Blue and Yellow Laccases from Alternaria sp. Strain HU: Characterization and Immobilization on Magnetic Nanoparticles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Media and Culture Conditions
2.3. Identification of Fungus
2.4. Purification of Laccases
2.5. Characterization of Laccases
2.6. Enzyme Activity Assay
2.7. Immobilization of Laccases on the Fe3O4 Nanoparticles
3. Results
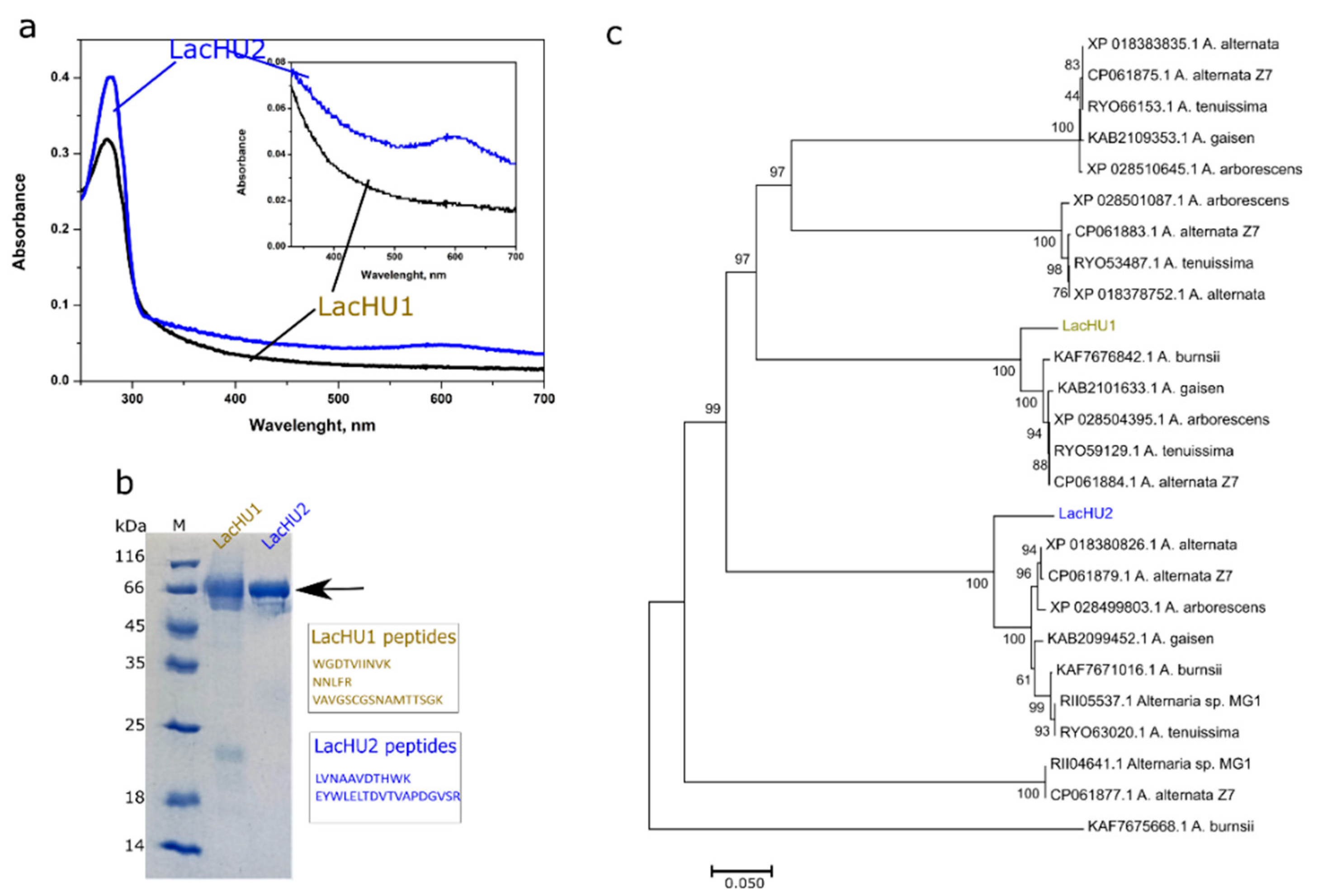
3.1. The Biosynthesis, Identification and Purification of Laccases
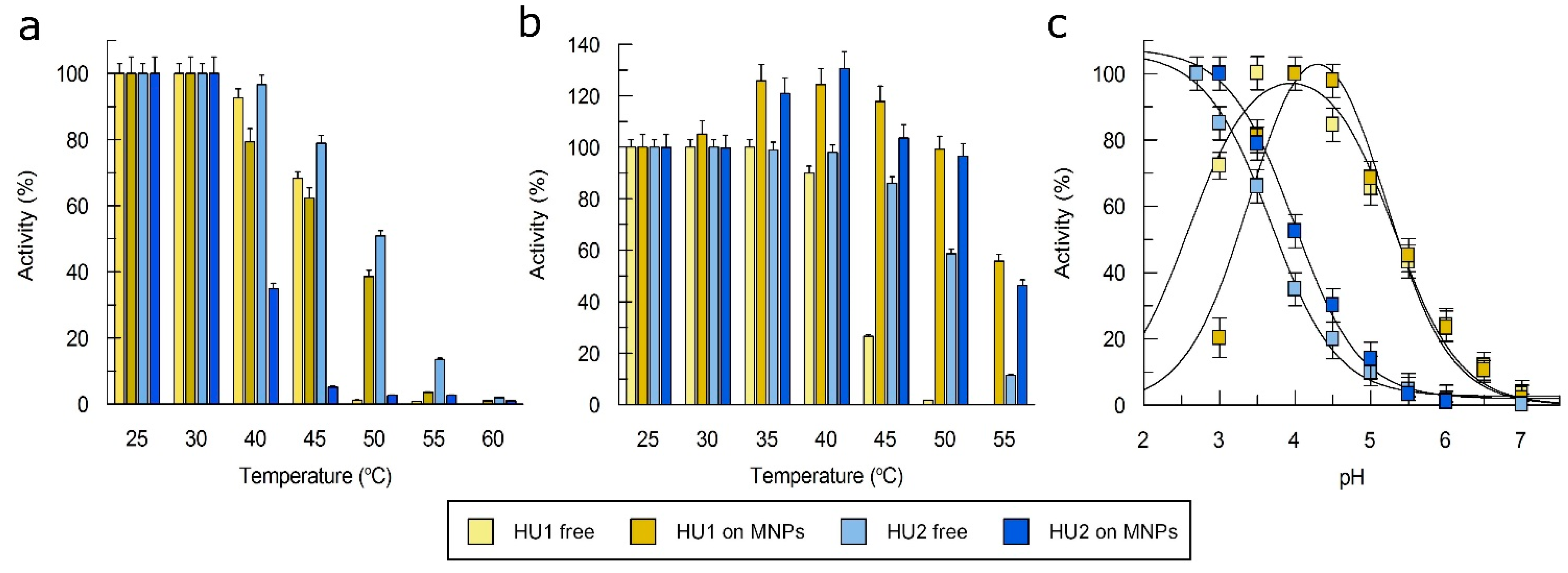
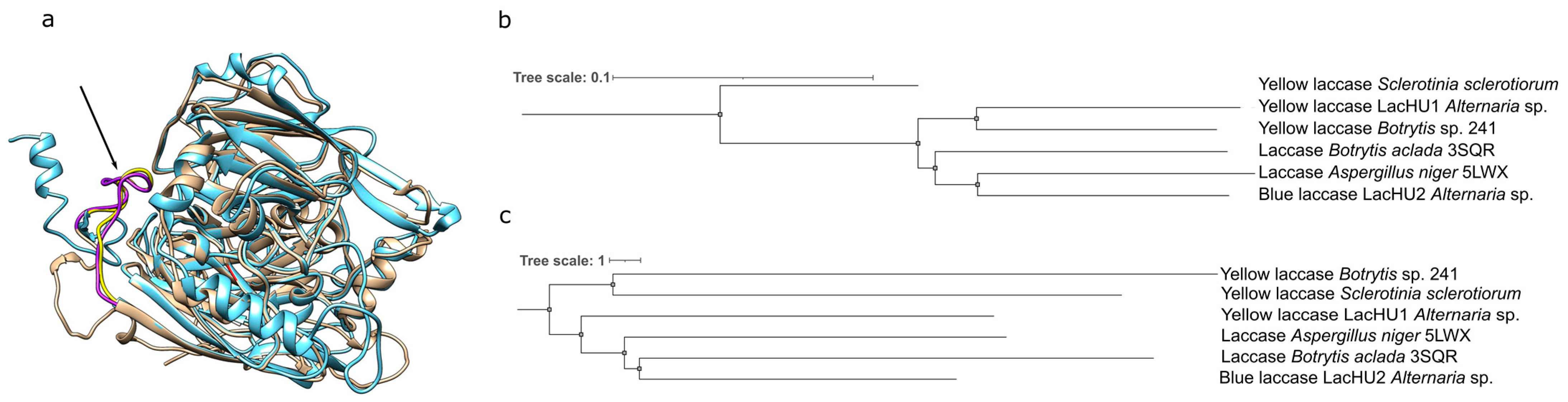
