Abstract
Marine ecosystems are important in discovering novel fungi with interesting metabolites that have shown great potential in pharmaceutical and biotechnological industries. Seagrasses, the sole submerged marine angiosperm, host diverse fungal taxa with mostly unknown metabolic capabilities. They are considered to be one of the least studied marine fungal habitats in the world. This review gathers and analyzes data from studies related to seagrasses-associated fungi, including taxonomy and biogeography, and highlights existing research gaps. The significance of the seagrass–fungal associations remains largely unknown, and current understanding of fungal diversity is limited to specific geographical regions such as the Tropical Atlantic, Mediterranean, and Indo-Pacific. Our survey yielded 29 culture-dependent studies on seagrass-associated endophytic and epiphytic fungi, and 13 miscellaneous studies, as well as 11 meta-studies, with no pathogenic true fungi described. There is a significant opportunity to expand existing studies and conduct multidisciplinary research into novel species and their potential applications, especially from understudied geographical locations. Future research should prioritize high-throughput sequencing and mycobiome studies, utilizing both culture-dependent and -independent approaches to effectively identify novel seagrass-associated fungal taxa.
1. Introduction
Fungi are a diverse group of organisms inhabiting a wide range of environments, playing significant functions in challenging ecosystems [1,2]. They can range from unicellular to filamentous taxa and exhibit various life modes, such as pathogens, saprobes, endophytes, and epiphytes [3]. All these different life modes of fungi contribute to the overall functioning of an ecosystem, though some may not always be beneficial, such as phytopathogenic fungi [4] and clinically important fungi [5]. Approximately, 156,000 species have been scientifically documented in Species Fungorum 2024 [6]. The global richness of fungi has been a popular topic among mycologists with diverse predictions made over the past three decades using various approaches [7,8,9,10,11,12]. Hawksworth and Lücking [10] estimated there to be between 2.2 to 3.8 million fungal species, but the most recent estimate by Niskanen et al. [11] predicts 2 to 3 million species to be more realistic, with over 90% of these species yet to be revealed. Marine fungi have been identified as a potential group for bridging gaps in missing species [11,13] and for discovering novel biological substances [14].
Microorganisms are considered ubiquitous in marine ecosystems, despite the environmental limitations posed by high salinity, low temperature, low water availability, and oligotrophic conditions [15]. Microorganisms represent 90% of the total oceanic biomass [16], making marine ecosystems a potential habitat for discovering novel microorganisms. This largely understudied habitat is rich in microorganisms producing bioactive natural products [17]. Marine fungi are one of the important groups of fungi that mycologists have continuously studied for over 50 years [18,19]. According to www.marinefungi.org (accession on 14 July 2024), 2041 species have been reported as marine fungi [19]. According to Calabon et al. [20] and Devadatha et al. [21], the majority of these taxa have been documented from mangroves and salt marshes. Additionally, substrates such as algae, silt, driftwood, and seagrasses have been researched globally [22]. While over 10,000 fungal species from different marine habitats have been predicted, less than 20% of them have been described [19]. Wijayawardene et al. [23] emphasized the need for investigating marine fungi from various microhabitats in understudied geographical regions and highlighted the role of multidisciplinary sciences in discovering novel fungi and understanding their potential uses.
For over 150 years, it has been known that plants and microorganisms have intimate relationships that affect overall plant fitness, growth, and survival [24]. Consequently, studying plant–microbe associations to uncover the elements of their interactions has become a topic of interest in modern times. As a component of the marine ecosystem, seagrasses host a diverse community of microorganisms, including bacteria, archaea, fungi, microalgae, and viruses [16]. These microbes have a fundamental impact on the physiology and well-being of seagrasses while playing a major role in controlling biogeochemical processes within entire seagrass meadows [16].
During our ongoing study on seagrass-associated fungi in the Puttalam lagoon, Sri Lanka, we isolated over 40 different fungal species from two seagrass species, namely, Enhalus acaroides (Linnaeus f.) Royle, 1839 and Oceana serrulata (R.Brown) Byng & Christenh. Concurrently, our literature survey revealed that a considerable number of studies on seagrass fungi (including fungus-like taxa) have been carried out, but the data (taxonomy, classification, applications, biogeography, and ecological data) are scattered. This review and opinion paper aims to compile the available data on seagrass-associated fungi (focusing on taxonomy, biogeography, and industrial applications) with the collaboration of fungal taxonomists, marine ecologists, and industry experts. Furthermore, we intend to highlight research gaps and emphasize the need for more research on seagrass-associated fungi to expand future studies in this field.
2. Marine Fungi
‘Marine fungi’ are defined in multiple ways in the literature. One of the earliest definitions is based on their requirement to grow in saline water [25]. The most quoted definition provided by Kohlmeyer and Kohlmeyer [26] restricts marine fungi to two ecological groups: obligate and facultative marine fungi. Obligate marine fungi exclusively grow and sporulate in marine environments, while facultative species sporulate in marine environments but can grow in freshwater or terrestrial environments [26,27]. Later, a broader definition was provided by Pang et al. [28], where marine fungi were defined as fungi that are repeatedly recovered from marine habitats and either: (1) grow and/or sporulate on substrates in marine environments; (2) form symbiotic relationships with other marine organisms; or (3) adapt and evolve at the genetic level or are metabolically active in marine environments. Recently, Pasqualetti et al. [15] defined fungi obtained from marine environments as “marine-derived” as they are isolated from marine sources. This definition includes obligates, facultatives, and fungi arising from dormant terrestrial or freshwater propagules. To address the confusion regarding the applicability of these terms, a revision of the taxonomy and a deeper understanding of the metabolomic basis of marine fungal life are necessary.
In general, marine fungi have been recovered from different habitats, including mangrove plants, macroalgae, coral reefs, drift and submerged wood, sponges, sea ice, sea garbage, seagrasses, deep-sea and benthic sediments, hydrothermal vents, oxygen-deficient environments, and the water column [29,30,31]. To provide a structured reference for marine fungal habitats, Wijayawardene et al. [23] defined these habitats as ‘coastal terrestrial ecosystems’, ‘semi-coastal marine ecosystems’, ‘coastal marine ecosystems’, and ‘deep marine ecosystems’. Despite this categorization, fungal associations in many marine habitats, especially seagrasses, remain understudied, which is the primary focus of this review.
3. Seagrasses
Seagrasses are flowering plants (angiosperms) found in shallow marine waters [32] and are adapted for fully submerged conditions [33]. They can exist as monospecific or multispecific meadows, often extending over a larger surface area covering the seabed [34].
Despite their crucial role as foundation species in coastal ecosystems [35], seagrass populations are declining at an alarming rate. Studies indicate that the extent of seagrass beds has been declining by 110 km2 per year since 1980, with 29% of their total extent lost since seagrass regions were first recorded in 1879 [36]. Consequently, there is an urgent necessity for focused worldwide conservation initiatives to protect the existing seagrass meadows [37].
Seagrass Distribution and Diversity
Seagrasses are widely distributed along tropical and temperate shallow coastal waters spanning 159 countries on six continents, making them one of the most widespread coastal habitats on Earth [38,39]. They are often associated with important marine habitats such as corals, bivalve reefs, and mangroves in tropics and marshes, and kelp forests in temperate regions [40].
According to the estimates from the UNEP World Conservation Monitoring Centre (WCMC), the spatial distribution of seagrasses was approximated at 177,000 km2 in 2001 [41]. However, a recent study by McKenzie et al. [42] revised this estimate, indicating a global seagrass distribution of 160,387 km2, with a high to moderate level of confidence. Furthermore, UNEP [39] highlighted that the estimated global extent of seagrass ranges between 300,000 and 600,000 km2. The depth limits of seagrasses are estimated to range from the intertidal zone to 90 m below mean sea level [43].
Seagrasses are currently divided into six families: Zosteraceae, Hydrocharitaceae, Posidoniaceae, Cymodoceaceae, Ruppiaceae, and Zannichelliaceae [44]. These six families contain twelve genera and 72 species worldwide [44]. The extinction risk assessment of the world’s seagrass species reveals that three species are Endangered, seven Vulnerable, five Near Threatened, 48 are categorized as Least Concern, and nine species are Data-Deficient [44]. According to Rasheed and Unsworth [45], nearly 10% of all coastal seabeds are covered with seagrass and they exhibit low taxonomic diversity.
Amongst these, the tropical Indo-Pacific represents one of the most highly diverse seagrass bioregions in the world, accounting for around 35% of the total species. However, this region also faces significant data scarcity, particularly for population spatial distribution data, accounting for around 24% of the data deficiency [38].
4. Seagrass-Associated Fungi
Compared to the extensive studies on seagrass ecology, less is known about the diversity and ecological roles played by the fungal communities associated with seagrasses [35]. The fungal community associated with land plants plays a crucial role in influencing plant health and survival. Similarly, in seagrass ecosystems, the fungal community can significantly impact overall seagrass functioning. Understanding these associations can be a valuable tool for future seagrass restoration and conservation projects [46,47].
4.1. Seagrass-Associated Endophytic Fungi
In this section, our focus is on seagrass endophytic fungi recovered through culture-dependent approaches. Fungi derived from culture-independent studies will be discussed in the section on seagrass mycobiome studies.
The term ‘endophyte’ (endon = within, phyte = plant) is used to define any organism found within living plant tissues. In mycology, this term often specifically refers to fungi that reside within (inside) the plant tissues for at least part of their lifecycle without causing apparent harm or disease symptoms to the host [48]. The association between endophytes and their host plants can range from “mutualistic to opportunistically pathogenic” in nature [49]. According to Schulz and Boyle [50], endophytes are considered as virulent pathogens when they produce enzymes that damage host tissues to aid in colonization. However, a clear distinction between a pathogenic and non-pathogenic nature is often challenging. Recently, Hardoim et al. [51] suggested that the term endophyte should include all microorganisms that colonize internal plant tissues for all or part of their lifetime. Nevertheless, the distinction between a pathogen and an endophyte becomes apparent when they engage with the plant’s defence system. A pathogen overcomes the plants’ defences, inflicting damage on the host, whereas endophytes overcome plant defences by masking themselves without causing apparent damage or symptoms.
In general, endophytic fungi are considered to enhance the hosts’ ability to tolerate environmental stresses, improve vigour, recycle nutrients, decrease susceptibility to pathogens and pests, and regulate the synthesis of phytohormones and metabolites [52,53,54,55]. In return, host plants provide them with organic nutrients, protection, and assurance of survival [56]. A recent study showed that two endophytic fungi, Trichoderma sp. and Diaporthe sp., isolated from a seagrass species, Thalassia testudinum, demonstrated significant bioactivity against pathogenic Labyrinthula infections, which have previously led to extensive seagrass die-offs in many parts of the world [57].
To the best of our knowledge, the first record of intercellular fungi in seagrass was reported by Kuo [58] in 1984, in the leaves of Zostera muelleri. Although they did not specifically use the term “endophytic”, the symbiotic and intercellular nature of fungal filaments was demonstrated using microscopy. The first direct evidence of endophytic fungi isolated from living, healthy seagrass tissues was reported by Wilson [59] through the screening of the leaf tissues of Thalassia testudinum, Halodule bennudensis, and Syringodium filiforme. Since then, numerous investigations on the isolation and identification of seagrass endophytic fungi have been published, and all culture-based studies as well as isolated taxa reported thus far are summarized in Table S1.
