Assessment of Genetic Diversity and the Population Structure of Species from the Fusarium fujikuroi Species Complex Causing Fusarium Stalk Rot of Maize
Abstract
1. Introduction
2. Materials and Methods
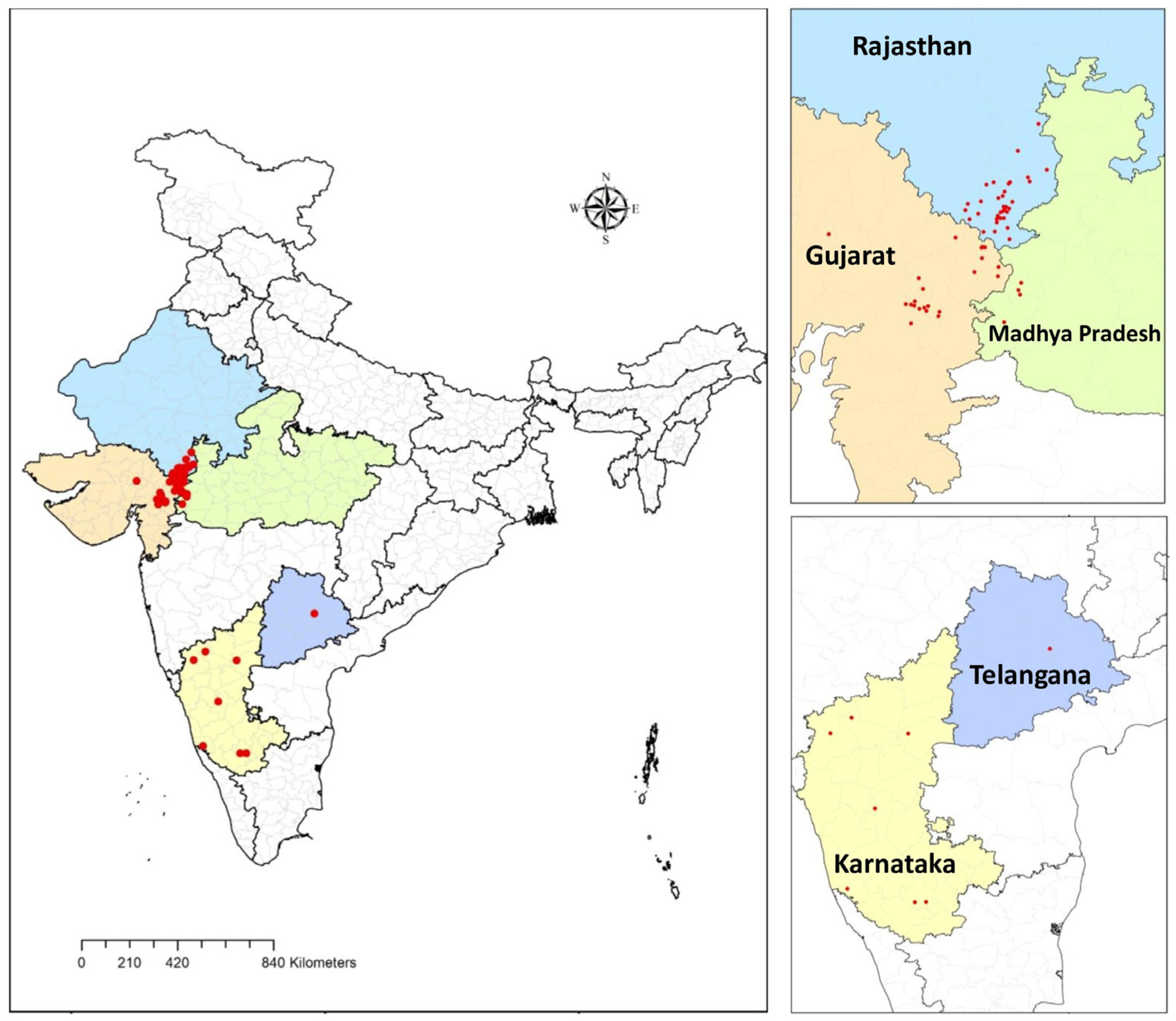
2.1. Study Area and Fungal Isolates Sampling
2.2. Fungal Isolation
2.3. DNA Isolation, PCR Amplification and Sequencing
2.4. Phylogenetic Analysis
2.5. DnaSP Analysis
2.6. Haplotype Network Analysis
2.7. AMOVA Analysis
2.8. Statistical Analysis
3. Results
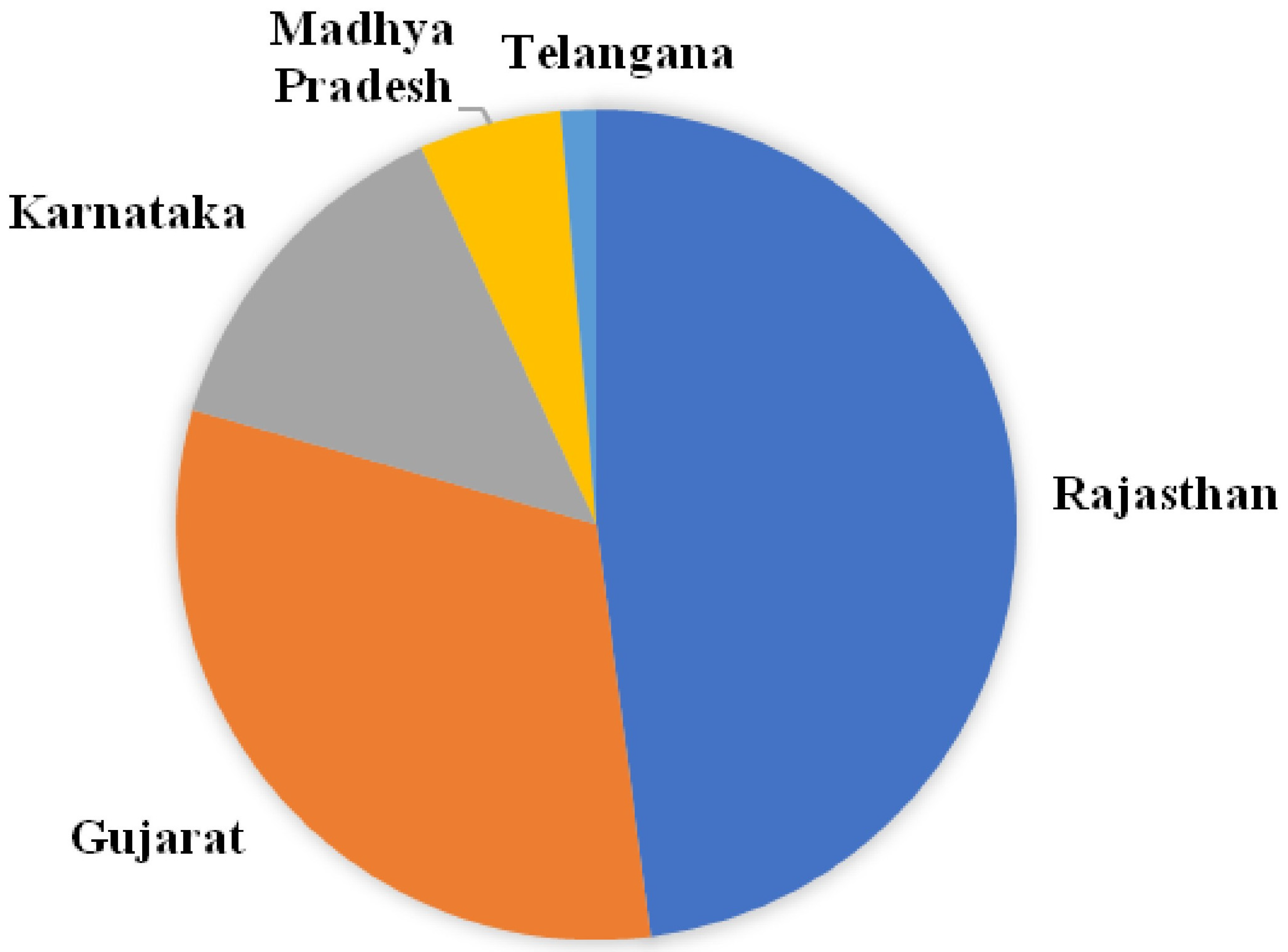
3.1. Collection of Isolates
3.2. Molecular Identification
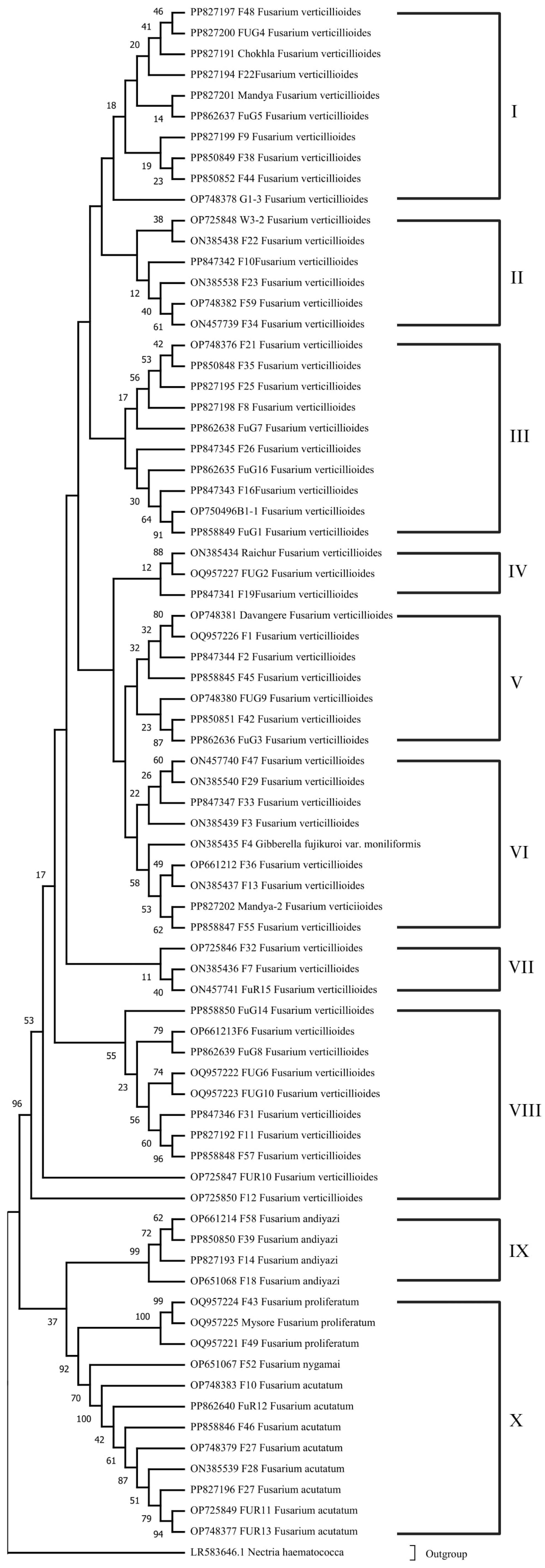
3.3. Phylogenetic Analysis and Evolutionary Relationship of Fusarium Strains Causing FSR
3.4. DNA Polymorphism and Genetic Differentiation Analysis
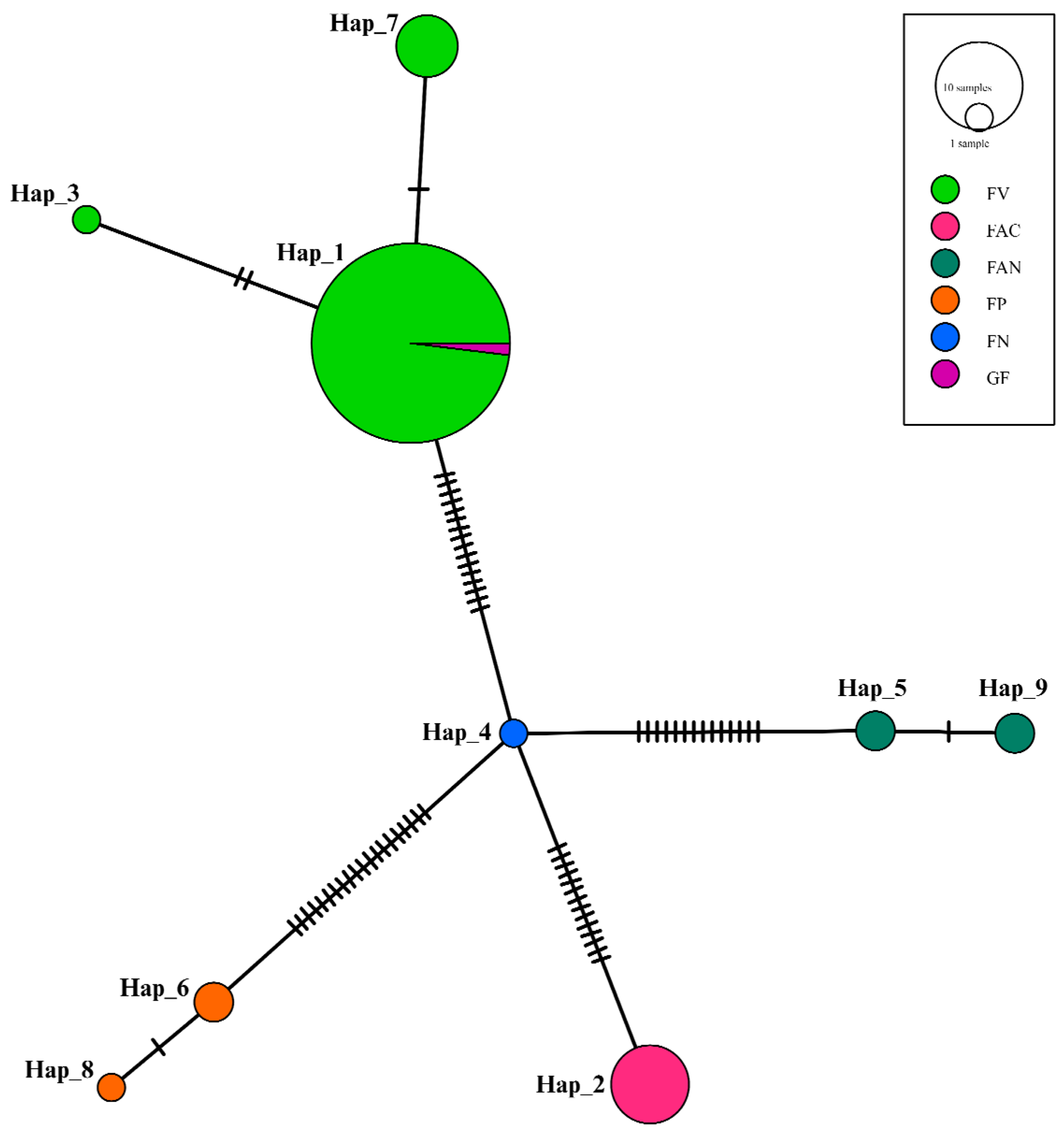
3.5. Haplotype Network Analysis Results
3.6. AMOVA Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munkvold, G.P.; Desjardins, A.E. Fumonisins in Maize: Can We Reduce Their Occurrence? Plant Dis. 1997, 81, 556–565. [Google Scholar] [CrossRef] [PubMed]
- Román, S.G. Caracterización de Genotipos de Maíz (Zea mays L.) a la Infección de Fusarium verticillioides en Diferentes Fases del Ciclo de Vida de la Planta y Su Correlación con Marcadores Moleculares de Tipo SNPs; Instituto Politécnico Nacional: Guasave, Mexico, 2017. [Google Scholar]
- Navale, V.D.; Sawant, A.M.; Vamkudoth, K.R. Genetic Diversity of Toxigenic Fusarium verticillioides Associated with Maize Grains, India. Genet. Mol. Biol. 2023, 46, e20220073. [Google Scholar] [CrossRef] [PubMed]
- Görtz, A.; Oerke, E.-C.; Steiner, U.; Waalwijk, C.; Vries, I.; Dehne, H.-W. Biodiversity of Fusarium Species Causing Ear Rot of Maize in Germany. Cereal Res. Commun. 2008, 36, 617–622. [Google Scholar] [CrossRef]
- Gai, X.; Dong, H.; Wang, S.; Liu, B.; Zhang, Z.; Li, X.; Gao, Z. Infection Cycle of Maize Stalk Rot and Ear Rot Caused by Fusarium Verticillioides. PLoS ONE 2018, 13, e0201588. [Google Scholar] [CrossRef] [PubMed]
- Harish, J.; Jambhulkar, P.P.; Bajpai, R.; Arya, M.; Babele, P.K.; Chaturvedi, S.K.; Kumar, A.; Lakshman, D.K. Morphological Characterization, Pathogenicity Screening, and Molecular Identification of Fusarium spp. Isolates Causing Post-Flowering Stalk Rot in Maize. Front. Microbiol. 2023, 14, 1121781. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Gardiner, D.M.; Manners, J.M. On the Trail of a Cereal Killer: Recent Advances in Fusarium graminearum Pathogenomics and Host Resistance. Mol. Plant Pathol. 2012, 13, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Saravanakumar, K.; Li, Y.; Yu, C.; Wang, Q.; Wang, M.; Sun, J.; Gao, J.; Chen, J. Effect of Trichoderma Harzianum on Maize Rhizosphere Microbiome and Biocontrol of Fusarium Stalk Rot. Sci. Rep. 2017, 7, 1771. [Google Scholar] [CrossRef] [PubMed]