3.2. Characterization of Laccases
3.3. Catalytic Properties of the LacHU1 and LacHU2
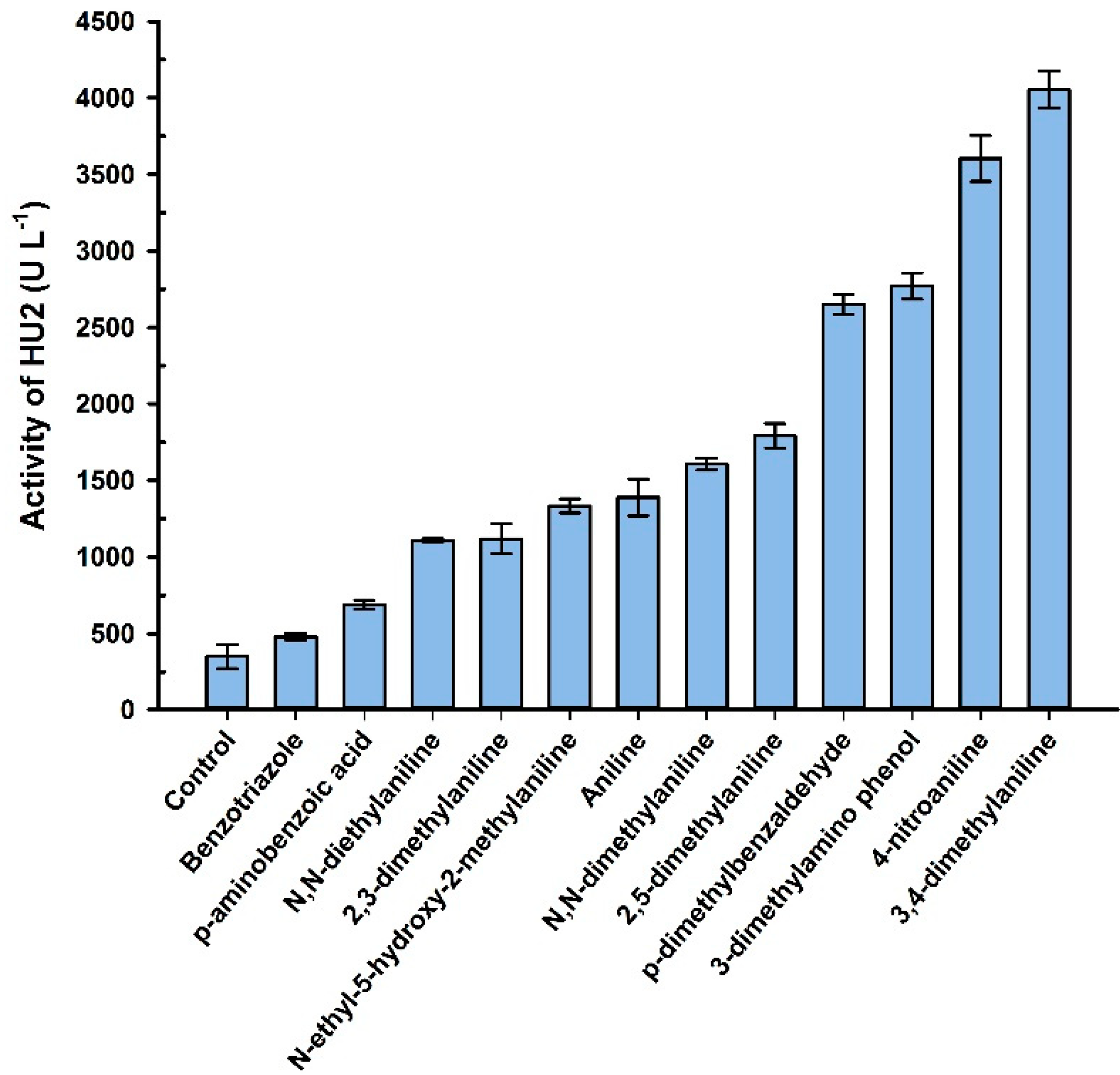
3.4. Influence of Metal Salts on the Activity of Laccase
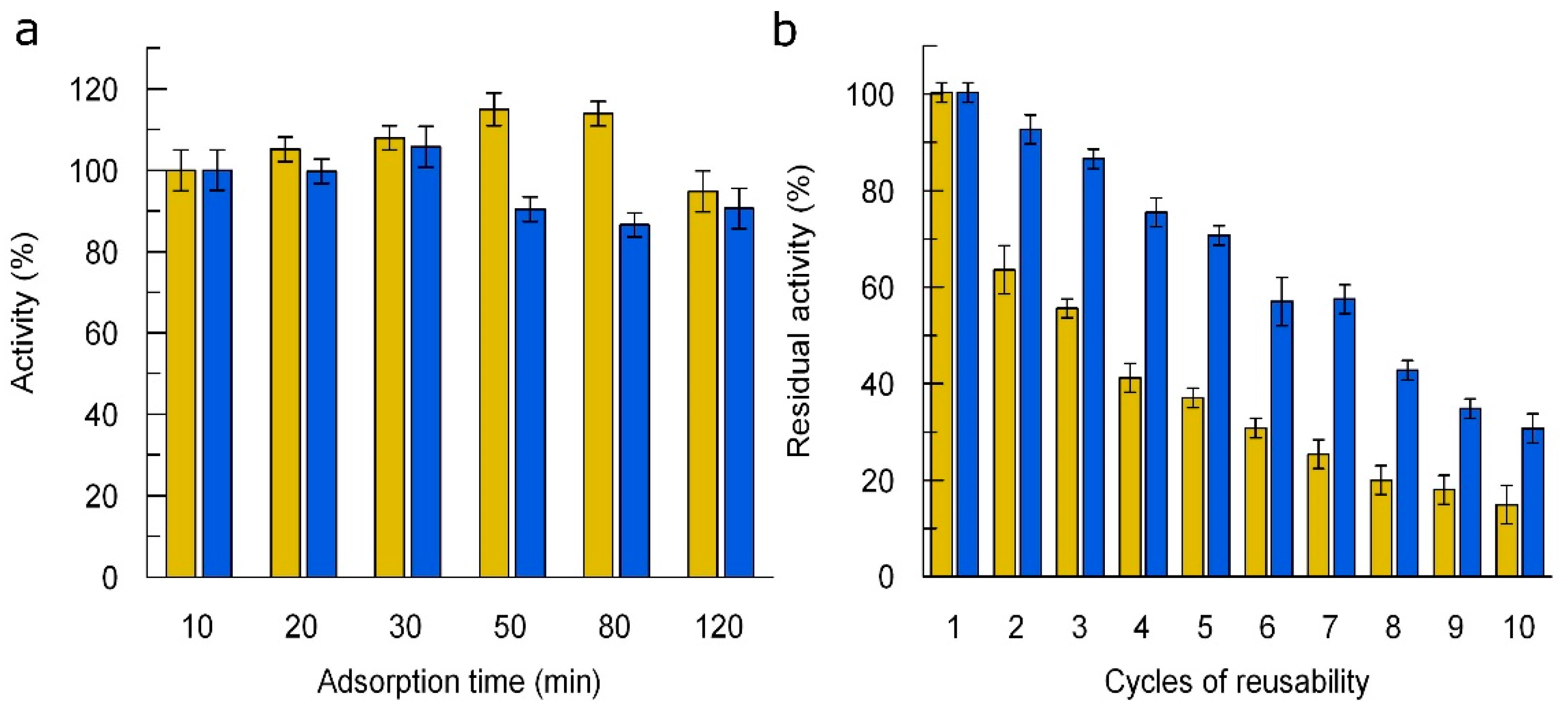
3.5. Immobilization of Laccases
4. Discussion
5. Conclusions
Supplementary Materials
 ) and LacHU2 (
) and LacHU2 ( ). (a) syringaldazine; (b) 1-naphthol; (c) ferulic acid and (d) myricetin as substrates in 60 mM Britton-Robinson buffer, 25 °C. The highest activity was equated to 100%.; Figure S3: Effect of pH on stability of LacHU1 (
). (a) syringaldazine; (b) 1-naphthol; (c) ferulic acid and (d) myricetin as substrates in 60 mM Britton-Robinson buffer, 25 °C. The highest activity was equated to 100%.; Figure S3: Effect of pH on stability of LacHU1 ( ) and LacHU2 (
) and LacHU2 ( ). Laccases were incubated for 20 hours in 60 mM Britton-Robinson buffer, at 25 °C, ABTS was used as a substrate. The highest activity was equated to 100%.; Figure S4: Dependence of enzymatic activity on laccase concentration used for immobilization.; Table S1: Comparison of the retained activity of laccase immobilized on different matrices after 10 cycles of reuse.
