. albicans | Derived from P2-67 | As P2-67 but RPS1/rps1∆:: pACT1-CTRG_05668 | This work |
| HMY213 | C. albicans | Derived from CAP2-2341 | As CAP2-2341 but RPS1/rps1∆::CIp10 | This work |
| HMY214 | C. albicans | Derived from CAP2-2341 | As CAP2-2341 but RPS1/rps1∆:: pACT1-CTRG_05668 | This work |
| ATCC® MYA-3404 | C. tropicalis | ATCC | Wild type | ATCC |
| HMY215 | C. tropicalis | MYA-3404 | As ATCC MYA-3404, but pmt2Δ::sat1/PMT2 | This work |
| HMY216 | C. tropicalis | HMY215 | As ATCC MYA-3404, but pmt2Δ/PMT2 | This work |
| HMY217 | C. tropicalis | HMY216 | As ATCC MYA-3404, but pmt2Δ/pmt2Δ::sat1 | This work |
| HMY218 | C. tropicalis | HMY217 | As ATCC MYA-3404, but pmt2Δ/pmt2Δ | This work |
| HMY219 | C. tropicalis | HMY218 | As ATCC MYA-3404, but pmt2Δ/pmt2Δ::sat1-PMT2 | This work |
| HMY220 | C. tropicalis | HMY219 | As ATCC MYA-3404, but pmt2Δ/pmt2Δ::PMT2v | This work |
2.3. Disruption of Candida tropicalis PMT2
2.4. Gene Expression Analysis
2.5. Analysis of the Cell Wall
2.6. Analysis of Enzyme Activity
2.7. Sensitivity to Antifungal Drugs
2.8. Analysis of Biofilm Formation
2.9. Adhesion Assays
2.10. Ethics Statement
2.11. Cytokine Stimulation by Human Peripheral Blood Mononuclear Cells
2.12. Phagocytosis Assays
2.13. Analysis of Virulence
2.14. Statistical Analysis
3. Results
3.1. Disruption of Candida tropicalis PMT2
3.2. The Candida albicans pmt2∆ Null Mutant Showed Defects in Cell Wall Composition and Organization
3.3. The Candida tropicalis pmt2∆ Null Mutant Showed Defects in the Sensitivity to Cell Wall-Perturbing Agents, Biofilm Formation, and Adhesion
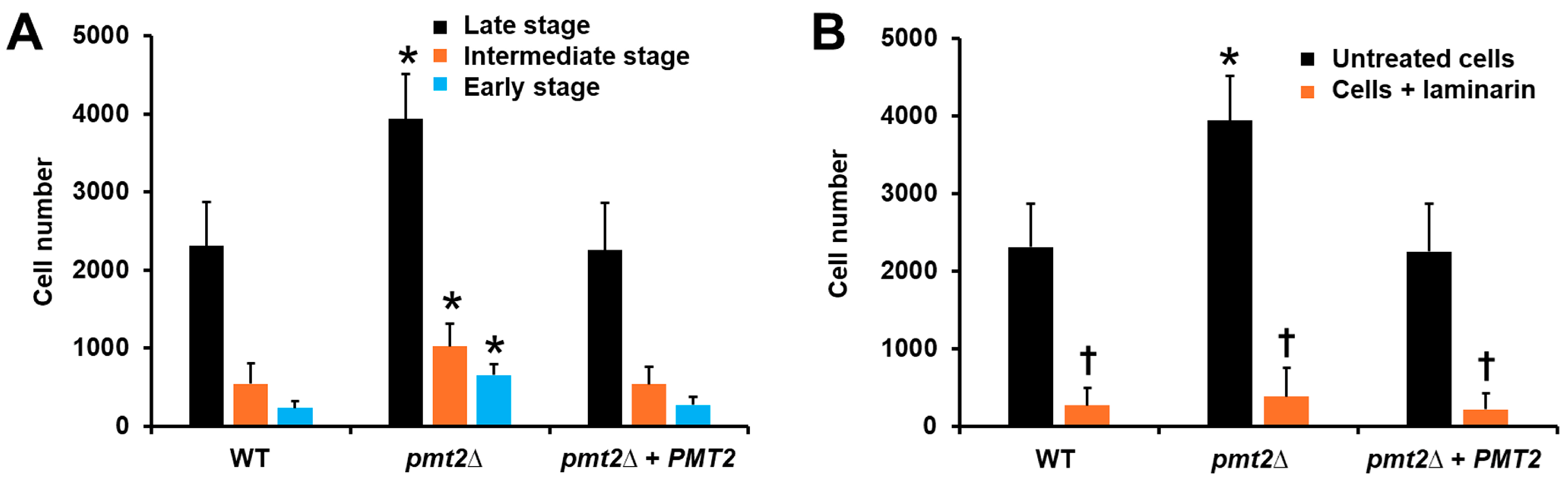
3.4. The PMT2 Disruption Affected the Candida tropicalis Interaction with Human Peripheral Blood Mononuclear Cells and Monocyte-Derived Macrophages
3.5. Candida tropicalis PMT2 Disruption Lead to Virulence Attenuation in Galleria mellonella
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kreulen, I.A.M.; de Jonge, W.J.; van den Wijngaard, R.M.; van Thiel, I.A.M. Candida spp. in human intestinal health and disease: More than a gut feeling. Mycopathologia 2023, 188, 845–862. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Apostolou, K.E.; Pappas, V.D. Attributable mortality of candidemia: A systematic review of matched cohort and case-control studies. Eur. J. Clin. Microbiol. Infect. Dis. 2006, 25, 419–425. [Google Scholar] [CrossRef]
- Lass-Flörl, C.; Kanj, S.S.; Govender, N.P.; Thompson, G.R., 3rd; Ostrosky-Zeichner, L.; Govrins, M.A. Invasive candidiasis. Nat. Rev. Dis. Primers 2024, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hamid, R.M.; El-Mahallawy, H.A.; Abdelfattah, N.E.; Wassef, M.A. The impact of increasing non-albicans Candida trends on diagnostics in immunocompromised patients. Braz. J. Microbiol. 2023, 54, 2879–2892. [Google Scholar] [CrossRef] [PubMed]
- Otto, C.; Babady, N.E. Epidemiology and outcomes of non-albicans Candida bloodstream infections in transplant recipients and cancer patients. Mycopathologia 2023, 188, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Kothavade, R.J.; Kura, M.M.; Valand, A.G.; Panthaki, M.H. Candida tropicalis: Its prevalence, pathogenicity and increasing resistance to fluconazole. J. Med. Microbiol. 2010, 59, 873–880. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, Y.C.; Hsueh, P.R. Epidemiology of candidemia and antifungal susceptibility in invasive Candida species in the Asia-Pacific region. Future Microbiol. 2016, 11, 1461–1477. [Google Scholar] [CrossRef] [PubMed]