- Douriet-Angulo, A.; López-Orona, C.A.; López-Urquídez, G.A.; Vega-Gutiérrez, T.A.; Tirado-Ramírez, M.A.; Estrada-Acosta, M.D.; Ayala-Tafoya, F.; Yáñez-Juárez, M.G. Maize Stalk Rot Caused by Fusarium Falciforme (FSSC 3 + 4) in Mexico. Plant Dis. 2019, 103, 2951. [Google Scholar] [CrossRef]
- Shin, J.-H.; Han, J.-H.; Lee, J.K.; Kim, K.S. Characterization of the Maize Stalk Rot Pathogens Fusarium Subglutinans and F. Temperatum and the Effect of Fungicides on Their Mycelial Growth and Colony Formation. Plant Pathol. J. 2014, 30, 397. [Google Scholar] [CrossRef]
- Leyva-Madrigal, K.Y.; Larralde-Corona, C.P.; Apodaca-Sánchez, M.A.; Quiroz-Figueroa, F.R.; Mexia-Bolaños, P.A.; Portillo-Valenzuela, S.; Ordaz-Ochoa, J.; Maldonado-Mendoza, I.E. Fusarium Species from the Fusarium fujikuroi Species Complex Involved in Mixed Infections of Maize in Northern Sinaloa, Mexico. J. Phytopathol. 2015, 163, 486–497. [Google Scholar] [CrossRef]
- Aguín, O.; Cao, A.; Pintos, C.; Santiago, R.; Mansilla, P.; Butrón, A. Occurrence of Fusarium Species in Maize Kernels Grown in Northwestern Spain. Plant Pathol. 2014, 63, 946–951. [Google Scholar] [CrossRef]
- Stumpf, R.; dos Santos, J.; Gomes, L.B.; Silva, C.N.; Tessmann, D.J.; Ferreira, F.D.; Machinski Junior, M.; Del Ponte, E.M. Fusarium Species and Fumonisins Associated with Maize Kernels Produced in Rio Grande Do Sul State for the 2008/09 and 2009/10 Growing Seasons. Braz. J. Microbiol. 2013, 44, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Mueller, D.S.; Wise, K.A.; Sisson, A.J.; Allen, T.W.; Bergstrom, G.C.; Bosley, D.B.; Bradley, C.A.; Broders, K.D.; Byamukama, E.; Chilvers, M.I.; et al. Corn Yield Loss Estimates Due to Diseases in the United States and Ontario, Canada from 2012 to 2015. Plant Health Prog. 2016, 17, 211–222. [Google Scholar] [CrossRef]
- Ortiz, C.S.; Richards, C.; Terry, A.; Parra, J.; Shim, W.-B. Genetic Variability and Geographical Distribution of Mycotoxigenic Fusarium verticillioides Strains Isolated from Maize Fields in Texas. Plant Pathol. J. 2015, 31, 203. [Google Scholar] [CrossRef]
- Sandoval-Denis, M.; Crous, P.W. Removing Chaos from Confusion: Assigning Names to Common Human and Animal Pathogens in Neocosmospora. Persoonia-Mol. Phylogeny Evol. Fungi 2018, 41, 109–129. [Google Scholar] [CrossRef]