). Laccases were incubated for 20 hours in 60 mM Britton-Robinson buffer, at 25 °C, ABTS was used as a substrate. The highest activity was equated to 100%.; Figure S4: Dependence of enzymatic activity on laccase concentration used for immobilization.; Table S1: Comparison of the retained activity of laccase immobilized on different matrices after 10 cycles of reuse.Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jeon, J.R.; Baldrian, P.; Murugesan, K.; Chang, Y.S. Laccase-catalysed oxidations of naturally occurring phenols: From in vivo biosynthetic pathways to green synthetic applications. Microb. Biotechnol. 2012, 5, 318–332. [Google Scholar] [CrossRef]
- Debnath, R.; Saha, T. An insight into the production strategies and applications of the ligninolytic enzyme laccase from bacteria and fungi. Biocatal. Agric. Biotechnol. 2020, 26, 101645. [Google Scholar] [CrossRef]
- Khatami, S.H.; Vakili, O.; Movahedpour, A.; Ghesmati, Z.; Ghasemi, H.; Taheri-Anganeh, M. Laccase: Various types and applications. Biotechnol. Appl. Biochem. 2022, 69, 2658–2672. [Google Scholar] [CrossRef] [PubMed]
- Curran, L.M.L.K.; Pham, L.T.M.; Sale, K.L.; Simmons, B.A. Review of advances in the development of laccases for the valorization of lignin to enable the production of lignocellulosic biofuels and bioproducts. Biotechnol. Adv. 2022, 54, 107809. [Google Scholar] [CrossRef]
- Piscitelli, A.; Pezzella, C.; Giardina, P.; Faraco, V.; Giovanni, S. Heterologous laccase production and its role in industrial applications. Bioeng. Bugs 2010, 1, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Marcinkevičiene, L.; Vidžiunaite, R.; Tauraite, D.; Rutkiene, R.; Bachmatova, I.; Morkunas, M.; Razumiene, J.; Časaite, V.; Meškiene, R.; Kulys, J.; et al. Characterization of laccase from Coriolopsis byrsina GRB13 and application of the enzyme for synthesis of redox mediators. Chemija 2013, 24, 48–58. [Google Scholar]
- Martínková, L.; Křístková, B.; Křen, V. Laccases and Tyrosinases in Organic Synthesis. Int. J. Mol. Sci. 2022, 23, 3462. [Google Scholar] [CrossRef] [PubMed]
- Laurinavičius, V.; Kurtinaitiene, B.; Liauksminas, V.; Jankauskaite, A.; Šimkus, R.; Meškys, R.; Boguslavsky, L.; Skotheim, T.; Tanenbaum, S. Reagentless biosensor based on PQQ-depended glucose dehydrogenase and partially hydrolyzed polyarbutin. Talanta 2000, 52, 485–493. [Google Scholar] [CrossRef]
- Bassanini, I.; Ferrandi, E.E.; Riva, S.; Monti, D. Biocatalysis with laccases: An updated overview. Catalysts 2021, 11, 26. [Google Scholar] [CrossRef]
- Dagys, M.; Laurynenas, A.; Ratautas, D.; Kulys, J.; Vidžiunaite, R.; Talaikis, M.; Niaura, G.; Marcinkevičiene, L.; Meškys, R.; Shleev, S. Oxygen electroreduction catalysed by laccase wired to gold nanoparticles via the trinuclear copper cluster. Energy Environ. Sci. 2017, 10, 498–502. [Google Scholar] [CrossRef]
- Kadam, A.A.; Saratale, G.D.; Ghodake, G.S.; Saratale, R.G.; Shahzad, A.; Magotra, V.K.; Kumar, M.; Palem, R.R.; Sung, J.S. Recent Advances in the Development of Laccase-Based Biosensors via Nano-Immobilization Techniques. Chemosensors 2022, 10, 58. [Google Scholar] [CrossRef]
- Zhang, Y.; Lv, Z.; Zhou, J.; Xin, F.; Ma, J.; Wu, H.; Fang, Y.; Jiang, M.; Dong, W. Application of eukaryotic and prokaryotic laccases in biosensor and biofuel cells: Recent advances and electrochemical aspects. Appl. Microbiol. Biotechnol. 2018, 102, 10409–10423. [Google Scholar] [CrossRef] [PubMed]