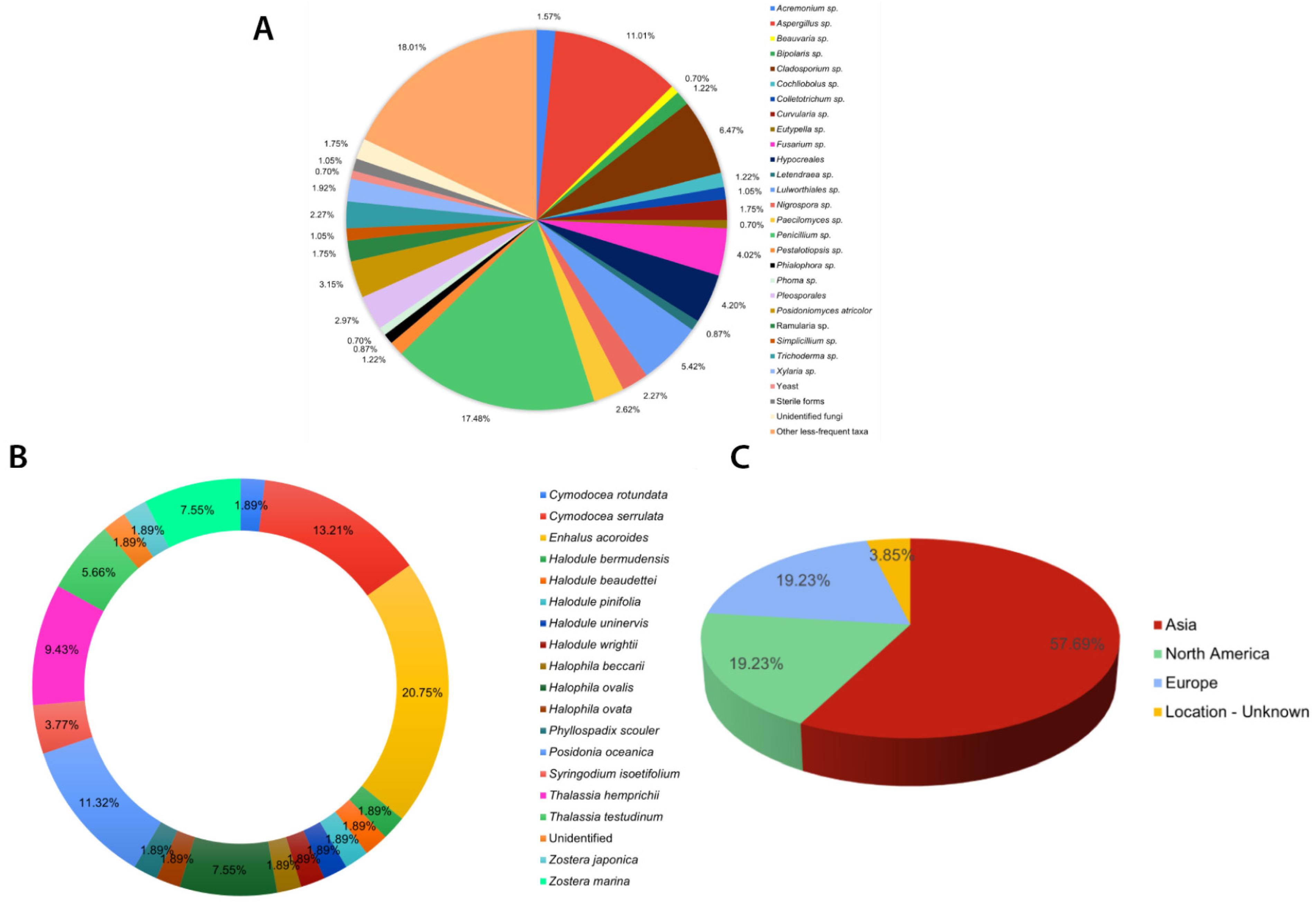
Eurotialean fungi, specifically Aspergillus and Penicillium, are reported as the dominant endophytic taxa in most culture-dependent studies [60,61,62,63,64] (see Table S1). These two genera, Penicillium (100/572) and Aspergillus (63/572), account for more than 25% of the listed taxa (Figure 1A). Nevertheless, in most of these publications, fungal identification is restricted only to the generic level without support from DNA sequencing. Therefore, the taxonomic placements may not be very accurate.
Figure 1.
Summary of culture-dependent seagrass endophytic fungal studies listed in Table S1: (A) composition of endophytic fungi from different seagrass studies; (B) percentage number of studies carried out for each seagrass species; (C) continent-wide distribution of sampling sites for seagrass fungal endophytes in the world.
Most of these dominant endophytic genera are reported to have a terrestrial origin [60,65,66,67]. While many studies report the absence of obligate marine fungi [66,67,68], some studies such as Cuomo et al. [69], Abdel-Wahab et al. [70], and Mata and Cebrián [61] mention the presence of obligate fungal species. According to Abdel-Wahab et al. [70], employing rigorous surface sterilization during endophytic isolations enhances the recovery of marine-derived fungal species, whereas a direct microscopic examination tends to identify obligate marine fungi. However, the validity of this statement is rather perplexing as the marine-derived, or obligatory, nature cannot be determined by morphological characters alone. In many studies, sterile forms are reported [61,63,66,67] and remain unidentified when morphological methods are used for identifications. In our ongoing study, over 15 endophytic fungi (out of a total of 40) isolated from two seagrass species, E. acaroides and O. serrulata, lacked any fruiting structures, necessitating molecular identification (Rajakaruna et al., unpublished). Gnavi et al. [71] highlight the need for molecular analyses to properly identify these sterile forms.
Seagrasses are reported to have a low diversity and density of fungal colonization compared to terrestrial plants [60,62,67,68]. However, Shoemaker and Wyllie-Echeverria [72] noted that the number of taxa isolated is “roughly similar” to that of land plants. While these comments appear contradictory, it is important to consider the diversity indices utilized by these authors, the climatic zones of the plants used for comparisons, and the total number of segments screened, before coming to a meaningful conclusion. Moreover, all the above predictions were based on culture-dependent techniques.
Nevertheless, it is stipulated that the low frequency of fungal colonization in seagrasses is attributed to multiple complex interactions between intrinsic and extrinsic factors. The endophytic fungal community in seagrasses is mainly influenced by intrinsic factors including tissue type (“district specificity”) [73], age [60,62], morphological characters [59,60], and the phytochemical composition of tissues [62,67]. The antifungal metabolites produced by seagrasses are reported to limit internal fungal colonization [74]. Additionally, other groups of microbes associated with seagrasses, which produce antifungal compounds, can further reduce endophytic colonization [75].
External factors such as the nutrient content in the water column, water temperature, wind and wave actions, seasonal variations [59,60,61], and other physicochemical factors at the sampling sites may affect the fungal colonization within seagrasses. A recent study by Solé et al. [76] showed that human-generated noise significantly impacts the degradation of fungal symbionts in Posidonia oceanica roots, subsequently affecting the normal root functions. Moreover, the absence of mycorrhizal associations in seagrasses is said to be associated with limitations posed by high salinity and oligotrophic conditions in the marine sediments [77]. Although seagrasses are adapted for nutrient uptake through leaves, having root–fungal associations (similar to mycorrhizal associations in land plants) could be beneficial for absorbing nutrients from recalcitrant material under oligotrophic conditions [78]. A summary of these symbiotic root–fungal associations is given in Section 4.1.1.
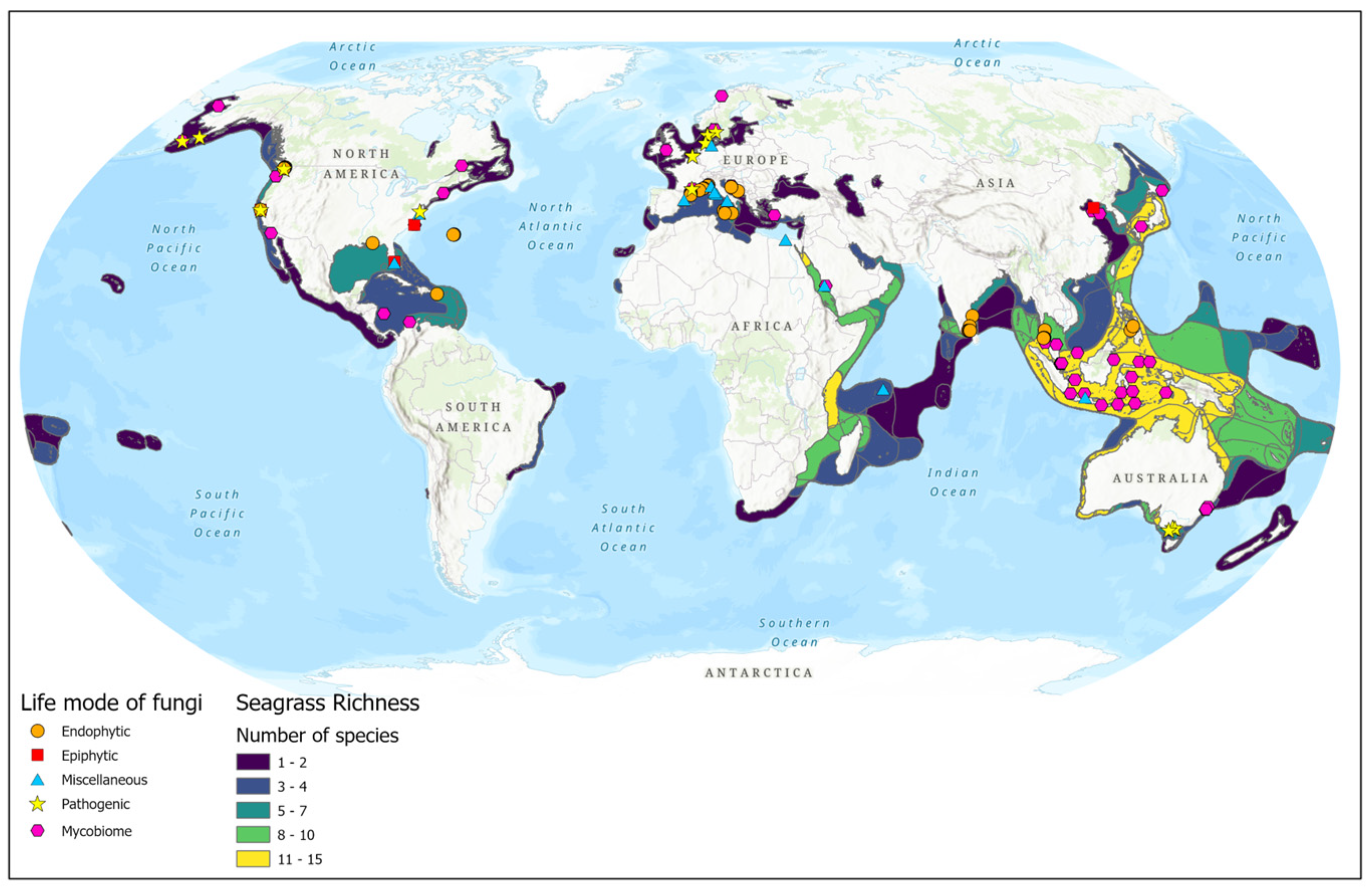
Most of the seagrass endophytic fungal research has been conducted in the Asian, European, and North American regions (Figure 2). More than half of the sampling sites are concentrated on the Asian continent (Figure 1C and Figure 2). The majority of studies are confined to a few seagrass species: Enhalus acaroides, Cymodocea serrulata, Posidonia oceanica, Thalassia hemprichii, and Zostera spp. (Figure 1B).
Figure 2.
Biogeographic distribution of seagrass fungal studies (based on the data from Tables S1–S4).
4.1.1. Root-Associated Endophytic Fungi
Some publications have explicitly investigated and uncovered interesting root–fungal relationships in seagrasses. Kuo et al. [79] reported the occurrence of fungi in the peripheral root tissues of two Australian seagrass species, Posidonia australis and P. sinuosa. To the best of our understanding, this is the first published investigation that specifically addresses the fungal inhabitants in seagrass roots.
Thereafter, for more than a decade, the topic remained unstudied. Nielsen et al. [77] attempted to find arbuscular mycorrhizal (AM) associations in the roots of Zostera marina and Thalassia testudinum. Since the majority of vascular plants contain these symbiotic associations, the authors aimed to address this gap in seagrass research. However, they were unable to observe AM associations in either seagrass species. The observed Zostera marina tissues showed unidentified fungal colonization that was not characteristic of AM. As they assumed, the lack of information on seagrass-associated root mycorrhizae in the 1900s is due to the inherent challenge of publishing negative results.
Torta et al. [80] conducted a study on root mycobiota of Posidonia oceanica and obtained a single species which they named Lulwoana sp. Without any supportive evidence, they claimed this species was a dark septate endophyte (DSE). Vohník and colleagues conducted numerous investigations to study fungi associated with seagrass roots but found no structures resembling mycorrhiza. However, in their study, Vohník et al. [81] described the presence of dark septate endophytes (DSE) with all typical structures, including extraradical and intraradical dark septate hyphae, dense melanized parenchymatous nets/hyphal sheaths on the root surface, and melanized intracellular microsclerotia for the first time in seagrasses. Subsequently, Vohník et al. [82] isolated three fungal species from the roots of the seagrass Posidonia oceanica with the dominant fungal species exhibiting characteristics of DSE. Sequencing data showed that this species belongs to the order Pleosporales, representing a new member in the Aigialaceae. A similar observation showing narrow endophytic diversity and Pleosporales dominance was reported from a culture-independent approach [83]. The mycobiont responsible for these dominant DSE associations in Posidonia oceanica was later described as Posidoniomyces atricolor [84].