- Al-Hatmi, A.; De Hoog, G.S.; Meis, J.F. Multiresistant Fusarium Pathogens on Plants and Humans: Solutions in (from) the Antifungal Pipeline? Infect. Drug Resist. 2019, 12, 3727–3737. [Google Scholar] [CrossRef]
- Wollenweber, H.W.; Sherbakoff, C.D.; Reinking, O.A.; Johann, H.; Bailey, A.A. Fundamentals for Taxonomic Studies of Fusarium. J. Agric. Res. 1925, 30, 833–843. [Google Scholar]
- O’Donnell, K.; Cigelnik, E.; Nirenberg, H.I. Molecular Systematics and Phylogeography of the Gibberella fujikuroi Species Complex. Mycologia 1998, 90, 465–493. [Google Scholar] [CrossRef]
- Geiser, D.M.; Lewis Ivey, M.L.; Hakiza, G.; Juba, J.H.; Miller, S.A. Gibberella xylarioides (Anamorph: Fusarium xylarioides), a Causative Agent of Coffee Wilt Disease in Africa, Is a Previously Unrecognized Member of the G. fujikuroi Species Complex. Mycologia 2005, 97, 191–201. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Tacke, B.K.; Casper, H.H. Gene Genealogies Reveal Global Phylogeographic Structure and Reproductive Isolation among Lineages of Fusarium graminearum, the Fungus Causing Wheat Scab. Proc. Natl. Acad. Sci. USA 2000, 97, 7905–7910. [Google Scholar] [CrossRef] [PubMed]
- Sandoval-Denis, M.; Guarnaccia, V.; Polizzi, G.; Crous, P.W. Symptomatic Citrus Trees Reveal a New Pathogenic Lineage in Fusarium and Two New Neocosmospora Species. Persoonia-Mol. Phylogeny Evol. Fungi 2018, 40, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, N.; Sandoval-Denis, M.; Lombard, L.; Visagie, C.M.; Wingfield, B.D.; Crous, P.W. Redefining Species Limits in the Fusarium fujikuroi Species Complex. Persoonia-Mol. Phylogeny Evol. Fungi 2021, 46, 129–162. [Google Scholar] [CrossRef] [PubMed]
- Jambhulkar, P.P.; Raja, M.; Singh, B.; Katoch, S.; Kumar, S.; Sharma, P. Potential Native Trichoderma Strains against Fusarium verticillioides Causing Post Flowering Stalk Rot in Winter Maize. Crop Prot. 2022, 152, 105838. [Google Scholar] [CrossRef]
- Harish, J.; Venkateshbabu, G.; Prasannakumar, M.K.; Devanna, P.; Mahesh, H.B.; Balasundara, D.C.; Swamy, S.D.; Kunjeti, S.G.; Manjunatha, C.; Puneeth, M.E.; et al. Stalk Rot Species Diversity and Molecular Phylogeny Associated with Diseased Maize in India. World J. Microbiol. Biotechnol. 2024, 40, 185. [Google Scholar] [CrossRef] [PubMed]
- Munkvold, G.P. Epidemiology of Fusarium Diseases and Their Mycotoxins in Maize Ears. Eur. J. Plant Pathol. 2003, 109, 705–713. [Google Scholar] [CrossRef]