- Won, E.J.; Sung, H.; Kim, M.N. Changing epidemiology of clinical isolates of Candida species during the Coronavirus Disease 2019 Pandemic: Data analysis from a Korean tertiary care hospital for 6 years (2017–2022). J. Fungi 2024, 10, 193. [Google Scholar] [CrossRef]
- Gow, N.A.; Hube, B. Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 2012, 15, 406–412. [Google Scholar] [CrossRef]
- Gómez-Gaviria, M.; García-Carnero, L.C.; Tamez-Castrellón, A.K.; Mora-Montes, H.M. The cell wall of medically relevant yeasts and molds. In Encyclopedia of Mycology; Zaragoza, Ó., Casadevall, A., Eds.; Elsevier: Oxford, UK, 2021; pp. 12–22. [Google Scholar]
- Hopke, A.; Brown, A.J.P.; Hall, R.A.; Wheeler, R.T. Dynamic fungal cell wall architecture in stress adaptation and immune evasion. Trends Microbiol. 2018, 26, 284–295. [Google Scholar] [CrossRef]
- Latgé, J.P. Tasting the fungal cell wall. Cell Microbiol. 2010, 12, 863–872. [Google Scholar] [CrossRef]
- Bizerra, F.C.; Melo, A.S.; Katchburian, E.; Freymüller, E.; Straus, A.H.; Takahashi, H.K.; Colombo, A.L. Changes in cell wall synthesis and ultrastructure during paradoxical growth effect of caspofungin on four different Candida species. Antimicrob. Agents Chemother. 2011, 55, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Mesa-Arango, A.C.; Forastiero, A.; Bernal-Martínez, L.; Cuenca-Estrella, M.; Mellado, E.; Zaragoza, O. The non-mammalian host Galleria mellonella can be used to study the virulence of the fungal pathogen Candida tropicalis and the efficacy of antifungal drugs during infection by this pathogenic yeast. Med. Mycol. 2013, 51, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Arias, M.J.; Hernández-Chávez, M.J.; García-Carnero, L.C.; Amezcua-Hernández, D.G.; Lozoya-Pérez, N.E.; Estrada-Mata, E.; Martínez-Duncker, I.; Franco, B.; Mora-Montes, H.M. Differential recognition of Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris by human innate immune cells. Infect. Drug Resist. 2019, 12, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Matsuda, K.; Ikeda, T.; Suzuki, M.; Takahashi, S.; Suzuki, A.; Shibata, N.; Suzuki, S. Structures of cell wall mannans of pathogenic Candida tropicalis IFO 0199 and IFO 1647 yeast strains. Infect. Immun. 1994, 62, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Chávez, M.J.; Clavijo-Giraldo, D.M.; Novák, Á.; Lozoya-Pérez, N.E.; Martínez-Álvarez, J.A.; Salinas-Marín, R.; Hernández, N.V.; Martínez-Duncker, I.; Gácser, A.; Mora-Montes, H.M. Role of protein mannosylation in the Candida tropicalis-host interaction. Front. Microbiol. 2019, 10, 2743. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.; Hall, R.A.; Cheetham, J.; Netea, M.G.; MacCallum, D.M.; Brown, A.J.; Odds, F.C.; Gow, N.A. Role of the Candida albicans MNN1 gene family in cell wall structure and virulence. BMC Res. Notes 2013, 6, 294. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.; Hughes, H.B.; Munro, C.A.; Thomas, W.P.; MacCallum, D.M.; Bertram, G.; Atrih, A.; Ferguson, M.A.; Brown, A.J.; Odds, F.C.; et al. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 2006, 281, 90–98. [Google Scholar] [CrossRef]
- Bates, S.; MacCallum, D.M.; Bertram, G.; Munro, C.A.; Hughes, H.B.; Buurman, E.T.; Brown, A.J.; Odds, F.C.; Gow, N.A. Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J. Biol. Chem. 2005, 280, 23408–23415. [Google Scholar] [CrossRef]
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Castillo, L.; Brand, A.; Buurman, E.T.; Diaz-Jimenez, D.F.; Jan Kullberg, B.; Brown, A.J.; Odds, F.C.; et al. A multifunctional mannosyltransferase family in Candida albicans determines cell wall mannan structure and host-fungus interactions. J. Biol. Chem. 2010, 285, 12087–12095. [Google Scholar] [CrossRef]