However, the results of Vohník et al. [84] conflicted with the findings of Torta et al. [80] regarding the dominant DSE fungal groups, form, and distribution of fungal colonization. To resolve this, a comprehensive study was later conducted by Vohník et al. [78], at the same localities previously investigated by Torta et al. [80] and the dominant DSE associations in Posidonia oceanica was confirmed again as Posidoniomyces atricolor and not Lulwoana sp., as described by Torta et al. [80] previously. Lefebvre et al. [85] claim the colonization of Posidoniomyces atricolor in the degrading tissue of Posidonia oceanica, showing its saprobic nature, which extends beyond a typical plant endophytic association. Overall, the findings of these studies indicate that roots of seagrasses are colonized by DSE, but, until now, as already foreseen, no mycorrhizal association has been described in the roots of seagrasses.
In addition to these reports, Marina Carrasco-Acosta recently presented the isolation of an obligate marine fungus, Cumulospora marina, from the root tissues of Cymodocea nodosa at an IUMBM conference on extremophilic fungi [86]. Furthermore, Wang et al. [87] identified a novel lulworthioid fungus, Halophilomyces hongkongensis, colonizing the roots and rhizomes of the seagrass Halophila ovalis. However, the contribution of these fungal species on seagrass health is still unknown.
4.2. Seagrass Epiphytic Fungi
Epiphytes are spatially different from endophytes and are defined as organisms that live upon the plant surfaces [32]. Similar to endophytes, seagrasses provide excellent habitats for the epiphytic organisms [32,88]. The physicochemical changes in the surrounding water column provide environmental variability, while the host characteristics (such as leaf area) create structural variability for a variety of epiphytic organisms to colonize seagrass surfaces [89]. These variations shape the composition of the epiphytic community and their interactions with the host.
The epiphytic community of seagrasses is primarily composed of algae dominated by the Rhodophyta, while fungi remain largely unstudied [90]. In a previous study, fungi were recorded as “rarely found” in the epiphytic community of Posidonia oceanica leaves [91]. However, recent studies indicate that epiphytic fungi are abundant in seagrasses, and their true diversity and potential are yet to be fully discovered.
Typically, endophytic fungal isolates are obtained following surface sterilization with sodium hypochlorite and ethanol, whereas epiphytes are isolated using less stringent surface sterilization methods [49]. In several studies, surface sterilization methods are not explicitly specified. Thus, it is reasonable to believe that these studies have isolated both endophytic and epiphytic fungi. Table S2 provides a summary of these miscellaneous forms reported from seagrasses worldwide.
Genera such as Cladosporium, Colletotrichum, and Penicillium, and the order Hypocreales, which are often recorded as endophytes, have also been reported as epiphytes in a few studies [92,93]. However, the possibility of contamination, deficiencies in surface sterilization stringency, misidentification of fungi, and human error should also be considered when making inferences from these outcomes. Recently, a novel epiphytic root–fungus symbiosis has been reported to be associated with the roots of the Indo-Pacific seagrass Thalassodendron ciliatum [94].
In comparison, studies on the culture-dependent isolation of seagrass endophytic fungi are more common than those on epiphytic fungi. However, several studies conducted thus far have identified bioactive compounds from epiphytic fungi with potential industrial applications (Table 1). Nonetheless, there exists a notable disparity between these findings and the published records on the initial isolation of these fungi.

Table 1.
Bioactivities of metabolites isolated from seagrass-derived fungi.
4.3. Pathogenic Phytophthora and Halophytophthora Species of Seagrasses
Studies indicate that seagrass meadows are gradually thinning and diminishing due to the increased occurrence of diseases caused by pathogenic microorganisms. Seagrasses are infected by four main groups of pathogens: Labyrinthula, Phytophthora, Halophytophthora, and Phytomyxea [122,123]. Members of Labyrinthula (Labyrinthulids) and Phytomyxea (Plasmodiophorids) are classified under the Kingdom Protista [124,125] while Phytophthora and Halophytophthora are in the class Oomycota, Kingdom Straminipila [126].
Thus far, our literature review has revealed no direct records of any major diseases caused by fungal pathogens in seagrasses. Hu et al. [127] reported the bioactivities of Aspergillus alabamensis, a “phytopathogenic fungus” isolated from the seagrass Enhalus acoroides. However, it is a speculation, and its pathogenicity to seagrass has not been established.
In contrast, considerable scientific attention has been dedicated to Labriyrnthula pathogens responsible for the “seagrass wasting disease”, which causes noticeable, extensive losses in many regions of the world. Little is known about the pathogenicity and disease ecology of other groups. Since the focus of this review is on fungal pathogens, we will only look at Phytophthora and Halophytophthora pathogens, which are considered as fungus-like Oomycetes.
Oomycetes are behaviourally similar but biologically distinct from other main groups belonging to Kingdom Fungi [126]. Historically, they were classified under ‘Phycomycetes’ or “lower fungi” [128]. Considering this early classification, a brief summary of Phytophthora and Halophytophthora species reported in seagrasses is given in Table S3. Relatively few studies have reported the discovery of pathogenic Phytophthora and Halophytophthora species in seagrasses. Phytophthora and Halophytophthora infections have been found to reduce the sexual reproduction of Zostera marina by sixfold [119]. Later, Govers et al. [129] demonstrated that treating Z. marina seeds with copper sulphate can effectively control Phytophthora and Halophytophthora infections, highlighting the effectiveness of this method in seed-based restoration projects.
Further, these pathogens are sensitive to annual and seasonal variations, and migratory bird species have been shown to impact disease dissemination [130]. Regardless of the significance of these pathogens in seagrass health, many studies are confined to Zostera spp. in the northern hemisphere. Comparable to Zostera spp., very little is known about the disease occurrence and pathogenicity of Phytophthora and Halophytophthora in other seagrass species.
4.4. Seagrass Mycobiome Studies
Conventional methods for investigating plant-associated fungi involve isolating and cultivating them on artificial media. Many investigations of seagrass-associated fungi have been undertaken using this standard methodology. However, culture-dependent approaches are often associated with inherent limitations, such as the inability of certain microorganisms to grow on culture media or under specific incubation conditions, as well as the masking of slow-growing microorganisms by fast growers [131]. Thus, these approaches often fail to capture the majority of microbial diversity within environmental samples [132].
As a result, culture-independent, high-throughput molecular methods have gained popularity in recent years to reveal the true diversity and abundance of microorganisms in complex environmental samples. A few meta-studies on seagrasses have been conducted focusing on the seagrass species such as Zostera marina, Z. muelleri, Posidonia oceanica, and Halophilia spp. Similar to the culture-dependent approaches, these studies report the dominance of Eurotiales fungi such as Aspergillus and Penicillium belonging to the phylum Ascomycota. However, unlike culture-dependent studies, the dominance of fungi belonging to the phylum Chytridiomycota is reported in a few studies [35,83,133,134]. This is the only phylum of true fungi that reproduces with zoospores (motile spores). Members of a relatively new order in Chytridiomycota, Lobulomycetales, have been reported in a few studies [35,83,133,134]. A concise summary of mycobiome studies reported thus far is given in Table S4. Designing new primer pairs and blocking oligonucleotides for fungal detection, especially for basal fungal lineages such as Cryptomycota and Chytridiomycota, can further refine these underrepresented groups associated with seagrasses [135,136].
5. Significance of Studying Seagrass-Associated Fungi
The significance of studying seagrass–fungal associations can be addressed from two perspectives. First, understanding seagrass-associated mycobionts can help to protect this vulnerable ecosystem. As previously noted, these relationships can have either a positive or negative impact on the overall health of seagrass beds. However, no studies have been conducted to investigate the molecular and biochemical mechanisms that govern the structure, activity, and function of these communities. A recent study demonstrated that endophytic fungi associated with seagrasses possess the ability to inhibit the growth of the devastative seagrass pathogen Labyrinthula spp. [57]. Therefore, comprehending these interactions and monitoring the compositional changes in fungal communities can serve as a crucial tool for seagrass transplant and restoration initiatives [46].
From a human-centric standpoint, seagrass-inhabiting fungi can be a novel reservoir of metabolites, useful in pharmaceutical and various other industrial applications. Since ‘marine drugs’ are becoming an appealing strategy for addressing antimicrobial resistance [95], seagrass-associated fungal communities can be studied for novel bioactive chemicals. However, previous reports on the screening and isolation of metabolites from fungi associated with seagrasses are limited, making it difficult to predict their real potential based on reliable scientific information. Table 1 lists some of the beneficial metabolites of seagrass-associated fungi and their bioactivities reported thus far. These bioactive metabolites are known to have antimicrobial, anticancer, anti-inflammatory, and antiviral properties. Some notable bioactive metabolites identified from seagrass mycoflora include Cladionol A [110], Sansalvamide [109], Malformin A1 [104], and Halovir A [106]. Peterson et al. [137] report the use of an omics-based high-throughput approach, a rather new approach for rapid bioactivity testing for seagrass-associated fungal metabolites.
Moreover, several publications report the production of lignocellulosic enzymes [114,115], xylan-degrading enzymes [116], and chitin-modifying enzymes [63] from seagrass-associated fungi. These enzymes have a wide range of biotechnological applications. For example, lignocellulosic and xylan-degrading enzymes are important in the biofuel industry, textile industry, paper and pulp industries, and in bioremediation [114,116]. However, to the best of our knowledge, no enzyme derived from a seagrass-associated fungus has reached mass-scale industrial production. This underscores the need for future studies in upscaling production following successful screening assays. Recently, a few studies have highlighted the potential of seagrass-associated fungi in agriculture, particularly as biocontrol agents and alleviating stress responses such as salinity stress [120,121,127].
6. Future Prospects
Currently, there is an emphasis on discovering new taxa from unexplored geographic regions that are quite promising and multifaceted. Nevertheless, it is essential that we incorporate the advances in mycology to expand fundamental studies (e.g., taxonomy based on polyphasic approaches and sequence-based nomenclature to name the Dark taxa). Hence, we recognize several key aspects to continue the research of fungi associated with seagrasses: 1. biodiversity exploration; 2. ecological role understanding; 3. biotechnological potential; and 4. use of emerging technologies.
6.1. Biodiversity Exploration
The exploration of biodiversity in the context of seagrass-associated fungi, particularly in uncharted regions such as parts of Southeast Asia and Northern and Southern America, presents an area rich with potential and complexity [22,46,135,138,139]. Seagrass ecosystems, known as biodiversity hotspots, are often under-researched regarding their fungal communities. These ecosystems may host unique fungal species adapted to specific seagrass environments, offering significant opportunities for discovering new fungal species and genera [80,138,140,141,142].
The geographical diversity of these regions suggests that the fungal diversity associated with seagrass beds could vary significantly, influenced by local environmental factors such as climate, water salinity, and the types of seagrasses present. This diversity is crucial for understanding the ecological roles these fungi play, from nutrient cycling and decomposition to aiding seagrasses in defence against pathogens and environmental stressors [16,93,143].
Advancements in molecular techniques, especially high-throughput DNA sequencing, have revolutionized the identification and cataloguing of fungal species. These methods are more efficient and accurate than traditional ones, capable of identifying even non-culturable fungi and uncovering cryptic species—different species previously thought to be the same due to similar appearances [70,84,133].
Exploring these ecosystems could also reveal new symbiotic relationships between fungi and seagrass or interactions with other microorganisms like bacteria and algae [35,134]. Additionally, understanding how these fungi have adapted to their specific environments can offer insights into their potential responses to global environmental changes, such as rising sea temperatures and ocean acidification [144,145].
Furthermore, each new fungal species discovered contributes to the global understanding of biodiversity, which is vital not just for academic knowledge but also for informing conservation strategies and understanding ecological balances. As seagrass meadows are among the most threatened ecosystems globally, knowing the full range of biodiversity within these areas is essential for their effective management and conservation. Overall, this exploration into the uncharted realms of seagrass-associated fungi holds the key to unlocking the complexities of these ecosystems and significantly contributes to our understanding of marine biodiversity and ecosystem health [22,146].