- Dekker, R.F.H.; Barbosa, A.M. The effects of aeration and veratryl alcohol on the production of two laccases by the ascomycete Botryosphaeria sp. Enzym. Microb. Technol. 2001, 28, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, Y.; Zhang, Y.; Yang, E.; Qu, Y.; Xu, H.; Chen, Y.; Irbis, C.; Yan, J. A Thermo-Active Laccase Isoenzyme From Trametes trogii and Its Potential for Dye Decolorization at High Temperature. Front. Microbiol. 2020, 11, 241. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.W.; Si, W.; Xue, H.P.; Yang, M.M.; Zhao, X. Molecular characterization and expression profiling of novel laccase isoenzyme genes from Fusarium solani X701, a wood-rotting ascomycete. Int. Biodeterior. Biodegrad. 2015, 104, 123–128. [Google Scholar] [CrossRef]
- Renfeld, Z.V.; Chernykh, A.M.; Egorova, A.D.; Baskunov, B.P.; Gaidina, A.S.; Myasoedova, N.M.; Moiseeva, O.V.; Kolomytseva, M.P. The Laccase of Myrothecium roridum VKM F-3565: A New Look at Fungal Laccase Tolerance to Neutral and Alkaline Conditions. ChemBioChem 2023, 24, e202200600. [Google Scholar] [CrossRef] [PubMed]
- De Paula, N.M.; da Silva, K.; Brugnari, T.; Haminiuk, C.W.I.; Maciel, G.M. Biotechnological potential of fungi from a mangrove ecosystem: Enzymes, salt tolerance and decolorization of a real textile effluent. Microbiol. Res. 2022, 254, 126899. [Google Scholar] [CrossRef]
- Du, W.; Sun, C.; Wang, J.; Wang, B.; Yao, Z.; Qu, F.; Xia, J.; Xie, W.; Sun, J.; Duan, D. Isolation, identification of a laccase-producing fungal strain and enzymatic properties of the laccase. 3 Biotech 2018, 8, 137. [Google Scholar] [CrossRef]
- Zhang, L.-B.; Yang, W.W.J.; Qiu, T.T. Genome-wide study of Cerrena unicolor 87613 laccase gene family and their mode prediction in association with substrate oxidation. BMC Genom. 2023, 24, 504. [Google Scholar] [CrossRef]
- Shankar, A.; Jain, K.K.; Kuhad, R.C.; Sharma, K.K. Comparison of lignocellulosic enzymes and CAZymes between ascomycetes (Trichoderma) and basidiomycetes (Ganoderma) species: A proteomic approach. Z. Naturforsch.-Sect. C J. Biosci. 2023. [Google Scholar] [CrossRef]
- Scott, C.J.R.; Leadbeater, D.R.; Oates, N.C.; James, S.R.; Newling, K.; Li, Y.; McGregor, N.G.S.; Bird, S.; Bruce, N.C. Whole genome structural predictions reveal hidden diversity in putative oxidative enzymes of the lignocellulose-degrading ascomycete Parascedosporium putredinis NO1. Microbiol. Spectr. 2023, 11, e0103523. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, G.; Cennamo, G.; Faraco, V.; Amoresano, A.; Sannia, G.; Giardina, P. Atypical laccase isoenzymes from copper supplemented Pleurotus ostreatus cultures. Enzym. Microb. Technol. 2003, 33, 220–230. [Google Scholar] [CrossRef]
- He, F.; Qin, X.; Zhang, H.; Yang, Y.; Zhang, X.; Yang, Y. Characterization of laccase isoenzymes from the white-rot fungus Ganoderma sp.En3 and synergistic action of isoenzymes for dye decolorization. J. Chem. Technol. Biotechnol. 2015, 90, 2265–2279. [Google Scholar] [CrossRef]
- Lisova, Z.A.; Lisov, A.V.; Leontievsky, A.A. Two laccase isoforms of the basidiomycete Cerrena unicolor VKMF-3196. Induction, isolation and properties. J. Basic Microbiol. 2010, 50, 72–82. [Google Scholar] [CrossRef]
- Cen, Q.; Wu, X.; Cao, L.; Lu, Y.; Lu, X.; Chen, J.; Fu, G.; Liu, Y.; Ruan, R. Green production of a yellow laccase by Coriolopsis gallica for phenolic pollutants removal. AMB Express 2022, 12, 96. [Google Scholar] [CrossRef]
- Chaurasia, P.K.; Bharati, S.L.; Singh, S.K. Comparative Studies on the Blue and Yellow Laccases. Res. Plant Sci. 2023, 1, 32–37. [Google Scholar] [CrossRef]
- Wong, J.K.H.; Tan, H.K.; Lau, S.Y.; Yap, P.S.; Danquah, M.K. Potential and challenges of enzyme incorporated nanotechnology in dye wastewater treatment: A review. J. Environ. Chem. Eng. 2019, 7, 103261. [Google Scholar] [CrossRef]
- Ahmad, R.; Sardar, M. Enzyme Immobilization: An Overview on Nanoparticles as Immobilization Matrix. Biochem. Anal. Biochem. 2015, 4, 178. [Google Scholar] [CrossRef]