- Mora-Montes, H.M.; Bates, S.; Netea, M.G.; Diaz-Jimenez, D.F.; Lopez-Romero, E.; Zinker, S.; Ponce-Noyola, P.; Kullberg, B.J.; Brown, A.J.; Odds, F.C.; et al. Endoplasmic reticulum alpha-glycosidases of Candida albicans are required for N glycosylation, cell wall integrity, and normal host-fungus interaction. Eukaryot. Cell 2007, 6, 2184–2193. [Google Scholar] [CrossRef] [PubMed]
- Munro, C.A.; Bates, S.; Buurman, E.T.; Hughes, H.B.; Maccallum, D.M.; Bertram, G.; Atrih, A.; Ferguson, M.A.; Bain, J.M.; Brand, A.; et al. Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J. Biol. Chem. 2005, 280, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, C.G.J.; Koser, U.; Lewis, L.E.; Bain, J.M.; Mora-Montes, H.M.; Barker, R.N.; Gow, N.A.R.; Erwig, L.P. Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect. Immun. 2010, 78, 1650–1658. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Mata, E.; Navarro-Arias, M.J.; Perez-Garcia, L.A.; Mellado-Mojica, E.; Lopez, M.G.; Csonka, K.; Gacser, A.; Mora-Montes, H.M. Members of the Candida parapsilosis complex and Candida albicans are differentially recognized by human peripheral blood mononuclear cells. Front. Microbiol. 2015, 6, 1527. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Brown, G.D.; Kullberg, B.J.; Gow, N.A. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 2008, 6, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Gow, N.A.; Munro, C.A.; Bates, S.; Collins, C.; Ferwerda, G.; Hobson, R.P.; Bertram, G.; Hughes, H.B.; Jansen, T.; et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J. Clin. Investig. 2006, 116, 1642–1650. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Joosten, L.A.; van der Meer, J.W.; Kullberg, B.J.; van de Veerdonk, F.L. Immune defence against Candida fungal infections. Nat. Rev. Immunol. 2015, 15, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Yadav, B.; Mora-Montes, H.M.; Wagener, J.; Cunningham, I.; West, L.; Haynes, K.; Brown, A.J.P.; Gow, N.A.R. Differences in fungal immune recognition by monocytes and macrophages: N-mannan can be a shield or activator of immune recognition. Cell Surf. 2020, 6, 100042. [Google Scholar] [CrossRef] [PubMed]
- Sheth, C.C.; Hall, R.; Lewis, L.; Brown, A.J.; Odds, F.C.; Erwig, L.P.; Gow, N.A. Glycosylation status of the C. albicans cell wall affects the efficiency of neutrophil phagocytosis and killing but not cytokine signaling. Med. Mycol. 2011, 49, 513–524. [Google Scholar] [CrossRef][Green Version]
- Gómez-Gaviria, M.; Vargas-Macías, A.P.; García-Carnero, L.C.; Martínez-Duncker, I.; Mora-Montes, H.M. Role of protein glycosylation in interactions of medically relevant fungi with the host. J. Fungi 2021, 7, 875. [Google Scholar] [CrossRef]
- Rosati, D.; Pradhan, A.; van Heck, J.I.P.; Helder, L.; Jaeger, M.; Gow, N.A.R.; Joosten, L.A.B.; Williams, D.L.; Brown, A.J.P.; Bruno, M.; et al. C. albicans N-linked mannans potentiate the induction of trained immunity via Dectin-2. J. Infect. Dis. 2024. [Google Scholar] [CrossRef] [PubMed]
- Ocaña-Ortega, A.; Pérez-Flores, G.; Torres-Tirado, D.; Pérez-García, L.A. O-Linked glycans of Candida albicans interact with specific GPCRs in the coronary endothelium and inhibit the cardiac response to agonists. J. Fungi 2023, 9, 141. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.; da Fonseca, D.M.; Walker, L.; Griffiths, J.S.; Taylor, P.R.; Gow, N.A.R.; Orr, S.J. Dependence on Mincle and Dectin-2 varies with multiple Candida species during systemic infection. Front. Microbiol. 2021, 12, 633229. [Google Scholar] [CrossRef] [PubMed]