6.2. Ecological Role Understanding
Understanding the ecological roles of fungi in seagrass ecosystems is vital. This includes their roles in nutrient cycling, decomposition processes, and interactions with other microbial communities. As we uncover more about these fungi, we can better understand the overall health and functioning of seagrass ecosystems.
In marine ecology, the ecological roles of fungi within seagrass ecosystems are of paramount importance, encompassing a diverse array of functions that are crucial for the health and functionality of these underwater habitats. Fungi play a vital role in nutrient cycling, breaking down organic matter and releasing essential nutrients back into the environment, thereby maintaining the nutrient balance in these often nutrient-poor marine settings [73,147]. Additionally, they are key players in the decomposition processes, aiding in the breakdown of dead plant materials such as seagrass leaves and roots. This decomposition not only contributes to the recycling of organic matter but also supports the detritus-based food web that is central to the health of the seagrass ecosystem [148,149].
The interactions of fungi with other microbial communities within these ecosystems, including bacteria, viruses, and algae, are also significant. These interactions, which can range from symbiotic to competitive, affect the distribution and abundance of various microbial species and play a crucial role in the microbial dynamics of the ecosystem [150,151]. The secondary metabolites produced by these microbes impact the fungal colonization, which, in turn, can affect the general health of seagrass [75,152,153,154]. Interestingly, Nerva et al. [155] demonstrated the importance of mycoviruses, a group of viruses which affect fungi, in influencing the overall health of Posidonia oceanica.
Moreover, the health of seagrasses themselves can be directly influenced by the fungi associated with them. While some fungi provide protective benefits, helping seagrasses withstand environmental stressors or deter pathogens, others might be pathogenic and detrimental to seagrass health [16,147]. Conversely, different antifungal metabolites produced by seagrasses can affect internal fungal colonization [75,156,157].
Furthermore, fungi within seagrass ecosystems can act as bioindicators of environmental changes. Variations in fungal communities can signal alterations in environmental conditions, such as pollution or changes in water temperature and salinity, providing valuable insights into the health and stability of these ecosystems [158,159]. For instance, the Seagrass Microbiome Project launched in 2014 aims to identify seagrass–microbe interactions that can reveal important information about seagrass ecology, evolution, and function [160]. Additionally, the role of fungi in the decomposition process has implications for carbon sequestration in seagrass meadows, a significant factor in global carbon dynamics [161,162].
6.3. Biotechnological Potential
The biotechnological potential of marine fungi, particularly those associated with seagrass ecosystems, is a burgeoning field of research with significant implications for various industries, including medicine, agriculture, and industrial processes [163,164,165]. Marine fungi are a largely untapped resource, known for their unique ability to produce novel bioactive compounds [166,167]. These compounds, often not found in terrestrial fungi, arise from the adaptation of fungi to the challenging marine environment, characterized by high salt concentrations, varying pressure conditions, and intense competition for resources [168,169].
In medicine, the unique bioactive compounds derived from marine fungi have shown promise in the development of new pharmaceuticals. These compounds can have diverse biological activities, including antibacterial, antiviral, antifungal, anti-inflammatory, and anticancer properties. For example, some compounds might inhibit the growth of cancer cells or bacteria resistant to current antibiotics, offering new avenues for treatment where traditional medicines are failing [166,170].
In agriculture, these fungi could be a source of new biopesticides or growth enhancers. Given their origin in a highly competitive and harsh environment, these fungi may produce substances that are effective in controlling agricultural pests or diseases, potentially reducing the reliance on synthetic chemicals that can be harmful to the environment [164,171].
The industrial sector could also benefit from enzymes and other molecules produced by seagrass-associated fungi. These enzymes might be particularly useful in processes that require tolerance to saline conditions, such as in certain bioremediation applications or in the processing of marine-derived materials [172,173,174]. As reported by Panno et al. [171], the test fungal isolates recovered from seagrasses exhibit no degradative properties in their enzyme activity in high salt concentrations. This indicates that these enzymes could be valuable for future biotechnological applications that operate under extreme physiochemical conditions. Further, Raghukumar et al. [117] demonstrated the use of a seagrass-derived fungus to remove and detoxify wastewater from molasses-based alcohol distilleries, demonstrating their ability for bioremediation while enhancing the sustainability in biorefinery processes.
Moreover, the exploration of these fungi for biotechnological applications also contributes to the understanding of their ecological roles and potential for sustainable utilization. By identifying and harnessing these bioactive compounds, not only can new, potentially groundbreaking products be developed, but it also encourages the conservation of marine ecosystems like seagrass meadows, which are vital for the health of the marine environment [93,175,176].
6.4. Use of Emerging Technologies
The use of emerging technologies such as remote sensing, artificial intelligence (AI), machine learning in ecological studies, and nanotechnology can revolutionize our understanding of seagrass-associated fungi. These advanced technologies offer powerful tools for gaining new insights into the distribution, health, and ecological roles of these fungi in marine ecosystems [177,178,179].
Remote sensing technology, including satellite imaging and aerial photography, can monitor the health and extent of seagrass meadows over large areas and over time. This technology enables scientists to detect changes in seagrass coverage and condition, which can be indicative of the health of the associated fungal communities [180]. For instance, a decline in seagrass health might suggest issues such as disease or environmental stress, potentially linked to changes in fungal communities. Remote sensing also allows for the mapping of seagrass habitats, providing valuable data for conservation and management efforts [181,182].
Artificial intelligence and machine learning are rapidly becoming indispensable in ecological research. These technologies can process and analyze vast amounts of data much faster and more accurately than traditional methods [179]. In the context of seagrass-associated fungi, AI can be used to analyze complex datasets from remote sensing, genetic sequencing, and ecological surveys to identify patterns and relationships that might be invisible to the human eye. For example, machine-learning algorithms can help in predicting the distribution of fungal species based on environmental variables or in identifying changes in fungal communities in response to environmental stressors [183,184].
Furthermore, the integration of AI with genomic studies, such as metagenomics and high-throughput sequencing, is particularly promising. This integration allows for the rapid identification and classification of fungal species, even those that are rare or previously unknown. AI algorithms can analyze genetic data to uncover relationships between fungal species, their adaptation strategies, and their interactions with seagrasses and other marine organisms [185].
The production and utilization of nanoparticles using fungi, which is also known as myconanotechnology, is also an emerging topic of research [186]. These fungal-derived nanoparticles have a low toxicity and are eco-friendly, compared to the conventional nanoparticles synthesized by chemical or physical means [187]. As seagrass-associated fungi produce unique metabolites, they can hold unique biochemical mechanisms to generate nanoparticles with diverse chemical characteristics. Further, silver nanoparticles synthesized by this biological method can simplify the synthesis process by eliminating the extra steps required to prevent particle aggregation [188]. According to Abdelrahman et al. [189], it is possible to screen and optimize seagrass-associated fungi for nanoparticle synthesis, leading to nanomaterials with different bioactive properties.
The application of these emerging technologies in the study of seagrass-associated fungi not only enhances research capabilities but also contributes to more effective conservation strategies. By providing a more comprehensive and nuanced understanding of these ecosystems, technology can help in predicting the impacts of environmental changes and human activities, thereby informing management and restoration efforts [95,133]. More importantly, understanding the significance of fungal genetic resources through these new approaches can be used as a valuable tool for in situ conservation and support seagrass reforestation.
7. Conclusions
Seagrass beds are distinct ecosystems that serve as a reservoir for a diverse array of microbes, including fungi. These fungi can engage in different relationships with their host, spanning from mutualistic to potentially pathogenic. Culture-dependent and culture-independent methods are currently employed to study the diversity of seagrass-associated fungi.
The majority of culture-based methods concentrate on seagrass inhabiting endophytic fungi. Epiphytic fungi are often overlooked, or it could be argued that they are not easily identified directly, due to the challenges associated with handling samples during initial isolation. Thus far, Eurotiales species such as Aspergillus spp. and Penicillium spp. dominate the fungal community associated with seagrasses. There have been no reports of mycorrhizal associations in seagrass roots so far, although other distinct fungal associations have been discovered. Infections by Phytophthora and Halophytophthora have been documented in seagrasses, which were formerly classified as lower fungi. However, diseases caused by true fungi in seagrasses remain largely unidentified.
Overall, the majority of culture-based studies are confined to Enhalus acaroides, Cymodocea serrulata, Posidonia oceanica, Thalassia hemprichii, and Zostera marina. Meanwhile, mycobiome studies have been carried out on Zostera spp., Halophilia spp., and Posidonia oceanica. With 72 seagrass species worldwide, there is a significant opportunity to expand both culture-dependent and -independent research on a global scale. Regions with rich diversity, such as the Tropical Indo-Pacifics along the east coast of Africa, lack sufficient data.
The harsh environment in marine ecosystems may lead to the accumulation of metabolites in seagrass-associated fungi, which could be employed in extreme physiological conditions. These fungi could also be investigated as a source of new pharmaceutical lead compounds for development. Recently, a few attempts to employ these fungi to alleviate stress responses in plants have been reported. However, there is a lack of studies on their applications, and their full potential is still unknown.
Rapidly disappearing seagrass beds highlight the importance of advancing research to preserve their valuable genetic resources before they are lost without documentation. Exploring the ecological significance and relationships of these seagrass–fungal associations can aid in future seagrass restoration and transplantation initiatives. The application of modern technologies such as omics-based methods and AI can enhance research on seagrass fungi even further.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof10090627/s1, Table S1: Previous records on endophytic and epiphytic fungi isolated from seagrasses using culture-dependent approaches. Table S2: Previous records on miscellaneous fungi isolated from seagrasses using culture-dependent approaches. Table S3: Previous records on pathogenic fungi-like organisms isolated from seagrasses using culture-dependent approaches. Table S4: Summary of the culture-independent studies on seagrass-associated fungi. References [15,35,47,57,59,60,61,62,64,65,66,67,68,69,70,71,72,78,80,82,83,84,92,93,101,102,105,118,119,133,134,135,138,144,145,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209] are cited in the supplementary tables.
Author Contributions
Conceptualization, N.N.W., O.R. and K.G.S.U.A.; software, O.R. and S.S.G.; data curation, O.R.; writing—original draft, O.R., N.N.W., S.U., P.K.J., S.S.G. and N.B.; writing—review and editing, O.R., N.N.W., K.G.S.U.A., S.U., P.K.J., S.S.G., N.B. and T.C.B.; visualization, O.R., S.S.G. and S.U.; supervision, N.N.W. and K.G.S.U.A.; project administration, K.G.S.U.A.; funding acquisition, T.C.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Science and Technology Human Resource Development Project, Ministry of Higher Education, Sri Lanka, funded by the Asian Development Bank (Grant number R3RJ2) and Tropical Microbiology Research Foundation (TMRF).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials; further inquiries can be directed to the corresponding authors.
Acknowledgments
Oshadi Rajakaruna would like to thank W. M. Ayesha Sanahari, Department of Plant Sciences, University of Colombo for the help in preparing image plates using Photoshop. Nattawut Boonyuen expresses gratitude to BIOTEC-NSTDA for supporting the fungal research in Thailand. Nalin N. Wijayawardene thanks the High-Level Talent Recruitment Plan of Yunnan Provinces (“Young Talents” Program and “High-End Foreign Experts” Program).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gladfelter, A.S.; James, T.Y.; Amend, A.S. Marine fungi. Curr. Biol. 2019, 29, R191–R195. [Google Scholar] [CrossRef] [PubMed]
- Amend, A.; Burgaud, G.; Cunliffe, M.; Edgcomb, V.P.; Ettinger, C.L.; Gutiérrez, M.H.; Heitman, J.; Hom, E.F.Y.; Ianiri, G.; Jones, A.C.; et al. Fungi in the marine environment: Open questions and unsolved problems. mBio 2019, 10, e01189-18. [Google Scholar] [CrossRef] [PubMed]
- Raghukumar, S. Fungi in Coastal and Oceanic Marine Ecosystems, 1st ed.; Springer International Publishing: New York, NY, USA, 2017; ISBN 978-3-319-54303-1. [Google Scholar]
- Salvatore, M.M.; Andolfi, A. Phytopathogenic fungi and toxicity. Toxins 2021, 13, 689. [Google Scholar] [CrossRef]
- Walsh, T.J.; Groll, A.; Hiemenz, J.; Fleming, R.; Roilides, E.; Anaissie, E. Infections due to emerging and uncommon medically important fungal pathogens. Clin. Microbiol. Infect. 2004, 10, 48–66. [Google Scholar] [CrossRef]
- Species Fungorum. Available online: http://www.speciesfungorum.org (accessed on 11 July 2024).