- Zhang, J.; Abdelraheem, A.; Zhu, Y.; Elkins-Arce, H.; Dever, J.; Whitelock, D.; Hake, K.; Wedegaertner, T.; Wheeler, T.A. Studies of Evaluation Methods for Resistance to Fusarium Wilt Race 4 (Fusarium oxysporum f. sp. vasinfectum) in Cotton: Effects of Cultivar, Planting Date, and Inoculum Density on Disease Progression. Front. Plant Sci. 2022, 13, 900131. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Rivera, M.G.; Rodríguez-Guerra, R.; Guerrero-Aguilar, B.Z.; González-Chavira, M.M.; Pons-Hernández, J.L.; Jiménez-Bremont, J.F.; Ramírez-Pimentel, J.G.; Andrio-Enríquez, E.; Mendoza-Elos, M. Caracterización de Especies de Fusarium Asociadas a La Pudrición de Raíz de Maíz En Guanajuato, México. Rev. Mex. Fitopatol. 2010, 28, 124–134. [Google Scholar]
- Ha, M.S.; Ryu, H.; Ju, H.J.; Choi, H.-W. Diversity and Pathogenic Characteristics of the Fusarium Species Isolated from Minor Legumes in Korea. Sci. Rep. 2023, 13, 22516. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Sutton, D.A.; Rinaldi, M.G.; Sarver, B.A.J.; Balajee, S.A.; Schroers, H.-J.; Summerbell, R.C.; Robert, V.A.R.G.; Crous, P.W.; Zhang, N.; et al. Internet-Accessible DNA Sequence Database for Identifying Fusaria from Human and Animal Infections. J. Clin. Microbiol. 2010, 48, 3708–3718. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple Evolutionary Origins of the Fungus Causing Panama Disease of Banana: Concordant Evidence from Nuclear and Mitochondrial Gene Genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- Ramdial, H.; Latchoo, R.K.; Hosein, F.N.; Rampersad, S.N. Phylogeny and Haplotype Analysis of Fungi within the Fusarium incarnatum-Equiseti Species Complex. Phytopathology 2017, 107, 109–120. [Google Scholar] [CrossRef]
- Ploetz, R.C. Fusarium Wilt of Banana Is Caused by Several Pathogens Referred to as Fusarium oxysporum f. sp. cubense. Phytopathology 2006, 96, 653–656. [Google Scholar] [CrossRef] [PubMed]
- Dobbs, J.T.; Kim, M.; Dudley, N.S.; Jones, T.C.; Yeh, A.; Dumroese, R.K.; Cannon, P.G.; Hauff, R.D.; Klopfenstein, N.B.; Wright, S.; et al. Fusarium spp. Diversity Associated with Symptomatic Acacia koa in Hawaiʽi. For. Pathol. 2021, 51, e12713. [Google Scholar] [CrossRef]
- Leslie, J.F.; Summerell, B.A. Fusarium Laboratory Workshops—A Recent History. Mycotoxin Res. 2006, 22, 73–74. [Google Scholar] [CrossRef] [PubMed]
- Liddell, C.M.; Burgess, L.W. Survival of Fusarium Moniliforme at Controlled Temperature and Relative Humidity. Trans. Br. Mycol. Soc. 1985, 84, 121–130. [Google Scholar] [CrossRef]