- Muthuvelu, K.S.; Rajarathinam, R.; Selvaraj, R.N.; Rajendren, V.B. A novel method for improving laccase activity by immobilization onto copper ferrite nanoparticles for lignin degradation. Int. J. Biol. Macromol. 2020, 152, 1098–1107. [Google Scholar] [CrossRef]
- Kulys, J.; Vidziunaite, R.; Janciene, R.; Palaima, A. Spectroelectrochemical study of N-substituted phenoxazines as electrochemical labels of biomolecules. Electroanalysis 2006, 18, 1771–1777. [Google Scholar] [CrossRef]
- Radveikienė, I.; Vidžiūnaitė, R.; Meškienė, R.; Meškys, R.; Časaitė, V. Characterization of a yellow laccase from Botrytis cinerea 241. J. Fungi 2021, 7, 143. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Rzhetsky, A.; Nei, M. A Simple Method for Estimating and Testing Minimum-Evolution Trees. Mol. Biol. Evol. 1992, 9, 945–967. [Google Scholar]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000; p. 352. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Brunak, S. Prediction of Glycosylation Across the Human Proteome and the Correlation to Protein Function. Pac. Symp. Biocomput. 2001, 7, 310–322. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Lowry, O.H.; Roserbrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Michałowska-Kaczmarczyk, A.M.; Michałowski, T. Dynamic Buffer Capacity in Acid-Base Systems. J. Solut. Chem. 2015, 44, 1256–1266. [Google Scholar] [CrossRef]
- Radveikienė, I.; Pilotaitė, I.; Dainytė, R.; Vidžiūnaitė, R. Biosynthesis, purification, characterization and immobilization of laccase from Lithothelium sp. Chemija 2020, 31, 178–190. [Google Scholar] [CrossRef]
- Ainiwaer, A.; Liang, Y.; Ye, X.; Gao, R. Characterization of a Novel Fe2+ Activated Non-Blue Laccase from Methylobacterium extorquens. Int. J. Mol. Sci. 2022, 23, 9804. [Google Scholar] [CrossRef]
- Ainiwaer, A.; Li, A.; Zhao, X.; Xu, Y.; Han, S.; Gao, R. Site-Specific Covalent Immobilization of Methylobacterium extorquens Non-Blue Laccse Melac13220 on Fe3O4 Nanoparticles by Aldehyde Tag. Catalysts 2022, 12, 1379. [Google Scholar] [CrossRef]
- Chernykh, A.; Myasoedova, N.; Kolomytseva, M.; Ferraroni, M.; Briganti, F.; Scozzafava, A.; Golovleva, L. Laccase isoforms with unusual properties from the basidiomycete Steccherinum ochraceum strain 1833. J. Appl. Microbiol. 2008, 105, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, R.; He, F.; Zhang, X.; Yang, Y. Characterization of a yeast recombinant laccase rLAC-EN3-1 and its application in decolorizing synthetic dye with the coexistence of metal ions and organic solvents. Biochem. Eng. J. 2015, 93, 63–72. [Google Scholar] [CrossRef]
- Mot, A.C.; Coman, C.; Hadade, N.; Damian, G.; Silaghi-Dumitrescu, R.; Heering, H. “Yellow” laccase from Sclerotinia sclerotiorum is a blue laccase that enhances its substrate affinity by forming a reversible tyrosyl-product adduct. PLoS ONE 2020, 15, e0225530. [Google Scholar] [CrossRef] [PubMed]
- Leontievsky, A.; Myasoedova, N.; Pozdnyakova, N.; Golovleva, L. ‘Yellow’ laccase of Panus tigrinus oxidizes non-phenolic substrates without electron-transfer mediators. FEBS Lett. 1997, 413, 446–448. [Google Scholar] [CrossRef] [PubMed]
- Chaoua, S.; Chaouche, N.K.; Songulashvili, G.; Gares, M.; Hiligsmann, S.; Flahaut, S. Yellow laccase produced by Trametes versicolor K1 on tomato waste: A comparative study with the blue one produced on semi-synthetic medium. J. Biotechnol. 2023, 361, 99–109. [Google Scholar] [CrossRef]
- Wang, S.; Chen, Q.; Zhu, M.; Xue, F.; Li, W.; Zhao, T.; Li, G.; Zhang, G. Biochimie An extracellular yellow laccase from white rot fungus Trametes sp. F1635 and its mediator systems for dye decolorization. Biochimie 2018, 148, 46–54. [Google Scholar] [CrossRef]
- Agrawal, K.; Verma, P. Production optimization of yellow laccase from Stropharia sp. ITCC 8422 and enzyme-mediated depolymerization and hydrolysis of lignocellulosic biomass for biorefinery application. Biomass Convers. Biorefin. 2022, 12, 4939–4958. [Google Scholar] [CrossRef]