- Hawksworth, D.L. The Fungal dimension of biodiversity: Magnitude, significance, and conservation. Mycol. Res. 1991, 95, 641–655. [Google Scholar] [CrossRef]
- Hawksworth, D.L. The Magnitude of fungal diversity: The 1·5 million species estimate revisited. Mycol. Res. 2001, 105, 1422–1432. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Villarreal Ruiz, L.; Vasco-Palacios, A.M.; Quang Thu, P.; Smith, M.E.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Lücking, R. Fungal diversity revisited: 2.2 to 3.8 Million species. Microbiol. Spectr. 2017, 5, 79–95. [Google Scholar] [CrossRef]
- Niskanen, T.; Lücking, R.; Dahlberg, A.; Gaya, E.; Suz, L.M.; Mikryukov, V.; Liimatainen, K.; Druzhinina, I.; Westrip, J.R.S.; Mueller, G.M.; et al. Pushing the frontiers of biodiversity research: Unveiling the global diversity, distribution, and conservation of Fungi. Annu. Rev. Environ. Resour. 2023, 48, 149–176. [Google Scholar] [CrossRef]
- Blackwell, M. The Fungi: 1, 2, 3 … 5.1 Million species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Rossman, A.Y. Where are all the undescribed fungi? Phytopathology 1997, 87, 881–891. [Google Scholar] [CrossRef] [PubMed]
- El-Bondkly, E.A.M.; El-Bondkly, A.A.M.; El-Bondkly, A.A.M. Marine endophytic fungal metabolites: A whole new world of pharmaceutical therapy exploration. Heliyon 2021, 7, e06362. [Google Scholar] [CrossRef]
- Pasqualetti, M.; Giovannini, V.; Barghini, P.; Gorrasi, S.; Fenice, M. Diversity and ecology of culturable marine fungi associated with Posidonia oceanica leaves and their epiphytic algae Dictyota dichotoma and Sphaerococcus coronopifolius. Fungal Ecol. 2020, 44, 100906. [Google Scholar] [CrossRef]
- Seymour, J.R.; Laverock, B.; Nielsen, D.A.; Trevathan-Tackett, S.M.; Macreadie, P.I. The microbiology of seagrasses. In Seagrasses of Australia; Larkum, A., Kendrick, G., Ralph, P., Eds.; Springer: Cham, Switzerland, 2018; pp. 343–392. ISBN 9783319713540. [Google Scholar]
- Sarasan, M.; Puthumana, J.; Job, N.; Han, J.; Lee, J.S.; Philip, R. Marine algicolous endophytic fungi-a promising drug resource of the era. J. Microbiol. Biotechnol. 2017, 27, 1039–1052. [Google Scholar] [CrossRef]
- Johnson, T.W.; Sparrow, F.K. Fungi in Oceans and Estuaries; J. Cramer: Weinheim, Germany, 1961. [Google Scholar]
- Jones, E.B.G.; Pang, K.L.; Abdel-Wahab, M.A.; Scholz, B.; Hyde, K.D.; Boekhout, T.; Ebel, R.; Rateb, M.E.; Henderson, L.; Sakayaroj, J.; et al. An online resource for marine fungi. Fungal Divers. 2019, 96, 347–433. [Google Scholar] [CrossRef]
- Calabon, M.S.; Jones, E.B.G.; Promputtha, I.; Hyde, K.D. Fungal biodiversity in salt marsh ecosystems. J. Fungi 2021, 7, 648. [Google Scholar] [CrossRef] [PubMed]
- Devadatha, B.; Jones, E.B.G.; Pang, K.L.; Abdel-Wahab, M.A.; Hyde, K.D.; Sakayaroj, J.; Bahkali, A.H.; Calabon, M.S.; Sarma, V.V.; Sutreong, S.; et al. Occurrence and geographical distribution of mangrove fungi. Fungal Divers. 2021, 106, 137–227. [Google Scholar] [CrossRef]
- Poli, A.; Varese, G.C.; Garzoli, L.; Prigione, V. Seagrasses, Seaweeds and Plant Debris: An extraordinary reservoir of fungal diversity in the Mediterranean Sea. Fungal Ecol. 2022, 60, 101156. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Dai, D.Q.; Jayasinghe, P.K.; Gunasekara, S.S.; Nagano, Y.; Tibpromma, S.; Suwannarach, N.; Boonyuen, N. Ecological and oceanographic perspectives in future marine fungal taxonomy. J. Fungi 2022, 8, 1141. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the Microbiome of the plant Holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Jones, E.B.G.; Jennings, D.H. The effect of salinity on the growth of marine fungi in comparison with non-marine species. Trans. Br. Mycol. Soc. 1964, 47, 619–625. [Google Scholar] [CrossRef]
- Kohlmeyer, J.; Kohlmeyer, E. Marine Mycology: The Higher Fungi; Academic Press: New York, NY, USA, 1979. [Google Scholar]
- Bugni, T.S.; Ireland, C.M. Marine-Derived Fungi: A chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 2004, 21, 143–163. [Google Scholar] [CrossRef]
- Pang, K.-L.; Overy, D.P.; Jones, E.B.G.; Calado, M.D.L.; Burgaud, G.; Walker, A.K.; Johnson, J.A.; Kerr, R.G.; Cha, H.-J.; Bills, G.F. ‘Marine Fungi’ and ‘Marine-Derived Fungi’ in natural product chemistry research: Toward a new consensual definition. Fungal Biol. Rev. 2016, 30, 163–175. [Google Scholar] [CrossRef]
- Jones, E.B.G. Are there more marine fungi to be described? Bot. Mar. 2011, 54, 343–354. [Google Scholar] [CrossRef]
- Gonçalves, M.F.M.; Esteves, A.C.; Alves, A. Marine fungi: Opportunities and challenges. Encyclopedia 2022, 2, 559–577. [Google Scholar] [CrossRef]
- Manohar, C.S.; Raghukumar, C. Fungal diversity from various marine habitats deduced through culture-independent studies. FEMS Microbiol. Lett. 2013, 341, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Larkum, A.W.D.; Orth, R.J.; Duarte, C.M. Seagrasses: Biology, Ecology and Conservation, 1st ed.; Springer: Dordrecht, The Netherlands, 2006; ISBN 978-1-4020-2942-4. [Google Scholar]
- Short, F.T.; Short, C.A.; Novak, A.B. Seagrasses. In The Wetland Book; Finlayson, C.M., Milton, G.R., Prentice, R.C., Davidson, N.C., Eds.; Springer: Dordrecht, The Netherlands, 2016; ISBN 9789400761735. [Google Scholar]
- Duarte, C.M. Seagrasses. In Encyclopedia of Biodiversity, 2nd ed.; Academic Press: Cambridge, MA, USA, 2001; ISBN 9780128225622. [Google Scholar]
- Ettinger, C.L.; Eisen, J.A. Characterization of the mycobiome of the seagrass, Zostera marina, reveals putative associations with marine chytrids. Front. Microbiol. 2019, 10, 2476. [Google Scholar] [CrossRef] [PubMed]
- Waycott, M.; Duarte, C.M.; Carruthers, T.J.B.; Orth, R.J.; Dennison, W.C.; Olyarnik, S.; Calladine, A.; Fourqurean, J.W.; Heck, K.L.; Hughes, A.R.; et al. Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proc. Natl. Aacad. Sci. USA 2009, 106, 12377–12381. [Google Scholar] [CrossRef]
- Orth, R.J.; Carruthers, T.J.B.; Dennison, W.C.; Duarte, C.M.; Fourqurean, J.W.; Heck, K.L.; Hughes, A.R.; Kendrick, G.A.; Kenworthy, W.J.; Olyarnik, S.; et al. A global crisis for seagrass ecosystems. Bioscience 2006, 56, 987–996. [Google Scholar] [CrossRef]
- Short, F.; Carruthers, T.; Dennison, W.; Waycott, M. Global seagrass distribution and diversity: A bioregional model. J. Exp. Mar. Biol. Ecol. 2007, 350, 3–20. [Google Scholar] [CrossRef]
- United Nations Environment Programme. Out of the Blue: The Value of Seagrasses to the Environment and to People; UNEP: Nairobi, Kenya, 2020. [Google Scholar]
- Björk, M.; Short, F.T.; Mcleod, E.; Beer, S. Managing Seagrasses for Resilience to Climate Change; IUCN: Gland, Switzerland, 2008; ISBN 9782831710891. [Google Scholar]
- Green, E.P.; Short, F.T. World Atlas of Seagrasses; University of California Press: Berkeley, CA, USA, 2003. [Google Scholar]
- McKenzie, L.J.; Nordlund, L.M.; Jones, B.L.; Cullen-Unsworth, L.C.; Roelfsema, C.; Unsworth, R.K.F. The Global Distribution of Seagrass Meadows. Environ. Res. Lett. 2020, 15, 074041. [Google Scholar] [CrossRef]
- Duarte, C.M. Seagrass Depth Limits. Aquat. Bot. 1991, 40, 363–377. [Google Scholar] [CrossRef]
- Short, F.T.; Polidoro, B.; Livingstone, S.R.; Carpenter, K.E.; Bandeira, S.; Sidik Bujang, J.; Calumpong, H.P.; Carruthers, T.J.B.; Coles, R.G.; Dennison, W.C.; et al. Extinction risk assessment of the world’s seagrass species. Biol. Conserv. 2011, 144, 1961–1971. [Google Scholar] [CrossRef]
- Rasheed, M.A.; Unsworth, R.K.F. Long-term climate-associated dynamics of a tropical seagrass meadow: Implications for the future. Mar. Ecol. Prog. Ser. 2011, 422, 93–103. [Google Scholar] [CrossRef]
- Wainwright, B.J.; Zahn, G.L.; Zushi, J.; Lee, N.L.Y.; Ooi, J.L.S.; Lee, J.N.; Huang, D. Seagrass-associated fungal communities show distance decay of similarity that has implications for seagrass management and restoration. Ecol. Evol. 2019, 9, 11288–11297. [Google Scholar] [CrossRef] [PubMed]
- Quek, Z.B.R.; Zahn, G.; Lee, N.L.Y.; Ooi, J.L.S.; Lee, J.N.; Huang, D.; Wainwright, B.J. Biogeographic structure of fungal communities in seagrass Halophilia ovalis across the Malay Peninsula. Environ. Microbiol. Rep. 2021, 13, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Petrini, O. Fungal Endophytes of Tree Leaves. In Microbial Ecology of Leaves; Springer: Berlin/Heidelberg, Germany, 1991; pp. 179–197. [Google Scholar] [CrossRef]
- Khiralla, A.; Spina, R.; Yagi, S.; Mohamed, I.; Laurain-Mattar, D. Endophytic fungi: Occurrence, classification, function and natural products. In Endophytic Fungi: Diversity, Characterization and Biocontrol; Nova Publishers: New York, NY, USA, 2016; ISBN 9781536103588. [Google Scholar]
- Schulz, B.; Boyle, C. The endophytic continuum. Mycol. Res. 2005, 109, 661–686. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Khare, E.; Mishra, J.; Arora, N.K. Multifaceted interactions between endophytes and plant: Developments and prospects. Front. Microbiol. 2018, 9, 2732. [Google Scholar] [CrossRef]
- Yan, L.; Zhu, J.; Zhao, X.; Shi, J.; Jiang, C.; Shao, D. Beneficial effects of endophytic fungi colonization on plants. Appl. Microbiol. Biotechnol. 2019, 103, 3327–3340. [Google Scholar] [CrossRef]
- Eid, A.M.; Salim, S.S.; Hassan, S.E.-D.; Ismail, M.A.; Fouda, A. Role of endophytes in plant health and abiotic stress management. In Microbiome in Plant Health and Disease; Springer: Singapore, 2019; pp. 119–144. [Google Scholar]
- Sodhi, G.K.; Saxena, S. Role of Endophytic Fungi in Promoting Plant Health. In Fungal Resources for Sustainable Economy; Springer Nature: Singapore, 2023; pp. 319–345. [Google Scholar]
- Rai, M.; Agarkar, G. Plant-Fungal Interactions: What triggers the fungi to switch among lifestyles? Crit. Rev. Microbiol. 2016, 42, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Ugarellia, K.; Jagels, A.; Choia, C.J.; Loesgenb, S.; Stingl, U. Fungal endophytes from Thalassia testudinum show bioactivity against seagrass pathogen Labyrinthula sp. Front. Mar. Sci. 2024, 11, 1359610. [Google Scholar]