- Neumann, A.K.; Jacobson, K. A novel pseudopodial component of the dendritic cell anti-fungal response: The fungipod. PLoS Pathog. 2010, 6, e1000760. [Google Scholar] [CrossRef] [PubMed]
- Clavijo-Giraldo, D.M.; Pérez-García, L.A.; Hernández-Chávez, M.J.; Martínez-Duncker, I.; Mora-Montes, H.M. Contribution of N-Linked Mannosylation Pathway to Candida parapsilosis and Candida tropicalis Biofilm Formation. Infect. Drug Resist. 2023, 16, 6843–6857. [Google Scholar] [CrossRef] [PubMed]
- Lengeler, K.B.; Tielker, D.; Ernst, J.F. Protein-O-mannosyltransferases in virulence and development. Cell Mol. Life Sci. 2008, 65, 528–544. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Jimenez, D.F.; Mora-Montes, H.M.; Hernandez-Cervantes, A.; Luna-Arias, J.P.; Gow, N.A.; Flores-Carreon, A. Biochemical characterization of recombinant Candida albicans mannosyltransferases Mnt1, Mnt2 and Mnt5 reveals new functions in O- and N-mannan biosynthesis. Biochem. Biophys. Res. Commun. 2012, 419, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Prill, S.K.; Klinkert, B.; Timpel, C.; Gale, C.A.; Schröppel, K.; Ernst, J.F. PMT family of Candida albicans: Five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 2005, 55, 546–560. [Google Scholar] [CrossRef] [PubMed]
- Gentzsch, M.; Immervoll, T.; Tanner, W. Protein O-glycosylation in Saccharomyces cerevisiae: The protein O-mannosyltransferases Pmt1p and Pmt2p function as heterodimer. FEBS Lett. 1995, 377, 128–130. [Google Scholar] [CrossRef]
- Liu, H.; Kohler, J.; Fink, G. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 1994, 266, 1723–1726. [Google Scholar] [CrossRef]
- Mukaremera, L.; Lee, K.K.; Mora-Montes, H.M.; Gow, N.A.R. Candida albicans yeast, pseudohyphal, and hyphal morphogenesis differentially affects immune recognition. Front. Immunol. 2017, 8, 629. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; Netea, M.G.; Munro, C.A.; Ferwerda, G.; Bates, S.; Mora-Montes, H.M.; Walker, L.; Jansen, T.; Jacobs, L.; Tsoni, V.; et al. Immune recognition of Candida albicans beta-glucan by dectin-1. J. Infect. Dis. 2007, 196, 1565–1571. [Google Scholar] [CrossRef] [PubMed]
- Barelle, C.J.; Manson, C.L.; MacCallum, D.M.; Odds, F.C.; Gow, N.A.; Brown, A.J. GFP as a quantitative reporter of gene regulation in Candida albicans. Yeast 2004, 21, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Timpel, C.; Zink, S.; Strahl-Bolsinger, S.; Schröppel, K.; Ernst, J. Morphogenesis, adhesive properties, and antifungal resistance depend on the Pmt6 protein mannosyltransferase in the fungal pathogen Candida albicans. J. Bacteriol. 2000, 182, 3063–3071. [Google Scholar] [CrossRef] [PubMed]
- Brand, A.; MacCallum, D.M.; Brown, A.J.; Gow, N.A.; Odds, F.C. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell 2004, 3, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Reuß, O.; Vik, Å.; Kolter, R.; Morschhäuser, J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 2004, 341, 119–127. [Google Scholar] [CrossRef]
- Contreras-Martínez, O.I.; Angulo-Ortíz, A.; Santafé-Patiño, G.; Aviña-Padilla, K.; Velasco-Pareja, M.C.; Yasnot, M.F. Transcriptional reprogramming of Candida tropicalis in response to isoespintanol treatment. J. Fungi 2023, 9, 1199. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Plaine, A.; Walker, L.; Da Costa, G.; Mora-Montes, H.M.; McKinnon, A.; Gow, N.A.; Gaillardin, C.; Munro, C.A.; Richard, M.L. Functional analysis of Candida albicans GPI-anchored proteins: Roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 2008, 45, 1404–1414. [Google Scholar] [CrossRef]