- Schenk, J.J.; Becklund, L.E.; Carey, S.J.; Fabre, P.P. What Is the “Modified” CTAB Protocol? Characterizing Modifications to the CTAB DNA Extraction Protocol. Appl. Plant Sci. 2023, 11, e11517. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit v7. 0.9: Biological Sequence Alignment Editor and Analysis Program for Win95/98/2K/XP/7. 2012. Available online: http://www.mbio.ncsu.edu/bioedit/bioedit.html (accessed on 8 July 2024).
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Macclade; Sinauer: Sunderland, MA, USA, 1992. [Google Scholar]
- Leigh, J.W.; Bryant, D.; Nakagawa, S. POPART: Full-Feature Software for Haplotype Network Construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Hudson, R.R. A New Statistic for Detecting Genetic Differentiation. Genetics 2000, 155, 2011–2014. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A.; Li, L.M.; Ward, R.; Pritchard, J.K. Informativeness of Genetic Markers for Inference of Ancestry. Am. J. Hum. Genet. 2003, 73, 1402–1422. [Google Scholar] [CrossRef] [PubMed]
- Davies, T.J.; Pedersen, A.B. Phylogeny and Geography Predict Pathogen Community Similarity in Wild Primates and Humans. Proc. R. Soc. B. 2008, 275, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.L. Divergence with Genetic Exchange; OUP Oxford: Oxford, UK, 2015. [Google Scholar]
- Richardson, E.J.; Bacigalupe, R.; Harrison, E.M.; Weinert, L.A.; Lycett, S.; Vrieling, M.; Robb, K.; Hoskisson, P.A.; Holden, M.T.; Feil, E.J. Gene Exchange Drives the Ecological Success of a Multi-Host Bacterial Pathogen. Nat. Ecol. Evol. 2018, 2, 1468–1478. [Google Scholar] [CrossRef] [PubMed]
- Wiedenbeck, J.; Cohan, F.M. Origins of Bacterial Diversity through Horizontal Genetic Transfer and Adaptation to New Ecological Niches. FEMS Microbiol. Rev. 2011, 35, 957–976. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A. The Shapes of Neutral Gene Genealogies in Two Species: Probabilities of Monophyly, Paraphyly, and Polyphyly in a Coalescent Model. Evolution 2003, 57, 1465–1477. [Google Scholar] [PubMed]
- Siepel, A.; Pollard, K.S.; Haussler, D. New Methods for Detecting Lineage-Specific Selection. In Research in Computational Molecular Biology; Apostolico, A., Guerra, C., Istrail, S., Pevzner, P.A., Waterman, M., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2006; Volume 3909, pp. 190–205. ISBN 978-3-540-33295-4. [Google Scholar]
- Singh-Babak, S.D.; Babak, T.; Fraser, H.B.; Johnson, A.D. Lineage-Specific Selection and the Evolution of Virulence in the Candida Clade. Proc. Natl. Acad. Sci. USA 2021, 118, e2016818118. [Google Scholar] [CrossRef] [PubMed]