- Kuo, J. Structural Aspects of apoplast fungal hyphae in a marine angiosperm, Zostera muelleri Irmisch Ex Aschers. (Zosteraceae). Protoplasma 1984, 121, 1–7. [Google Scholar] [CrossRef]
- Wilson, W.L. Isolation of Endophytes from Seagrasses from Bermuda. Master’s Thesis, The University of New Brunswick, Saint John, NB, Canada, 1998. [Google Scholar]
- Sakayaroj, J.; Preedanon, S.; Supaphon, O.; Jones, E.B.G.; Phongpaichit, S. Phylogenetic diversity of endophyte assemblages associated with the tropical seagrass Enhalus acoroides in Thailand. Fungal Divers. 2010, 42, 27–45. [Google Scholar] [CrossRef]
- Mata, J.L.; Cebrián, J. Fungal endophytes of the seagrasses Halodule wrightii and Thalassia testudinum in the Northcentral Gulf of Mexico. Bot. Mar. 2013, 56, 541–545. [Google Scholar] [CrossRef]
- Supaphon, P.; Phongpaichit, S.; Rukachaisirikul, V.; Sakayaroj, J. Diversity and antimicrobial activity of endophytic fungi isolated from the seagrass Enhalus acoroides. Indian J. Geo-Mar. Sci. 2014, 43, 785–797. [Google Scholar]
- Venkatachalam, A.; Govinda Rajulu, M.; Thirunavukkarasu, N.; Suryanarayanan, T. Endophytic fungi of marine algae and seagrasses: A novel source of chitin modifying enzymes. Mycosphere 2015, 6, 345–355. [Google Scholar] [CrossRef]
- Subrmaniyan, R.; Ponnambalam, S.; Thirunavukarassu, T. Inter species variations in cultivable endophytic fungal diversity among the tropical seagrasses. Proc. Natl. Acad. Sci. India Sect B—Biol. Sci. 2018, 88, 849–857. [Google Scholar] [CrossRef]
- Supaphon, P.; Phongpaichit, S.; Rukachaisirikul, V.; Sakayaroj, J. Antimicrobial potential of endophytic fungi derived from three seagrass species: Cymodocea serrulata, Halophila ovalis and Thalassia hemprichii. PLoS ONE 2013, 8, e72520. [Google Scholar] [CrossRef]
- Supaphon, P.; Phongpaichit, S.; Sakayaroj, J.; Rukachaisirikul, V.; Kobmoo, N.; Spatafora, J.W. Phylogenetic community structure of fungal endophytes in seagrass species. Bot. Mar. 2017, 60, 489–501. [Google Scholar] [CrossRef]
- Devarajan, P.T.; Suryanarayanan, T.S.; Geetha, V. Endophytic fungi associated with the tropical seagrass Halophila ovalis (Hydrocharitaceae). Indian J. Mar. Sci. 2002, 31, 73–74. [Google Scholar]
- Venkatachalam, A.; Thirunavukkarasu, N.; Suryanarayanan, T.S. Distribution and Diversity of Endophytes in Seagrasses. Fungal Ecol. 2015, 13, 60–65. [Google Scholar] [CrossRef]
- Cuomo, V.; Vanzanella, F.; Fresi, E.; Cinelli, F.; Mazzella, L. Fungal flora of Posidonia oceanica and its ecological significance. Trans. Br. Mycol. Soc. 1985, 84, 35–40. [Google Scholar] [CrossRef]
- Abdel-Wahab, M.A.; Bahkali, A.H.; Elgorban, A.M.; Jones, E.B.G. High-throughput amplicon sequencing of fungi and microbial eukaryotes associated with the seagrass Halophila stipulacea (Forssk.) Asch. from Al-Leith Mangroves, Saudi Arabia. Mycol. Prog. 2021, 20, 1365–1381. [Google Scholar] [CrossRef]
- Gnavi, G.; Ercole, E.; Panno, L.; Vizzini, A.; Varese, G.C. Dothideomycetes and Leotiomycetes sterile mycelia isolated from the italian seagrass Posidonia oceanica based on RDNA Data. SpringerPlus 2014, 3, 508. [Google Scholar] [CrossRef]
- Shoemaker, G.; Wyllie-Echeverria, S. Occurrence of rhizomal endophytes in three temperate northeast pacific seagrasses. Aquat. Bot. 2013, 111, 71–73. [Google Scholar] [CrossRef]
- Panno, L.; Bruno, M.; Voyron, S.; Anastasi, A.; Gnavi, G.; Miserere, L.; Varese, G.C. Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia oceanica. New Biotechnol. 2013, 30, 685–694. [Google Scholar] [CrossRef]
- Ross, C.; Puglisi, M.P.; Paul, V.J. Antifungal defenses of seagrasses from the Indian River Lagoon, Florida. Aquat. Bot. 2008, 88, 134–141. [Google Scholar] [CrossRef]
- Bibi, F.; Naseer, M.I.; Hassan, A.M.; Yasir, M.; Al-Ghamdi, A.A.K.; Azhar, E.I. Diversity and antagonistic potential of bacteria isolated from marine grass Halodule uninervis. 3 Biotech 2018, 8, 48. [Google Scholar] [CrossRef]
- Solé, M.; Lenoir, M.; Durfort, M.; Fortuño, J.M.; van der Schaar, M.; De Vreese, S.; André, M. Seagrass Posidonia is impaired by human-generated noise. Commun. Biol. 2021, 4, 743. [Google Scholar] [CrossRef]
- Nielsen, S.L.; Thingstrup, I.; Wigand, C. Apparent Lack of Vesicular-Arbuscular Mycorrhiza (VAM) in the seagrasses Zostera marina L. and Thalassia testudinum Banks Ex Konig. Aquat. Bot. 1999, 63, 261–266. [Google Scholar] [CrossRef]
- Vohník, M. Are Lulworthioid Fungi dark septate endophytes of the Dominant Mediterranean Seagrass Posidonia oceanica? Plant Biol. 2022, 24, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Kuo, J.; McComb, A.J.; Cambridge, M.L. Ultrastructure of the Seagrass Rhizosphere. New Phytol. 1981, 89, 139–143. [Google Scholar] [CrossRef]
- Torta, L.; Lo Piccolo, S.; Piazza, G.; Burruano, S.; Colombo, P.; Ottonello, D.; Perrone, R.; Di Maida, G.; Pirrotta, M.; Tomasello, A.; et al. Lulwoana sp., a Dark Septate Endophyte in roots of Posidonia oceanica (L.) Delile seagrass. Plant Biol. 2015, 17, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Vohník, M.; Borovec, O.; Župan, I.; Vondrášek, D.; Petrtýl, M.; Sudová, R. Anatomically and morphologically unique dark septate endophytic association in the roots of the Mediterranean endemic seagrass Posidonia oceanica. Mycorrhiza 2015, 25, 663–672. [Google Scholar] [CrossRef]
- Vohník, M.; Borovec, O.; Kolařík, M. Communities of Cultivable Root mycobionts of the seagrass Posidonia oceanica in the Northwest Mediterranean Sea are dominated by a hitherto undescribed Pleosporalean Dark Septate Endophyte. Microb. Ecol. 2016, 71, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Vohník, M.; Borovec, O.; Župan, I.; Kolařík, M.; Sudová, R. Fungal root symbionts of the seagrass Posidonia oceanica in the Central Adriatic Sea revealed by Microscopy, Culturing and 454-Pyrosequencing. Mar. Ecol. Prog. Ser. 2017, 583, 107–120. [Google Scholar] [CrossRef]
- Vohník, M.; Borovec, O.; Kolaříková, Z.; Sudová, R.; Réblová, M. Extensive sampling and high-throughput sequencing reveal Posidoniomyces atricolor gen. Et sp. nov. (Aigialaceae, Pleosporales) as the dominant root mycobiont of the dominant Mediterranean seagrass Posidonia oceanica. MycoKeys 2019, 55, 59–86. [Google Scholar] [CrossRef]
- Lefebvre, L.; Compère, P.; Gobert, S. The formation of aegagropiles from the Mediterranean seagrass Posidonia oceanica (L.) Delile (1813): Plant tissue sources and colonisation by melanised fungal mycelium. Mar. Biol. 2023, 170, 19. [Google Scholar] [CrossRef]
- Carrasco-Acosta, M.; Poli, A.; Garcia-Jimenez, P.; Prigione, V.P.; Varese, G.C. The cultivable mycobiota associated with the seagrass Cymodocea nodosa for ecological and biotechnological purposes. In Proceedings of the IUBMB Focused Meeting on Extremophilic Fungi, Ljubljana, Slovenia, 19–22 September 2023; Biotechnical Faculty, University of Ljubljana: Ljubljana, Slovenia, 2023; p. 127. [Google Scholar]
- Wang, X.; Pecoraro, L.; Chen, J.; Tang, Y.; Lee, S.; Chen, S.; Liu, H. Halophilomyces hongkongensis, a novel species and genus in the Lulworthiaceae with antibacterial potential, colonizing the roots and rhizomes of the seagrass Halophila ovalis. J. Fungi 2024, 10, 474. [Google Scholar] [CrossRef]
- Michael, T.S.; Shin, H.W.; Hanna, R.; Spafford, D.C. A review of epiphyte community development: Surface interactions and settlement on seagrass. J. Environ. Biol. 2008, 98, 629–638. [Google Scholar]
- Trevizan Segovia, B.; Sanders-Smith, R.; Adamczyk, E.M.; Forbes, C.; Hessing-Lewis, M.; O’Connor, M.I.; Parfrey, L.W. Microeukaryotic communities associated with the seagrass Zostera marina are spatially structured. J. Eukaryot. Microbiol. 2021, 68, e12827. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, T.G.; Komarpant, D.S.; Rodrigues, R.S. Status of a seagrass ecosystem: An ecologically sensitive wetland habitat from India. Wetlands 2003, 23, 161–170. [Google Scholar] [CrossRef]
- Novak, R. A Study in Ultra-Ecology: Microorganisms on the seagrass Posidonia oceanica (L.) Delile. Mar. Ecol. 1984, 5, 143–190. [Google Scholar] [CrossRef]
- Ettinger, C.L.; Eisen, J.A. Fungi, Bacteria and Oomycota opportunistically isolated from the seagrass, Zostera marina. PLoS ONE 2020, 15, e0236135. [Google Scholar] [CrossRef]
- Tasdemir, D.; Scarpato, S.; Utermann-Thüsing, C.; Jensen, T.; Blümel, M.; Wenzel-Storjohann, A.; Welsch, C.; Echelmeyer, V.A. Epiphytic and endophytic microbiome of the seagrass Zostera marina: Do they contribute to pathogen reduction in seawater? Sci. Total Environ. 2024, 908, 168422. [Google Scholar] [CrossRef]
- Vohník, M.; Josefiová, J. Novel epiphytic root-fungus symbiosis in the Indo-Pacific Seagrass Thalassodendron ciliatum from the Red Sea. Mycorrhiza 2024, 1–15. [Google Scholar] [CrossRef]
- Zhang, Y.; Mu, J.; Feng, Y.; Kang, Y.; Zhang, J.; Gu, P.J.; Wang, Y.; Ma, L.F.; Zhu, Y.H. Broad-spectrum antimicrobial epiphytic and endophytic fungi from marine organisms: Isolation, Bioassay and Taxonomy. Mar. Drugs 2009, 7, 97–112. [Google Scholar] [CrossRef]
- Arunpanichlert, J.; Rukachaisirikul, V.; Tadpetch, K.; Phongpaichit, S.; Hutadilok-Towatana, N.; Supaphon, O.; Sakayaroj, J. A Dimeric Chromanone and a Phthalide: Metabolites from the seagrass-derived fungus Bipolaris sp. PSU-ES64. Phytochem. Lett. 2012, 5, 604–608. [Google Scholar] [CrossRef]
- Alfattani, A.; Queiroz, E.F.; Marcourt, L.; Leoni, S.; Stien, D.; Hofstetter, V.; Gindro, K.; Perron, K.; Wolfender, J.-L. One-Step bio-guided isolation of secondary metabolites from the endophytic fungus Penicillium crustosum using high-resolution semi-preparative HPLC. Comb. Chem. High Throughput Screen. 2024, 27, 573–583. [Google Scholar] [CrossRef]
- Arunpanichlert, J.; Rukachaisirikul, V.; Sukpondma, Y.; Phongpaichit, S.; Supaphon, O.; Sakayaroj, J. A β-resorcylic macrolide from the seagrass-derived fungus Fusarium sp. PSU-ES73. Arch. Pharm. Res. 2011, 34, 1633–1637. [Google Scholar] [CrossRef] [PubMed]