- Mora-Montes, H.M.; McKenzie, C.; Bain, J.M.; Lewis, L.E.; Erwig, L.P.; Gow, N.A. Interactions between macrophages and cell wall oligosaccharides of Candida albicans. Methods Mol. Biol. 2012, 845, 247–260. [Google Scholar]
- Mora-Montes, H.M.; Netea, M.G.; Ferwerda, G.; Lenardon, M.D.; Brown, G.D.; Mistry, A.R.; Kullberg, B.J.; O’Callaghan, C.A.; Sheth, C.C.; Odds, F.C.; et al. Recognition and blocking of innate immunity cells by Candida albicans chitin. Infect. Immun. 2011, 79, 1961–1970. [Google Scholar] [CrossRef] [PubMed]
- Graham, L.M.; Tsoni, S.V.; Willment, J.A.; Williams, D.L.; Taylor, P.R.; Gordon, S.; Dennehy, K.; Brown, G.D. Soluble Dectin-1 as a tool to detect beta-glucans. J. Immunol. Methods 2006, 314, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Perez-Garcia, L.A.; Csonka, K.; Flores-Carreon, A.; Estrada-Mata, E.; Mellado-Mojica, E.; Nemeth, T.; Lopez-Ramirez, L.A.; Toth, R.; Lopez, M.G.; Vizler, C.; et al. Role of protein glycosylation in Candida parapsilosis cell wall integrity and host interaction. Front. Microbiol. 2016, 7, 306. [Google Scholar] [CrossRef] [PubMed]
- De Nobel, J.G.; Klis, F.M.; Munnik, T.; Priem, J.; Van Den Ende, H. An assay of relative cell wall porosity in Saccharomyces cerevisiae, Kluyveromyces lactis and Schizosaccharomyces pombe. Yeast 1990, 6, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Hobson, R.P.; Munro, C.A.; Bates, S.; MacCallum, D.M.; Cutler, J.E.; Heinsbroek, S.E.; Brown, G.D.; Odds, F.C.; Gow, N.A. Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J. Biol. Chem. 2004, 279, 39628–39635. [Google Scholar] [CrossRef] [PubMed]
- Smolenski, G.; Sullivan, P.A.; Cutfield, S.M.; Cutfield, J.F. Analysis of secreted aspartic proteinases from Candida albicans: Purification and characterization of individual Sap1, Sap2 and Sap3 isoenzymes. Microbiology 1997, 143, 349–356. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mora-Montes, H.M.; Lopez-Romero, E.; Zinker, S.; Ponce-Noyola, P.; Flores-Carreon, A. Hydrolysis of Man9GlcNAc2 and Man8GlcNAc2 oligosaccharides by a purified alpha-mannosidase from Candida albicans. Glycobiology 2004, 14, 593–598. [Google Scholar] [CrossRef]
- Papp, C.; Kocsis, K.; Tóth, R.; Bodai, L.; Willis, J.R.; Ksiezopolska, E.; Lozoya-Pérez, N.E.; Vágvölgyi, C.; Mora Montes, H.; Gabaldón, T.; et al. Echinocandin-induced microevolution of Candida parapsilosis influences virulence and abiotic stress tolerance. mSphere 2018, 3, e00518–e00547. [Google Scholar] [CrossRef]
- Peeters, E.; Nelis, H.J.; Coenye, T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 2008, 72, 157–165. [Google Scholar] [CrossRef]
- Pierce, C.G.; Thomas, D.P.; López-Ribot, J.L. Effect of tunicamycin on Candida albicans biofilm formation and maintenance. J. Antimicrob. Chemother. 2009, 63, 473–479. [Google Scholar] [CrossRef][Green Version]
- Al-Fattani, M.A.; Douglas, L.J. Biofilm matrix of Candida albicans and Candida tropicalis: Chemical composition and role in drug resistance. J. Med. Microbiol. 2006, 55, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Lima, O.C.; Figueiredo, C.C.; Pereira, B.A.; Coelho, M.G.; Morandi, V.; Lopes-Bezerra, L.M. Adhesion of the human pathogen Sporothrix schenckii to several extracellular matrix proteins. Braz. J. Med. Biol. Res. 1999, 32, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Endres, S.; Ghorbani, R.; Lonnemann, G.; van der Meer, J.W.; Dinarello, C.A. Measurement of immunoreactive interleukin-1 beta from human mononuclear cells: Optimization of recovery, intrasubject consistency, and comparison with interleukin-1 alpha and tumor necrosis factor. Clin. Immunol. Immunopathol. 1988, 49, 424–438. [Google Scholar] [CrossRef] [PubMed]