- Amos, W.; Harwood, J. Factors Affecting Levels of Genetic Diversity in Natural Populations. Philos. Trans. R. Soc. Lond. B 1998, 353, 177–186. [Google Scholar] [CrossRef]
- Charlesworth, B.; Jensen, J.D. Effects of Selection at Linked Sites on Patterns of Genetic Variability. Annu. Rev. Ecol. Evol. Syst. 2021, 52, 177–197. [Google Scholar] [CrossRef]
- Lynch, M.; Ackerman, M.S.; Gout, J.-F.; Long, H.; Sung, W.; Thomas, W.K.; Foster, P.L. Genetic Drift, Selection and the Evolution of the Mutation Rate. Nat. Rev. Genet. 2016, 17, 704–714. [Google Scholar] [CrossRef]
- Hemstrom, W.; Dauwalter, D.; Peacock, M.M.; Leasure, D.; Wenger, S.; Miller, M.R.; Neville, H. Population Genomic Monitoring Provides Insight into Conservation Status but No Correlation with Demographic Estimates of Extinction Risk in a Threatened Trout. Evol. Appl. 2022, 15, 1449–1468. [Google Scholar] [CrossRef] [PubMed]
- Mondal, D.; Dutta, S.; Mallik, A.; Mandal, N. Mitochondrial DNA Diversity: Insight into Population Diversity, Structure and Demographic History of Penaeus monodon along the Entire Coastal Region of India. Aquat. Res. 2020, 51, 4649–4680. [Google Scholar] [CrossRef]
- Labbe, F.; Fontaine, M.C.; Robin, C.; Dutech, C. Genetic Signatures of Variation in Population Size in a Native Fungal Pathogen after the Recent Massive Plantation of Its Host Tree. Heredity 2017, 119, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Rivas, G.; Zapater, M.; Abadie, C.; Carlier, J. Founder Effects and Stochastic Dispersal at the Continental Scale of the Fungal Pathogen of Bananas Mycosphaerella fijiensis. Mol. Ecol. 2004, 13, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Croll, D.; Lendenmann, M.H.; Stewart, E.; McDonald, B.A. The Impact of Recombination Hotspots on Genome Evolution of a Fungal Plant Pathogen. Genetics 2015, 201, 1213–1228. [Google Scholar] [CrossRef] [PubMed]
- Drenth, A.; McTaggart, A.R.; Wingfield, B.D. Fungal Clones Win the Battle, but Recombination Wins the War. IMA Fungus 2019, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Stalder, L.; Oggenfuss, U.; Mohd-Assaad, N.; Croll, D. The Population Genetics of Adaptation through Copy Number Variation in a Fungal Plant Pathogen. Mol. Ecol. 2023, 32, 2443–2460. [Google Scholar] [CrossRef]
- Bömke, C.; Rojas, M.C.; Hedden, P.; Tudzynski, B. Loss of Gibberellin Production in Fusarium verticillioides (Gibberella fujikuroi MP-A) Is Due to a Deletion in the Gibberellic Acid Gene Cluster. Appl. Environ. Microbiol. 2008, 74, 7790–7801. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barrès, B.; Halkett, F.; Dutech, C.; Andrieux, A.; Pinon, J.; Frey, P. Genetic Structure of the Poplar Rust Fungus Melampsora Larici-Populina: Evidence for Isolation by Distance in Europe and Recent Founder Effects Overseas. Infect. Genet. Evol. 2008, 8, 577–587. [Google Scholar] [CrossRef]