- Linn, K.T. Screening of marine endophytic fungi isolated from some seagrasses leaves and their antibacterial activities on Micrococcus luteus NITE83297. J. Myanmar Acad. Arts Sci. 2018, 17, 139–154. [Google Scholar]
- Botta, L.; Saladino, R.; Barghini, P.; Fenice, M.; Pasqualetti, M. Production and identification of two antifungal terpenoids from the Posidonia oceanica epiphytic Ascomycota Mariannaea humicola IG100. Microb. Cell Fact. 2020, 19, 184. [Google Scholar] [CrossRef] [PubMed]
- Qader, M.M.; Hamed, A.A.; Soldatou, S.; Abdelraof, M.; Elawady, M.E.; Hassane, A.S.I.; Belbahri, L.; Ebel, R.; Rateb, M.E. Antimicrobial and antibiofilm activities of the fungal metabolites isolated from the marine endophytes Epicoccum nigrum M13 and Alternaria alternata 13A. Mar. Drugs 2021, 19, 232. [Google Scholar] [CrossRef] [PubMed]
- Hamed, A.A.; Soldatou, S.; Mallique Qader, M.; Arjunan, S.; Miranda, K.J.; Casolari, F.; Pavesi, C.; Diyaolu, O.A.; Thissera, B.; Eshelli, M.; et al. Screening fungal endophytes derived from under-explored Egyptian marine habitats for antimicrobial and antioxidant properties in factionalised textiles. Microorganisms 2020, 8, 1617. [Google Scholar] [CrossRef] [PubMed]
- Setyati, W.A.; Sedjati, S.; Samudra, A.; Ariyanto, D. Investigation of seagrass-associated fungi as antifouling candidates with anti-bacterial properties. Jordan J. Biol. Sci. 2023, 16, 323–327. [Google Scholar] [CrossRef]
- Notarte, K. Trypanocidal activity, cytotoxicity and histone modifications induced by Malformin A1 isolated from the marine-derived fungus Aspergillus Tubingensis IFM 63452. Mycosphere 2017, 8, 111–120. [Google Scholar] [CrossRef]
- Morell-Rodríguez, G. Potential of Fungal Endophytes from Thalassia testudinum Bank Ex KD Koenig as Producers of Bioactive compounds. Ph.D. Thesis, University of Puerto Rico, Mayaguez, Puerto Rico, 2008. [Google Scholar]
- Fenical, W.; Jensen, P.R.; Rowley, D.C. Halovir, an Antiviral Marine Natural Product, and Derivatives Thereof. U.S. Patent 983 6,458,766 B1, 1 October 2002. [Google Scholar]
- Kaushik, N.; Murali, T.; Sahal, D.; Suryanarayanan, T. A search for antiplasmodial metabolites among fungal endophytes of terrestrial and marine plants of Southern India. Acta Parasitol. 2014, 59, 745–757. [Google Scholar] [CrossRef]
- Notarte, K.I.; Yaguchi, T.; Suganuma, K.; dela Cruz, T.E. Antibacterial, cytotoxic and trypanocidal activities of marine-derived fungi isolated from Philippine macroalgae and seagrasses. Acta Bot. Croat. 2018, 77, 141–151. [Google Scholar] [CrossRef]
- Belofsky, G.N.; Jensen, P.R.; Fenical, W. Sansalvamide: A new cytotoxic cyclic depsipeptide produced by a marine fungus of the genus Fusarium. Tetrahedron Lett. 1999, 40, 2913–2916. [Google Scholar] [CrossRef]
- Kasai, Y.; Komatsu, K.; Shigemori, H.; Tsuda, M.; Mikami, Y.; Kobayashi, J. Cladionol A, a polyketide glycoside from marine-derived fungus Gliocladium species. J. Nat. Prod. 2005, 68, 777–779. [Google Scholar] [CrossRef] [PubMed]
- Afiyatullov, S.S.; Leshchenko, E.V.; Sobolevskaya, M.P.; Denisenko, V.A.; Kirichuk, N.N.; Khudyakova, Y.V.; Hoai, T.P.; Dmitrenok, P.S.; Menchinskaya, E.S.; Pislyagin, E.A.; et al. New eudesmane sesquiterpenes from the marine-derived fungus Penicillium thomii. Phytochem. Lett. 2015, 14, 209–214. [Google Scholar] [CrossRef]
- Afiyatullov, S.S.; Leshchenko, E.V.; Sobolevskaya, M.P.; Antonov, A.S.; Denisenko, V.A.; Popov, R.S.; Khudyakova, Y.V.; Kirichuk, N.N.; Kuz’mich, A.S.; Pislyagin, E.A.; et al. New Thomimarine E from marine isolate of the fungus Penicillium thomii. Chem. Nat. Compd. 2017, 53, 290–294. [Google Scholar] [CrossRef]
- Cicatiello, P.; Gravagnuolo, A.M.; Gnavi, G.; Varese, G.C.; Giardina, P. Marine fungi as source of new hydrophobins. Int. J. Biol. Macromol. 2016, 92, 1229–1233. [Google Scholar] [CrossRef] [PubMed]
- Raghukumar, C.; D’souza, T.; Thorn, R.; Reddy, C. Lignin-modifying enzymes of Flavodon flavus a Basidiomycete isolated from a coastal marine environment. Appl. Environ. Microbiol. 1999, 65, 2103–2111. [Google Scholar] [CrossRef]
- Mtui, G.; Nakamura, Y. Lignocellulosic enzymes from Flavodon flavus, a fungus isolated from Western Indian ocean off the coast of Dar Es Salaam, Tanzania. Afr. J. Biotechnol. 2008, 7, 3066–3072. [Google Scholar]
- Thirunavukkarasu, N.; Jahnes, B.; Broadstock, A.; Govinda Rajulu, M.B.; Murali, T.S.; Gopalan, V.; Suryanarayanan, T.S. Screening marine-derived endophytic fungi for xylan-degrading enzymes. Curr. Sci. 2015, 109, 112–120. [Google Scholar]
- Raghukumar, C.; Mohandass, C.; Kamat, S.; Shailaja, M.S. Simultaneous detoxification and decolorization of molasses spent wash by the immobilized white-rot fungus Flavodon flavus isolated from a marine habitat. Enzyme Microb. Technol. 2004, 35, 197–202. [Google Scholar] [CrossRef]
- Arunpanichlert, J.; Rukachaisirikul, V.; Phongpaichit, S.; Supaphon, O.; Sakayaroj, J. Xylariphilone: A new azaphilone derivative from the seagrass-derived fungus Xylariales sp. PSU-ES163. Nat. Prod. Res. 2016, 30, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Govers, L.L.; Man In ‘T Veld, W.A.; Meffert, J.P.; Bouma, T.J.; van Rijswick, P.C.J.; Heusinkveld, J.H.T.; Orth, R.J.; van Katwijk, M.M.; van der Heide, T. Marine Phytophthora species can hamper conservation and restoration of vegetated coastal ecosystems. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160812. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, J.; Liu, Q.; Wu, Q.; Chen, S.; Wang, J.; Li, J.; Liu, L.; Gao, Z. Cyclohexenone derivative and drimane sesquiterpenes from the seagrass-derived fungus Aspergillus insuetus. Chem. Biodivers. 2023, 20, e202300424. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Montesinos, B.; Diánez, F.; Moreno-Gavira, A.; Gea, F.J.; Santos, M. Plant growth promotion and biocontrol of Pythium ultimum by saline tolerant Trichoderma isolates under salinity stress. Int. J. Environ. Res. Public Health 2019, 16, 2053. [Google Scholar] [CrossRef]
- Sullivan, B.K.; Trevathan-Tackett, S.M.; Neuhauser, S.; Govers, L.L. Review: Host-pathogen dynamics of seagrass diseases under future global change. Mar. Pollut. Bull. 2018, 134, 75–88. [Google Scholar] [CrossRef]
- Hua Tan, M.; Loke, S.; Croft, L.J.; Gleason, F.H.; Lange, L.; Trevathan-Tackett, S.M. First genome of Labyrinthula, an opportunistic seagrass pathogen, reveals novel insight into marine protist phylogeny, ecology and CAZyme cell-wall degradation. Microb. Ecol. 2021, 82, 498–511. [Google Scholar] [CrossRef]
- Raghukumar, S. Ecology of the marine Protists, the Labyrinthulomycetes (Thraustochytrids and Labyrinthulids). Eur. J. Protistol. 2002, 38, 127–145. [Google Scholar] [CrossRef]
- Neuhauser, S.; Kirchmair, M.; Gleason, F.H. Ecological roles of the parasitic phytomyxids (plasmodiophorids) in marine ecosystems—A Review. Mar. Freshw. Res. 2011, 62, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Cooke, D.E.L.; Drenth, A.; Duncan, J.M.; Wagels, G.; Brasier, C.M. A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet. Biol. 2000, 30, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhu, Y.; Chen, J.; Li, C.; Gao, Z.; Li, J.; Liu, L. Sesquiterpenoids with phytotoxic and antifungal activities from a pathogenic fungus Aspergillus alabamensis. J. Agric. Food Chem. 2022, 70, 12065–12073. [Google Scholar] [CrossRef] [PubMed]
- Fry, W.E.; Grünwald, N.J. Introduction to Oomycetes. Plant Health Instr. 2010, 10. [Google Scholar] [CrossRef]
- Govers, L.L.; Van Der Zee, E.M.; Meffert, J.P.; Van Rijswick, P.C.J.; Man In T Veld, W.A.; Heusinkveld, J.H.T.; Van Der Heide, T. Copper treatment during storage reduces Phytophthora and Halophytophthora infection of Zostera marina seeds used for restoration. Sci. Rep. 2017, 7, 43172. [Google Scholar] [CrossRef]
- Menning, D.M.; Ward, D.H.; Wyllie-Echeverria, S.; Sage, G.K.; Gravley, M.C.; Gravley, H.A.; Talbot, S.L. Are migratory waterfowl vectors of seagrass pathogens? Ecol. Evol. 2020, 10, 2062–2073. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.R.; Randolph, K.C.; Osborn, S.L.; Tyler, H.L. Culture dependent and independent analysis of bacterial communities associated with commercial salad leaf vegetables. BMC Microbiol. 2013, 13, 274. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.R.; James, E.K.; Poole, P.S. The Plant Microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef]
- Ettinger, C.L.; Vann, L.E.; Eisen, J.A. Global diversity and biogeography of the Zostera marina mycobiome. Appl. Environ. Microbiol. 2021, 87, e02795-20. [Google Scholar] [CrossRef]
- Chen, J.; Zang, Y.; Yang, Z.; Qu, T.; Sun, T.; Liang, S.; Zhu, M.; Wang, Y.; Tang, X. Composition and functional diversity of epiphytic bacterial and fungal communities on marine macrophytes in an intertidal zone. Front. Microbiol. 2022, 13, 839465. [Google Scholar] [CrossRef]
- Banos, S.; Lentendu, G.; Kopf, A.; Wubet, T.; Glöckner, F.O.; Reich, M. A Comprehensive Fungi-specific 18S rRNA gene sequence primer toolkit suited for diverse research issues and sequencing platforms. BMC Microbiol. 2018, 18, 190. [Google Scholar] [CrossRef]
- Ishii, N.; Ishida, S.; Kagami, M. PCR Primers for assessing community structure of aquatic fungi including Chytridiomycota and Cryptomycota. Fungal Ecol. 2015, 13, 33–43. [Google Scholar] [CrossRef]
- Petersen, L.-E.; Marner, M.; Labes, A.; Tasdemir, D. Rapid metabolome and bioactivity profiling of fungi associated with the leaf and rhizosphere of the baltic seagrass Zostera marina. Mar. Drugs 2019, 17, 419. [Google Scholar] [CrossRef] [PubMed]
- Castro González, M.; Gómez-López, D.I. Endophyte mycobiota of Thalassia testudinum König in the Colombian Caribbean [Micobiota Endófita de Thalassia Testudinum König En El Caribe Colombiano]. Bol. Investig. Mar. Y Costeras 2022, 52, 185–192. [Google Scholar] [CrossRef]