- Bhoyar, S.S.; Chaudhari, A.U.; Desai, M.A.; Latpate, R.V.; Sartale, S.D.; Kodam, K.M. Wheat bran as an efficient agro-process waste for enhanced yellow laccase production by Lentinus tigrinus SSB_W2 and its application in anthraquinone dye degradation. 3 Biotech 2024, 14, 33. [Google Scholar] [CrossRef] [PubMed]
- Buzzo, B.B.; Lima, N.S.M.; Pereira, P.A.M.; Gomes-Pepe, E.S.; Sartini, C.C.F.; Lemos, E.G. de M. Lignin degradation by a novel thermophilic and alkaline yellow laccase from Chitinophaga sp. Microbiol. Spectr. 2024, 12, e0401323. [Google Scholar] [CrossRef] [PubMed]
- Holm, L. Dali server: Structural unification of protein families. Nucleic Acids Res. 2022, 50, W210–W215. [Google Scholar] [CrossRef] [PubMed]
- Gianfreda, L.; Xu, F.; Bollag, J.-M. Laccases: A Useful Group of Oxidoreductive Enzymes. Bioremediat. J. 1999, 3, 1–26. [Google Scholar] [CrossRef]
- Viswanath, B.; Rajesh, B.; Janardhan, A.; Kumar, A.P.; Narasimha, G. Fungal laccases and their applications in bioremediation. Enzym. Res. 2014, 2014, 163242. [Google Scholar] [CrossRef]
- Soden, D.M.; Dobson, A.D.W. Differential regulation of laccase gene expression in Pleurotus sajor-caju. Microbiology 2001, 147, 1755–1763. [Google Scholar] [CrossRef]
- Eggert, C.; Temp, U.; Eriksson, K.E. ; The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: Purification and characterization of the laccase. Appl. Environ. Microbiol. 1996, 62, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Galhaup, C.; Goller, S.; Peterbauer, C.K.; Strauss, J.; Haltrich, D. Characterization of the major laccase isoenzyme from Trametes pubescens and regulation of its synthesis by metal ions. Microbiology 2002, 148, 2159–2169. [Google Scholar] [CrossRef]
- Ibrahim, V.; Mendoza, L.; Mamo, G.; Hatti-Kaul, R. Blue laccase from Galerina sp.: Properties and potential for Kraft lignin demethylation. Process Biochem. 2011, 46, 379–384. [Google Scholar] [CrossRef]
- Kim, S.; Moldes, D.; Cavaco-Paulo, A. Laccases for enzymatic colouration of unbleached cotton. Enzym. Microb. Technol. 2007, 40, 1788–1793. [Google Scholar] [CrossRef]
- Shraddha; Shekher, R.; Sehgal, S.; Kamthania, M.; Kumar, A. Laccase: Microbial sources, production, purification, and potential biotechnological applications. Enzym. Res. 2011, 2011, 217861. [Google Scholar] [CrossRef] [PubMed]
- Bayramoglu, G.; Karagoz, B.; Arica, M.Y. Cyclic-carbonate functionalized polymer brushes on polymeric microspheres: Immobilized laccase for degradation of endocrine disturbing compounds. J. Ind. Eng. Chem. 2017, 60, 407–417. [Google Scholar] [CrossRef]
- Bankeeree, W.; Prasongsuk, S.; Imai, T.; Lotrakul, P.; Punnapayak, H. A novel xylan-polyvinyl alcohol hydrogel bead with laccase entrapment for decolorization of Reactive Black 5. BioResources 2016, 11, 6984–7000. [Google Scholar] [CrossRef]
- Wang, F.; Owusu-Fordjour, M.; Xu, L.; Ding, Z.; Gu, Z. Immobilization of Laccase on Magnetic Chelator Nanoparticles for Apple Juice Clarification in Magnetically Stabilized Fluidized Bed. Front. Bioeng. Biotechnol. 2020, 8, 589. [Google Scholar] [CrossRef]


| Purification Step | Volume, mL | Total Activity, u | Total Protein, mg | Specific Activity, U/mg | Purification Factor, Fold | Yield, % |
|---|---|---|---|---|---|---|
| LacHU1 | ||||||
| Culture liquid | 4000 | 2210 | 3200 | 0.7 | 1 | 100 |
| Ultrafiltration | 262 | 1540 | 330 | 4.7 | 7 | 69.6 |
| PHE FF | 4 | 1160 | 61 | 19 | 27 | 52.5 |
| Source 15Q | 15 | 960 | 10.5 | 91 | 132 | 43.4 |
| Source15PHE | 1 | 566 | 4.0 | 142 | 205 | 25.6 |
| LacHU2 | ||||||
| Culture liquid | 960 | 7104 | 701 | 10 | 1 | 100 |
| Ultrafiltration | 248 | 6795 | 221 | 31 | 3 | 96 |