- Ene, I.V.; Farrer, R.A.; Hirakawa, M.P.; Agwamba, K.; Cuomo, C.A.; Bennett, R.J. Global Analysis of Mutations Driving Microevolution of a Heterozygous Diploid Fungal Pathogen. Proc. Natl. Acad. Sci. USA 2018, 115, E8688–E8697. [Google Scholar] [CrossRef]
- Giraud, T. Patterns of within Population Dispersal and Mating of the Fungus Microbotryum Violaceum Parasitising the Plant Silene Latifolia. Heredity 2004, 93, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Laine, A. Resistance Variation within and among Host Populations in a Plant–Pathogen Metapopulation: Implications for Regional Pathogen Dynamics. J. Ecol. 2004, 92, 990–1000. [Google Scholar] [CrossRef]
- Wright, S. Genic and Organismic Selection. Evolution 1980, 34, 825–843. [Google Scholar] [CrossRef] [PubMed]
- Gladieux, P.; Feurtey, A.; Hood, M.E.; Snirc, A.; Clavel, J.; Dutech, C.; Roy, M.; Giraud, T. The Population Biology Of Fungal Invasions. In Invasion Genetics; Barrett, S.C.H., Colautti, R.I., Dlugosch, K.M., Rieseberg, L.H., Eds.; Wiley: Hoboken, NJ, USA, 2016; pp. 81–100. ISBN 978-1-118-92216-3. [Google Scholar]
- Sletvold, N.; Grindeland, J.M.; Zu, P.; Ågren, J. Strong Inbreeding Depression and Local Outbreeding Depression in the Rewarding Orchid Gymnadenia Conopsea. Conserv. Genet. 2012, 13, 1305–1315. [Google Scholar] [CrossRef]



| Analysis Type | Number of Sites Analyzed | Number of Polymorphic Sites, S | Average Number of Differences | Nucleotide Diversity, Pi | Theta-W, Per Sequence | Theta-W, Per Site |
|---|---|---|---|---|---|---|
| Pairwise Comparisons | 509.38 (average) | N/A | 12.544 | 0.02471 | N/A | N/A |
| Individual Sites (column by column) | 628.00 | 109 | 17.202 | 0.02739 | 24.10441 | 0.03838 |
| Population | Number of Sequences | Number of Segregating Sites, S | Number of Haplotypes, h | Haplotype Diversity, Hd | Average Number of Differences, K | Nucleotide Diversity, Pi | Nucleotide Diversity with JC, PiJC |
|---|---|---|---|---|---|---|---|
| FV | 57 | 3 | 3 | 0.19486 | 0.23308 | 0.00064 | 0.00064 |
| FAC | 8 | 0 | 1 | 0.0 | 0.0 | 0.0 | 0.0 |
| FAN | 4 | 1 | 2 | 0.66667 | 0.66667 | 0.00184 | 0.00184 |
| FP | 3 | 1 | 2 | 0.66667 | 0.66667 | 0.00184 | 0.00184 |
| Total Data Estimates | 72 | 44 | 8 | 0.48513 | 7.64358 | 0.02106 | N/A |