- Tamez-Castrellón, A.K.; van der Beek, S.L.; López-Ramírez, L.A.; Martínez-Duncker, I.; Lozoya-Pérez, N.E.; van Sorge, N.M.; Mora-Montes, H.M. Disruption of protein rhamnosylation affects the Sporothrix schenckii-host interaction. Cell Surf. 2021, 7, 100058. [Google Scholar] [CrossRef] [PubMed]
- Clavijo-Giraldo, D.M.; Matinez-Alvarez, J.A.; Lopes-Bezerra, L.M.; Ponce-Noyola, P.; Franco, B.; Almeida, R.S.; Mora-Montes, H.M. Analysis of Sporothrix schenckii sensu stricto and Sporothrix brasiliensis virulence in Galleria mellonella. J. Microbiol. Methods 2016, 122, 73–77. [Google Scholar] [CrossRef] [PubMed]
- García-Carnero, L.C.; Clavijo-Giraldo, D.M.; Gómez-Gaviria, M.; Lozoya-Pérez, N.E.; Tamez-Castrellón, A.K.; López-Ramírez, L.A.; Mora-Montes, H.M. Early virulence predictors during the Candida species-Galleria mellonella interaction. J. Fungi 2020, 6, 152. [Google Scholar] [CrossRef]
- Loibl, M.; Strahl, S. Protein O-mannosylation: What we have learned from baker’s yeast. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 2438–2446. [Google Scholar] [CrossRef]
- Marakalala, M.J.; Vautier, S.; Potrykus, J.; Walker, L.A.; Shepardson, K.M.; Hopke, A.; Mora-Montes, H.M.; Kerrigan, A.; Netea, M.G.; Murray, G.I.; et al. Differential adaptation of Candida albicans in vivo modulates immune recognition by dectin-1. PLoS Pathog. 2013, 9, e1003315. [Google Scholar] [CrossRef]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef]
- Dichtl, K.; Samantaray, S.; Wagener, J. Cell wall integrity signalling in human pathogenic fungi. Cell Microbiol. 2016, 18, 1228–1238. [Google Scholar] [CrossRef]
- Martín, H.; Molina, M. Special issue “The Fungal Cell Wall Integrity Pathway”. J. Fungi 2023, 9, 293. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.J.; Wu, C.Y.; Yu, S.J.; Chen, Y.L. Protein kinase A governs growth and virulence in Candida tropicalis. Virulence 2018, 9, 331–347. [Google Scholar] [CrossRef] [PubMed]
- Contreras Martínez, O.I.; Angulo Ortíz, A.; Santafé Patiño, G.; Peñata-Taborda, A.; Berrio Soto, R. Isoespintanol antifungal activity involves mitochondrial dysfunction, inhibition of biofilm formation, and damage to cell wall integrity in Candida tropicalis. Int. J. Mol. Sci. 2023, 24, 10187. [Google Scholar] [CrossRef] [PubMed]
- Ramos Lde, S.; Barbedo, L.S.; Braga-Silva, L.A.; dos Santos, A.L.; Pinto, M.R.; Sgarbi, D.B. Protease and phospholipase activities of Candida spp. isolated from cutaneous candidiasis. Rev. Iberoam. Micol. 2015, 32, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Staniszewska, M. Virulence factors in Candida species. Curr. Protein Pept. Sci. 2020, 21, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.J.; Mohamad Ali, M.S.; Sabri, S.; Muhd Noor, N.D.; Salleh, A.B.; Oslan, S.N. Opportunistic yeast pathogen Candida spp.: Secreted and membrane-bound virulence factors. Med. Mycol. 2021, 59, 1127–1144. [Google Scholar] [CrossRef] [PubMed]
- Gantner, B.N.; Simmons, R.M.; Underhill, D.M. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. Embo J. 2005, 24, 1277–1286. [Google Scholar] [CrossRef]
- Smith, A.J.; Graves, B.; Child, R.; Rice, P.J.; Ma, Z.; Lowman, D.W.; Ensley, H.E.; Ryter, K.T.; Evans, J.T.; Williams, D.L. Immunoregulatory activity of the natural product laminarin varies widely as a result of its physical properties. J. Immunol. 2018, 200, 788–799. [Google Scholar] [CrossRef]
- Gómez-Gaviria, M.; Lozoya-Pérez, N.E.; Staniszewska, M.; Franco, B.; Niño-Vega, G.A.; Mora-Montes, H.M. Loss of Kex2 affects the Candida albicans cell wall and interaction with innate immune cells. J. Fungi 2020, 6, 57. [Google Scholar] [CrossRef]