| PHE FF | 10 | 5403 | 12.4 | 436 | 43 | 76 |
| Source 15Q | 0.7 | 5247 | 3.2 | 1630 | 160 | 74 |
| Substrate | Kinetic Parameters | |||||
|---|---|---|---|---|---|---|
| KM, µM | kcat, S−1 | kcat/KM, M−1·s−1 | ||||
| LacHU1 | LacHU2 | LacHU1 | LacHU2 | LacHU1 | LacHU2 | |
| Non-phenolic compounds | ||||||
| ABTS | 60.0 ± 10.0 | (5.6 ± 0.6) × 103 | 88.0 ± 5.0 | 440.0 ± 43.0 | (1.4 ± 0.1) × 106 | (8.0 ± 1.0) × 104 |
| N,N’-Dimethylamine-4-(4-morpholine)benzene | 90.0 ± 10.0 | (1.0 ± 0.2) × 104 | 100.0 ± 4.0 | 130.0 ± 20.0 | (1.1 ± 0.1) × 106 | (1.3 ± 0.4) × 104 |
| 2-(10H-phenoxazin-10-yl)ethanol | 40.0 ± 10.0 | 200.0 ± 50.0 | 34.0 ± 3.0 | 55.0 ± 3.0 | (9.0 ± 0.1) × 105 | (3.0 ± 1.0) × 105 |
| p-Phenylenediamine | 340.0 ± 40.0 | (3.6 ± 0.8) × 103 | 30.0 ± 1.0 | 5.4 ± 0.3 | (9.0 ± 0.1) × 104 | (1.5 ± 0.4) × 103 |
| Promazine hydrochloride | (3.8 ± 0.5) × 103 | (2.6 ± 0.6) × 103 | 10.0 ± 3.0 | 3.3 ± 0.1 | (3.0 ± 1.0) × 103 | (1.3 ± 0.3) × 103 |
| Potassium hexacyanoferrate (II) | nd 1 | (5.9 ± 1.7) × 105 | nd | 152.0 ± 21.0 | (1.7 ± 0.1) × 103 | (3.0 ± 1.0) × 102 |
| Phenolic compounds | ||||||
| Syringaldazine | 1.0 ± 0.1 | 13.0 ± 3.0 | 36.0 ± 1.0 | 5.8 ± 0.7 | (5.7 ± 0.5) × 107 | (4.5 ± 1.0) × 104 |
| Quercetin | 2.0 ± 0.4 | 80.0 ± 3.0 | 24.0 ± 1.0 | 500.0 ± 140.0 | (1.3 ± 0.2) × 107 | (1.5 ± 0.1) × 105 |
| Myricetin | 2.0 ± 0.3 | 15.0 ± 3.0 | 28.0 ± 5.0 | 200.0 ± 0.07 | (1.3 ± 0.2) × 107 | 1.2 ± 0.04) × 104 |
| 2,6-DMP | 10.0 ± 2.0 | (4.3 ± 0.3) × 103 | 33.0 ± 1.0 | 204.0 ± 6.0 | (3.4 ± 0.4) × 106 | (5.0 ± 1.0) × 104 |
| Fisetin | 15.0 ± 1.0 | 27.0 ± 2.0 | 21.0 ± 2.0 | 128.0 ± 13.0 | (1.4 ± 0.1) × 106 | (5.5 ± 0.3) × 104 |
| 1-naphthol | 100.0 ± 10.0 | nd | 130.0 ± 9.0 | nd | (1.2 ± 0.1) × 106 | (1.0 ± 0.1) × 102 |
| Caffeic acid | 50.0 ± 3.0 | 400.0 ± 30.0 | 27.0 ± 1.0 | 161.0 ± 33.0 | (6.0 ± 0.1) × 105 | (4.0 ± 1.0) × 105 |
| Ferulic acid | 50.0 ± 4.0 | 230.0 ± 30.0 | 28.0 ± 1.5 | 9.0 ± 1.0 | (6.0 ± 0.1) × 105 | (4.3 ± 1.0) × 104 |
| Syringic acid | 40.0 ± 0.5 | nox 2 | 20.0 ± 1.0 | nox | (5.0 ± 0.1) × 105 | nox |
| Kaempferol | 110.0 ± 22.0 | 120.0 ± 10.0 | 38.0 ± 14.0 | 26.0 ± 4.0 | (3.0 ± 1.0) × 105 | (7.0 ± 1.0) × 104 |
| Gallic acid | nd | nox | nd | nox | (3.0 ± 0.1) × 105 | nox |
| Chlorogenic acid | 100.0 ± 1.0 | 90.0 ± 40.0 | 13.5 ± 0.2 | 38.0 ± 12.0 | (1.3 ± 0.1) × 105 | (4.3 ± 1.0) × 105 |
| Hydroquinone | 540.0 ± 10.0 | (1.3 ± 0.2) × 104 | 62.0 ± 1.0 | 400.0 ± 46.0 | (1.1 ± 0.3) × 105 | (3.0 ± 1.0) × 104 |
| Catechol | (1.8 ± 0.2) × 103 | (2.0 ± 0.4) × 104 | 105.0 ± 5.0 | 900.0 ± 160.0 | (6.0 ± 0.1) × 104 | (4.4 ± 1.0) × 104 |
| Laccase | KM, μM | kcat, s−1 | kcat/KM, M−1S−1 |
|---|---|---|---|
| Free LacHU1 | 60 ± 10 | 88.0 ± 5.0 | (1.4 ± 0.1) × 106 |
| Immobilized LacHU1 | 65 ± 9.0 | 3.0 ± 0.14 | (4.6 ± 1.0) × 104 |
| Free LacHU2 | (5.6 ± 0.6) × 103 | 440.0 ± 43.0 | (8.0 ± 1.0) × 104 |
| Immobilized LacHU2 | (2.2 ± 0.5) × 103 | 100.0 ± 15.0 | (4.6 ± 3.0) × 104 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radveikienė, I.; Vidžiūnaitė, R.; Meškys, R.; Časaitė, V. Blue and Yellow Laccases from Alternaria sp. Strain HU: Characterization and Immobilization on Magnetic Nanoparticles. J. Fungi 2024, 10, 559. https://doi.org/10.3390/jof10080559
Radveikienė I, Vidžiūnaitė R, Meškys R, Časaitė V. Blue and Yellow Laccases from Alternaria sp. Strain HU: Characterization and Immobilization on Magnetic Nanoparticles. Journal of Fungi. 2024; 10(8):559. https://doi.org/10.3390/jof10080559
Chicago/Turabian StyleRadveikienė, Ingrida, Regina Vidžiūnaitė, Rolandas Meškys, and Vida Časaitė. 2024. "Blue and Yellow Laccases from Alternaria sp. Strain HU: Characterization and Immobilization on Magnetic Nanoparticles" Journal of Fungi 10, no. 8: 559. https://doi.org/10.3390/jof10080559