| Population 1 | Population 2 | Hs | Ks | Kxy | Gst | DeltaSt | GammaSt | Nst | Fst | Dxy | Da |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FV | FAC | 0.17570 | 0.20440 | 19.12281 | 0.53712 | 0.01130 | 0.95334 | 0.99411 | 0.99391 | 0.05268 | 0.05236 |
| FV | FAN | 0.21142 | 0.26151 | 19.62281 | 0.26440 | 0.00650 | 0.90534 | 0.97787 | 0.97707 | 0.05406 | 0.05282 |
| FV | FP | 0.20329 | 0.25476 | 26.45029 | 0.24162 | 0.00683 | 0.91187 | 0.98380 | 0.98299 | 0.07287 | 0.07163 |
| FAC | FAN | 0.16667 | 0.22222 | 17.50000 | 0.57333 | 0.02112 | 0.97872 | 0.98154 | 0.98095 | 0.04821 | 0.04729 |
| FAC | FP | 0.09524 | 0.18182 | 25.33333 | 0.59648 | 0.02744 | 0.98798 | 0.98744 | 0.98684 | 0.06979 | 0.06887 |
| FAN | FP | 0.66667 | 0.66667 | 22.83333 | 0.19864 | 0.03017 | 0.95833 | 0.97199 | 0.97080 | 0.06290 | 0.06107 |
| Source of Variation | Sum of Squares | Variance Components | Percentage Variation |
|---|---|---|---|
| Among populations | 2170.96 | 21.51 | 10.37 |
| Within populations | 26,389.34 | 185.85 | 89.63 |
| Total | 28,560.29 | 207.35 | 100.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jambhulkar, P.P.; Bajpai, R.; Reddy, H.J.; Tripathy, P.S.; Varun, P.; Rout, A.K.; Behera, B.K.; Lakshman, D.K.; Nanjundappa, M. Assessment of Genetic Diversity and the Population Structure of Species from the Fusarium fujikuroi Species Complex Causing Fusarium Stalk Rot of Maize. J. Fungi 2024, 10, 574. https://doi.org/10.3390/jof10080574
Jambhulkar PP, Bajpai R, Reddy HJ, Tripathy PS, Varun P, Rout AK, Behera BK, Lakshman DK, Nanjundappa M. Assessment of Genetic Diversity and the Population Structure of Species from the Fusarium fujikuroi Species Complex Causing Fusarium Stalk Rot of Maize. Journal of Fungi. 2024; 10(8):574. https://doi.org/10.3390/jof10080574
Chicago/Turabian StyleJambhulkar, Prashant P., Ruchira Bajpai, Harish Jayarama Reddy, Partha Sarathi Tripathy, Priyanka Varun, Ajaya Kumar Rout, Bijay Kumar Behera, Dilip K. Lakshman, and Mallikarjuna Nanjundappa. 2024. "Assessment of Genetic Diversity and the Population Structure of Species from the Fusarium fujikuroi Species Complex Causing Fusarium Stalk Rot of Maize" Journal of Fungi 10, no. 8: 574. https://doi.org/10.3390/jof10080574
APA StyleJambhulkar, P. P., Bajpai, R., Reddy, H. J., Tripathy, P. S., Varun, P., Rout, A. K., Behera, B. K., Lakshman, D. K., & Nanjundappa, M. (2024). Assessment of Genetic Diversity and the Population Structure of Species from the Fusarium fujikuroi Species Complex Causing Fusarium Stalk Rot of Maize. Journal of Fungi, 10(8), 574. https://doi.org/10.3390/jof10080574