- Zuza-Alves, D.L.; Silva-Rocha, W.P.; Chaves, G.M. An update on Candida tropicalis based on basic and clinical approaches. Front. Microbiol. 2017, 8, 1927. [Google Scholar] [CrossRef]
- Butler, G.; Rasmussen, M.D.; Lin, M.F.; Santos, M.A.S.; Sakthikumar, S.; Munro, C.A.; Rheinbay, E.; Grabherr, M.; Forche, A.; Reedy, J.L.; et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009, 459, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, S.S.; Jofre, F.M.; Bianchini, I.A.; Boaes, T.D.S.; Bordini, F.W.; Chandel, A.K.; Felipe, M. Current advances in Candida tropicalis: Yeast overview and biotechnological applications. Biotechnol. Appl. Biochem. 2023, 70, 2069–2087. [Google Scholar] [CrossRef] [PubMed]
- de Souza, C.M.; Dos Santos, M.M.; Furlaneto-Maia, L.; Furlaneto, M.C. Adhesion and biofilm formation by the opportunistic pathogen Candida tropicalis: What do we know? Can. J. Microbiol. 2023, 69, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gaviria, M.; Ramírez-Sotelo, U.; Mora-Montes, H.M. Non-albicans Candida species: Immune response, evasion mechanisms, and new plant-derived alternative therapies. J. Fungi 2022, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Lussier, M.; Gentzsch, M.; Sdicu, A.M.; Bussey, H.; Tanner, W. Protein O-glycosylation in yeast. The PMT2 gene specifies a second protein O-mannosyltransferase that functions in addition to the PMT1-encoded activity. J. Biol. Chem. 1995, 270, 2770–2775. [Google Scholar] [CrossRef]
- Willger, S.D.; Ernst, J.F.; Alspaugh, J.A.; Lengeler, K.B. Characterization of the PMT gene family in Cryptococcus neoformans. PLoS ONE 2009, 4, e6321. [Google Scholar] [CrossRef]
- Girrbach, V.; Strahl, S. Members of the evolutionarily conserved PMT family of protein O-mannosyltransferases form distinct protein complexes among themselves. J. Biol. Chem. 2003, 278, 12554–12562. [Google Scholar] [CrossRef] [PubMed]
- Strahl-Bolsinger, S.; Gentzsch, M.; Tanner, W. Protein O-mannosylation. Biochim. Biophys. Acta 1999, 1426, 297–307. [Google Scholar] [CrossRef]
- Cantero, P.D.; Ernst, J.F. Damage to the glycoshield activates PMT-directed O-mannosylation via the Msb2-Cek1 pathway in Candida albicans. Mol. Microbiol. 2011, 80, 715–725. [Google Scholar] [CrossRef]
- Peltroche-Llacsahuanga, H.; Goyard, S.; d’Enfert, C.; Prill, S.K.; Ernst, J.F. Protein O-mannosyltransferase isoforms regulate biofilm formation in Candida albicans. Antimicrob. Agents Chemother. 2006, 50, 3488–3491. [Google Scholar] [CrossRef][Green Version]
- Kozik, A.; Karkowska-Kuleta, J.; Zajac, D.; Bochenska, O.; Kedracka-Krok, S.; Jankowska, U.; Rapala-Kozik, M. Fibronectin-, vitronectin- and laminin-binding proteins at the cell walls of Candida parapsilosis and Candida tropicalis pathogenic yeasts. BMC Microbiol. 2015, 15, 197. [Google Scholar] [CrossRef] [PubMed]
- Corbucci, C.; Cenci, E.; Skrzypek, F.; Gabrielli, E.; Mosci, P.; Ernst, J.F.; Bistoni, F.; Vecchiarelli, A. Immune response to Candida albicans is preserved despite defect in O-mannosylation of secretory proteins. Med. Mycol. 2007, 45, 709–719. [Google Scholar] [CrossRef][Green Version]
- Heinsbroek, S.E.; Taylor, P.R.; Martinez, F.O.; Martinez-Pomares, L.; Brown, G.D.; Gordon, S. Stage-specific sampling by pattern recognition receptors during Candida albicans phagocytosis. PLoS Pathog. 2008, 4, e1000218. [Google Scholar] [CrossRef] [PubMed]
- Rouabhia, M.; Schaller, M.; Corbucci, C.; Vecchiarelli, A.; Prill, S.K.; Giasson, L.; Ernst, J.F. Virulence of the fungal pathogen Candida albicans requires the five isoforms of protein mannosyltransferases. Infect. Immun. 2005, 73, 4571–4580. [Google Scholar] [CrossRef] [PubMed]
- Wächtler, B.; Wilson, D.; Haedicke, K.; Dalle, F.; Hube, B. From attachment to damage: Defined genes of Candida albicans mediate adhesion, invasion and damage during interaction with oral epithelial cells. PLoS ONE 2011, 6, e17046. [Google Scholar] [CrossRef] [PubMed]
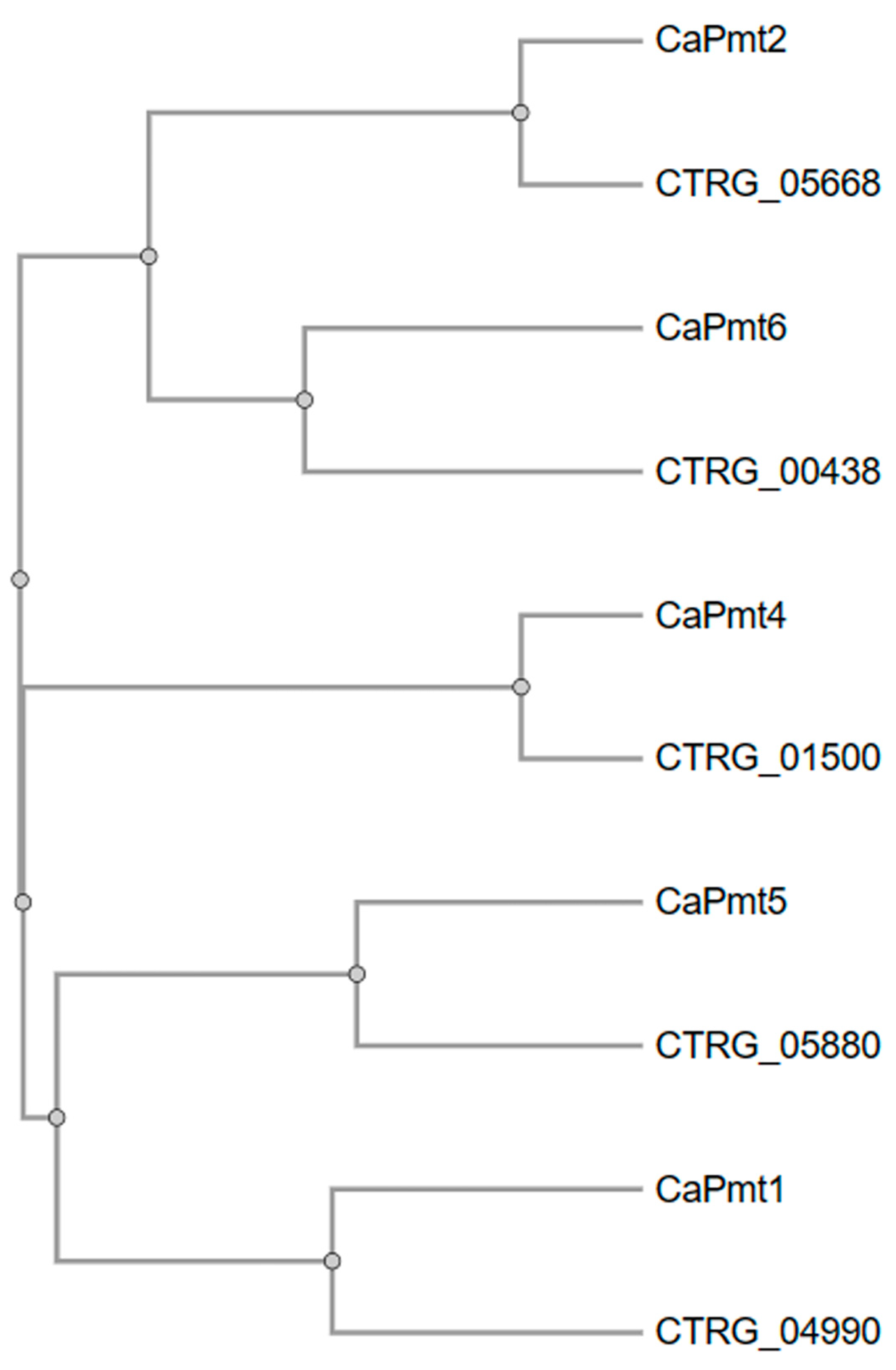
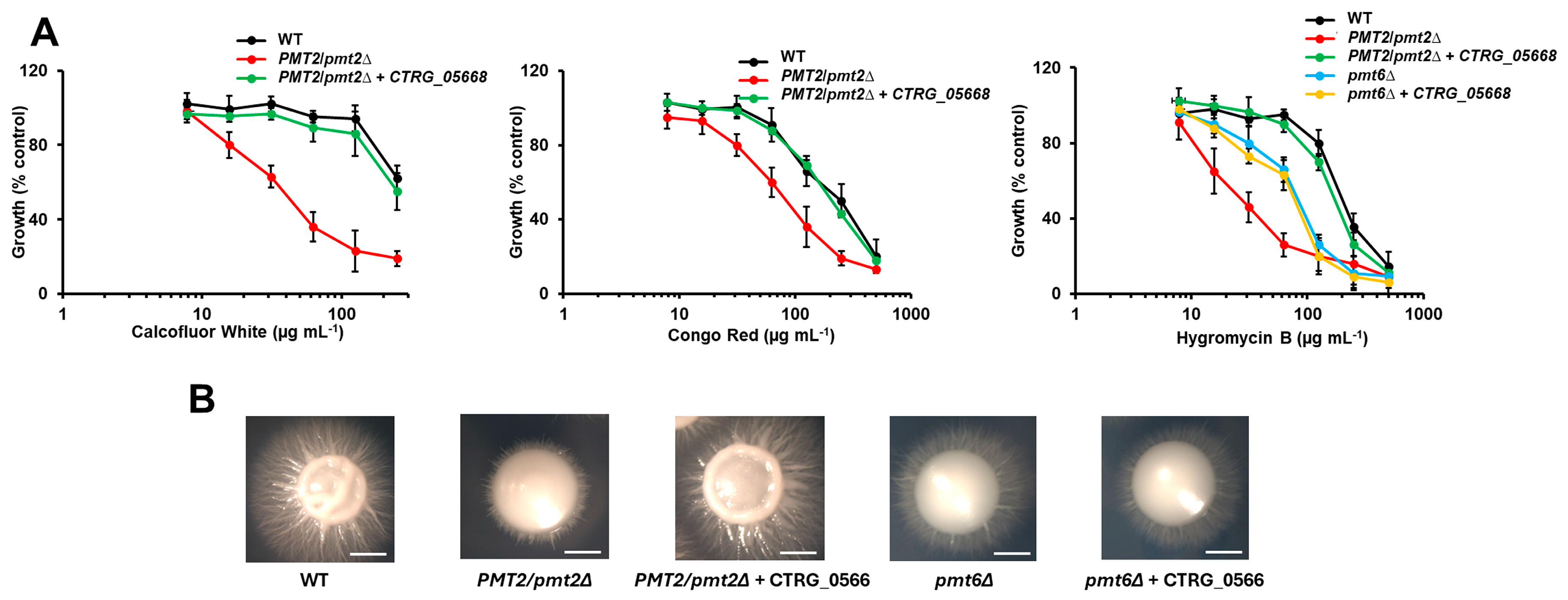


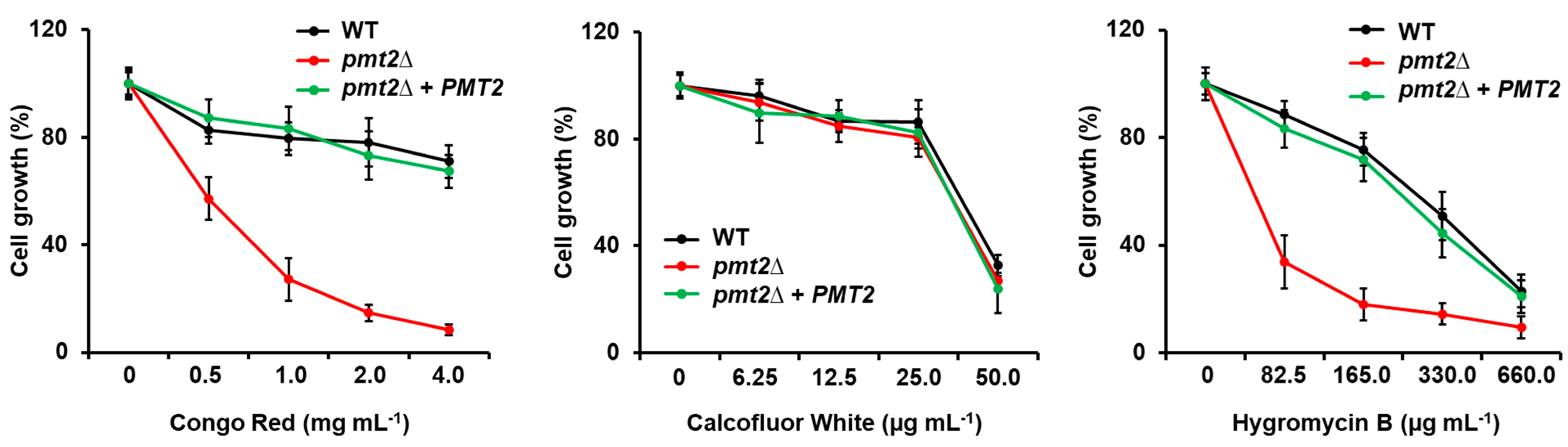
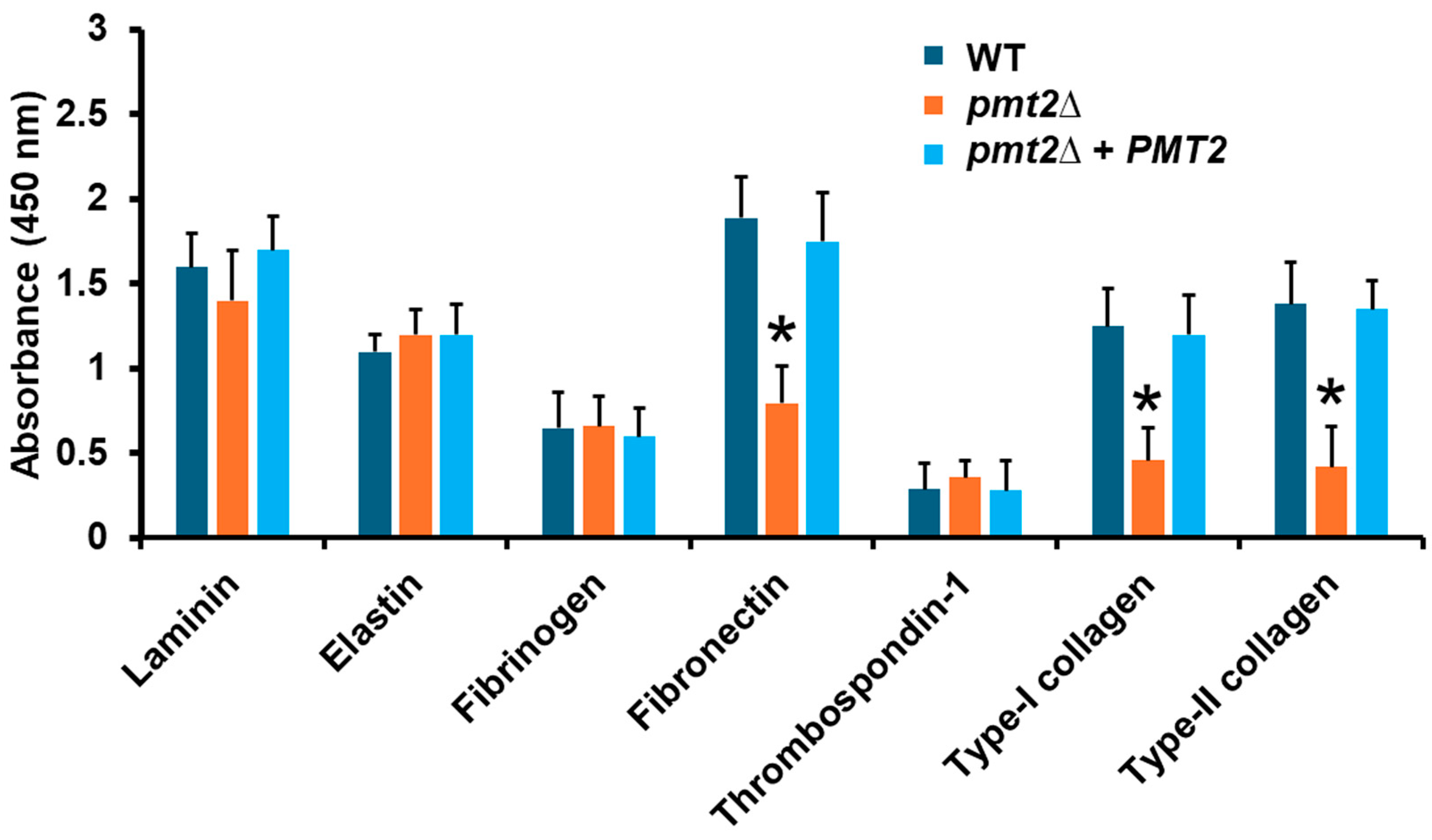


| Strain | PMT1 | PMT2 | PMT4 | PMT5 | PMT6 |
|---|---|---|---|---|---|
| YPD a | |||||
| Wild-type | 1.0 ± 0.2 | 3.3 ± 0.4 | 2.4 ± 0.6 | 0.8 ± 0.3 | 0.6 ± 0.3 |
| pmt2∆ | 0.9 ± 0.4 | N.D. d | 2.8 ± 0.3 | 0.9 ± 0.5 | 0.5 ± 0.4 |
| pmt2∆ + PMT2 | 0.9 ± 0.3 | 3.1 ± 0.3 | 2.6 ± 0.7 | 0.6 ± 0.4 | 0.5 ± 0.4 |
| SC b | |||||
| Wild-type | 1.6 ± 0.4 | 2.9 ± 0.4 | 3.9 ± 0.7 | 0.9 ± 0.5 | 1.1 ± 0.5 |
| pmt2∆ | 1.8 ± 0.3 | N.D. | 4.2 ± 0.8 | 1.1 ± 0.5 | 0.8 ± 0.4 |
| pmt2∆ + PMT2 | 1.8 ± 1.6 | 2.6 ± 0.5 | 4.0 ± 0.9 | 1.0 ± 0.6 | 0.9 ± 0.5 |
| RPMI + FCS c | |||||
| Wild-type | 2.4 ± 0.4 | 5.2 ± 0.6 | 5.2 ± 0.9 | 1.1 ± 0.2 | 3.1 ± 0.4 |
| pmt2∆ | 2.6 ± 0.5 | N.D. | 4.9 ± 0.8 | 1.4 ± 0.4 | 3.5 ± 0.6 |
| pmt2∆ + PMT2 | 2.2 ± 0.5 | 5.0 ± 0.5 | 5.3 ± 1.1 | 1.0± 0.5 | 3.0 ± 0.5 |
| Strain | PKC1 a | MKC1 a | Secreted Protease (U) b | Secreted Lipase (U) c |
|---|---|---|---|---|
| Wild-type | 1.2 ± 0.5 | 1.0 ± 0.3 | 1122.4 ± 223.4 | 395.5 ± 66.5 |
| pmt2∆ | 3.8 ± 0.8 * | 4.6 ± 0.6 * | 589.5 ± 179.6 * | 99.2 ± 49.7 * |
| pmt2∆ + PMT2 | 1.0 ± 0.4 | 1.3 ± 0.5 | 1098.2 ± 186.4 | 402.5± 63.0 |
| Strain | Biofilm Formation a | CFU (×107 cells) b | Glucose (µg mL−1) c | Glucosamine (µg mL−1) c | Protein (µg mL−1) c |
|---|---|---|---|---|---|
| Wild-type | 4.1 ± 0.2 | 1.4 ± 0.6 | 40.4 ± 6.2 | 563.8 ± 45.6 | 65.5 ± 9.8 |
| pmt2∆ | 2.7 ± 0.2 * | 0.9 ± 0.2 * | 18.5 ± 5.6 * | 265.6 ± 33.5 * | 22.8 ± 15.6 * |
| pmt2∆ + PMT2 | 4.0 ± 0.3 | 1.5 ± 0.3 | 37.7 ± 86.4 | 532.6 ± 33.8 | 61.5 ± 11.6 |
| Strain | CFUs (×107) a | Cytotoxicity (%) b | Hemocytes (×106) mL−1 | Melanin c | Phenoloxidase d |
|---|---|---|---|---|---|
| PBS e | 0.0 ± 0.0 | 9.8 ± 4.2 | 5.4 ± 0.4 | 1.1 ± 0.2 | 0.3 ± 0.1 |
| WT | 1.3 ± 0.4 | 78.9 ± 9.6 | 2.2 ± 0.5 | 4.2 ± 0.4 | 2.9 ± 0.3 |
| pmt2 ∆ | 1.4 ± 0.3 | 26.4 ± 11.5 * | 4.9 ± 0.2 * | 2.3 ± 0.5 * | 1.1 ± 0.2 * |
| pmt2∆ + PMT2 | 1.3 ± 0.5 | 81.2 ± 15.4 | 2.5 ± 0.6 | 4.4 ± 0.2 | 3.2 ± 0.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Chávez, M.J.; Martínez-Duncker, I.; Clavijo-Giraldo, D.M.; López-Ramirez, L.A.; Mora-Montes, H.M. Candida tropicalis PMT2 Is a Dispensable Gene for Viability but Required for Proper Interaction with the Host. J. Fungi 2024, 10, 502. https://doi.org/10.3390/jof10070502
Hernández-Chávez MJ, Martínez-Duncker I, Clavijo-Giraldo DM, López-Ramirez LA, Mora-Montes HM. Candida tropicalis PMT2 Is a Dispensable Gene for Viability but Required for Proper Interaction with the Host. Journal of Fungi. 2024; 10(7):502. https://doi.org/10.3390/jof10070502
Chicago/Turabian StyleHernández-Chávez, Marco J., Iván Martínez-Duncker, Diana M. Clavijo-Giraldo, Luz A. López-Ramirez, and Héctor M. Mora-Montes. 2024. "Candida tropicalis PMT2 Is a Dispensable Gene for Viability but Required for Proper Interaction with the Host" Journal of Fungi 10, no. 7: 502. https://doi.org/10.3390/jof10070502
APA StyleHernández-Chávez, M. J., Martínez-Duncker, I., Clavijo-Giraldo, D. M., López-Ramirez, L. A., & Mora-Montes, H. M. (2024). Candida tropicalis PMT2 Is a Dispensable Gene for Viability but Required for Proper Interaction with the Host. Journal of Fungi, 10(7), 502. https://doi.org/10.3390/jof10070502