- Bantoto-Kinamot, V.; Monotilla, A. New report of endophytic Sordariomycetes from the seagrasses of Cebu, Central Philippines. J. Trop. Life Sci. 2023, 13, 553–562. [Google Scholar] [CrossRef]
- Man in ’t Veld, W.A.; Rosendahl, K.C.H.M.; Brouwer, H.; de Cock, A.W.A.M. Phytophthora gemini sp. Nov., a new species isolated from the halophilic plant Zostera marina in the Netherlands. Fungal Biol. 2011, 115, 724–732. [Google Scholar] [CrossRef]
- Poli, A.; Bovio, E.; Ranieri, L.; Varese, G.C.; Prigione, V. News from the Sea: A new genus and seven new species in the pleosporalean families Roussoellaceae and Thyridariaceae. Diversity 2020, 12, 144. [Google Scholar] [CrossRef]
- Poli, A.; Bovio, E.; Verkley, G.; Prigione, V.; Varese, G.C. Elbamycella rosea gen. et sp. nov. (Juncigenaceae, Torpedosporales) isolated from the Mediterranean Sea. MycoKeys 2019, 55, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Yujiao, Z.; Junjie, C.; Jun, C.; Chunyuan, L.; Zhizeng, G.; Jing, L.; Lan, L. Discovery of novel bactericides from Aspergillus alabamensis and their antibacterial activity against fish pathogens. J. Agric. Food Chem. 2023, 71, 4298–4305. [Google Scholar] [CrossRef] [PubMed]
- Camp, E.F.; Suggett, D.J.; Gendron, G.; Jompa, J.; Manfrino, C.; Smith, D.J. Mangrove and seagrass beds provide different biogeochemical services for corals threatened by climate change. Front. Mar. Sci. 2016, 3, 52. [Google Scholar] [CrossRef]
- Viana, I.G.; Artika, S.R.; Moreira-Saporiti, A.; Teichberg, M. Limited trait responses of a tropical seagrass to the combination of increasing pCO2 and Warming. J. Exp. Bot. 2023, 74, 472–488. [Google Scholar] [CrossRef] [PubMed]
- Tisthammer, K.H.; Cobian, G.M.; Amend, A.S. Global biogeography of marine fungi is shaped by the environment. Fungal Ecol. 2016, 19, 39–46. [Google Scholar] [CrossRef]
- Hurtado-McCormick, V.; Kahlke, T.; Petrou, K.; Jeffries, T.; Ralph, P.J.; Seymour, J.R. Regional and microenvironmental scale characterization of the Zostera muelleri seagrass microbiome. Front. Microbiol. 2019, 10, 1011. [Google Scholar] [CrossRef]
- Liu, S.; Trevathan-Tackett, S.M.; Jiang, Z.; Cui, L.; Wu, Y.; Zhang, X.; Li, J.; Luo, H.; Huang, X. Nutrient loading decreases blue carbon by mediating fungi activities within seagrass meadows. Environ. Res. 2022, 212, 113280. [Google Scholar] [CrossRef]
- Bongiorni, L. Thraustochytrids, a neglected component of organic matter decomposition and food webs in marine sediments. In Biology of Marine Fungi; Raghukumar, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–13. [Google Scholar]
- Menaa, F.; Wijesinghe, P.A.U.I.; Thiripuranathar, G.; Uzair, B.; Iqbal, H.; Khan, B.A.; Menaa, B. Ecological and industrial implications of dynamic seaweed-associated microbiota interactions. Mar. Drugs 2020, 18, 641. [Google Scholar] [CrossRef]
- Hebert, T.A.; Kuehn, K.A.; Halvorson, H.M. Land use differentially alters microbial interactions and detritivore feeding during leaf decomposition in headwater streams. Freshw. Biol. 2023, 68, 1386–1399. [Google Scholar] [CrossRef]
- Siro, G.; Pipite, A. Mini-Review on the antimicrobial potential of actinobacteria associated with seagrasses. Explor. Drug Sci. 2024, 2, 117–125. [Google Scholar] [CrossRef]
- Boontanom, P.; Chantarasiri, A. Short Communication: Diversity of culturable epiphytic bacteria isolated from seagrass (Halodule uninervis) in Thailand and their preliminary antibacterial activity. Biodiversitas J. Biol. Divers. 2020, 21, 2907–2913. [Google Scholar] [CrossRef]
- Srinivasan, R.; Kannappan, A.; Shi, C.; Lin, X. Marine bacterial secondary metabolites: A treasure house for structurally unique and effective antimicrobial compounds. Mar. Drugs 2021, 19, 530. [Google Scholar] [CrossRef]
- Nerva, L.; Ciuffo, M.; Vallino, M.; Margaria, P.; Varese, G.C.; Gnavi, G.; Turina, M. Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res. 2016, 219, 22–38. [Google Scholar] [CrossRef]
- Gono, C.M.P.; Ahmadi, P.; Hertiani, T.; Septiana, E.; Putra, M.Y.; Chianese, G. A comprehensive update on the bioactive compounds from seagrasses. Mar. Drugs 2022, 20, 406. [Google Scholar] [CrossRef]
- Vasarri, M.; De Biasi, A.M.; Barletta, E.; Pretti, C.; Degl’Innocenti, D. An overview of new insights into the benefits of the seagrass Posidonia oceanica for human health. Mar. Drugs 2021, 19, 476. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Sahoo, R.; Samantaray, S.S.; Apte, D.; Chowk, S.A.; Singh, S.B. Seagrass ecosystems of India as bioindicators of trace elements. In Coastal Ecosystems; Madhav, S., Nazneen, S., Singh, P., Eds.; Springer: Cham, Switzerland, 2020; Volume 38, pp. 45–65. [Google Scholar]
- Stipcich, P.; Balmas, V.; Jimenez, C.E.; Oufensou, S.; Ceccherelli, G. Cultivable mycoflora on bleached, decaying and healthy Posidonia oceanica leaves in a warm-edge Mediterranean location. Mar. Environ. Res. 2023, 192, 106188. [Google Scholar] [CrossRef]
- The Seagrass Microbiome Project. Available online: https://seagrassmicrobiome.org/ (accessed on 11 July 2024).
- Han, Q.; Qiu, C.; Zeng, W.; Chen, S.; Zhao, M.; Shi, Y.; Zhang, X. Sediment carbon sequestration and driving factors in seagrass beds from Hainan Island and the Xisha Islands. Processes 2023, 11, 456. [Google Scholar] [CrossRef]
- Fu, C.; Frappi, S.; Havlik, M.N.; Howe, W.; Harris, S.D.; Laiolo, E.; Gallagher, A.J.; Masqué, P.; Duarte, C.M. Substantial blue carbon sequestration in the world’s largest seagrass meadow. Commun. Earth Environ. 2023, 4, 474. [Google Scholar] [CrossRef]
- Behera, A.D.; Das, S. Ecological Insights and Potential Application of Marine Filamentous Fungi in Environmental Restoration. Rev. Environ. Sci. Biotechnol. 2023, 22, 281–318. [Google Scholar] [CrossRef]
- Noorjahan, A.; Aiyamperumal, B.; Anantharaman, P. Fungal endophytes from seaweeds and bio-potential applications in agriculture. In Fungi Bio-Prospects in Sustainable Agriculture, Environment and Nano-Technology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 83–95. [Google Scholar]
- Pang, K.-L.; Jones, E.B.G.; Abdel-Wahab, M.A.; Adams, S.J.; Alves, A.; Azevedo, E.; Bahkali, A.H.; Barata, M.; Burgaud, G.; Caeiro, M.F.; et al. Recent progress in marine mycological research in different countries, and prospects for future developments worldwide. Bot. Mar. 2023, 66, 239–269. [Google Scholar] [CrossRef]
- dela Cruz, T.E.E.; Notarte, K.I.R.; Apurillo, C.C.S.; Tarman, K.; Bungihan, M.E. Biomining fungal endophytes from tropical plants and seaweeds for drug discovery. In Biodiversity and Biomedicine: Our Future; Elsevier: Amsterdam, The Netherlands, 2020; pp. 51–62. ISBN 9780128195413. [Google Scholar]
- Tarman, K. Marine Fungi as a Source of Natural Products. In Encyclopedia of Marine Biotechnology; Wiley: Hoboken, NJ, USA, 2020; pp. 2147–2160. ISBN 9781119143802. [Google Scholar]
- Varrella, S.; Barone, G.; Tangherlini, M.; Rastelli, E.; Dell’Anno, A.; Corinaldesi, C. Diversity, ecological role and biotechnological potential of Antarctic marine fungi. J. Fungi 2021, 7, 391. [Google Scholar] [CrossRef]
- Bhatia, S.; Makkar, R.; Behl, T.; Sehgal, A.; Singh, S.; Rachamalla, M.; Mani, V.; Iqbal, M.S.; Bungau, S.G. Biotechnological innovations from ocean: Transpiring role of marine drugs in management of chronic disorders. Molecules 2022, 27, 1539. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qin, Y.; Lin, M.; Song, Y.; Lu, H.; Xu, X.; Liu, Y.; Zhou, X.; Gao, C.; Luo, X. Marine natural products from the Beibu Gulf: Sources, chemistry, and bioactivities. Mar. Drugs 2023, 21, 63. [Google Scholar] [CrossRef]
- Thatoi, H.; Behera, B.C.; Mishra, R.R. Ecological role and biotechnological potential of mangrove fungi: A Review. Mycology 2013, 4, 54–71. [Google Scholar] [CrossRef]
- Chen, H.Y.; Xue, D.S.; Feng, X.Y.; Yao, S.J. Screening and production of ligninolytic enzyme by a marine-derived fungal Pestalotiopsis sp. J63. Appl. Biochem. Biotechnol. 2011, 165, 1754–1769. [Google Scholar] [CrossRef]
- Morlighem, J.É.R.L.; Radis-Baptista, G. The place for enzymes and biologically active peptides from marine organisms for application in industrial and pharmaceutical biotechnology. Curr. Protein Pept. Sci. 2019, 20, 334–355. [Google Scholar] [CrossRef]
- Poli, A.; Bovio, E.; Perugini, I.; Varese, G.C.; Prigione, V. Corollospora mediterranea: A novel species complex in the Mediterranean Sea. Appl. Sci. 2021, 11, 5452. [Google Scholar] [CrossRef]
- Heckwolf, M.J.; Peterson, A.; Jänes, H.; Horne, P.; Künne, J.; Liversage, K.; Sajeva, M.; Reusch, T.B.H.; Kotta, J. From ecosystems to socio-economic benefits: A systematic review of coastal ecosystem services in the Baltic Sea. Sci. Total Environ. 2021, 755, 142565. [Google Scholar] [CrossRef]
- Setiawan, E.; Chodiantoro, M.R.; Insany, G.F.; Subagio, I.B.; Dewi, N.K.; Muzaki, F.K. Short Communication: Diversity of sponges associated in seagrass meadows at coastal area of Pacitan District, East Java, Indonesia. Biodiversitas J. Biol. Divers. 2021, 22, 3105–3112. [Google Scholar] [CrossRef]
- Hossain, M.S.; Hashim, M. Potential of Earth Observation (EO) technologies for seagrass ecosystem service assessments. Int. J. Appl. Earth Obs. Geoinf. 2019, 77, 15–29. [Google Scholar] [CrossRef]
- Kamal, A.H.M.; Asif, A.-A.; Idris, M.H.; Bhuiyan, M.K.A.; Rahman, A.A. Trends in seagrass research and conservation in Malaysian waters. J. Trop. Life Sci. 2023, 13, 93–114. [Google Scholar] [CrossRef]
- Pham, T.D.; Ha, N.T.; Saintilan, N.; Skidmore, A.; Phan, D.C.; Le, N.N.; Viet, H.L.; Takeuchi, W.; Friess, D.A. Advances in earth observation and machine learning for quantifying blue carbon. Earth Sci. Rev. 2023, 243, 104501. [Google Scholar] [CrossRef]
- Araya-Lopez, R.; de Paula Costa, M.D.; Wartman, M.; Macreadie, P.I. Trends in the application of remote sensing in blue carbon science. Ecol. Evol. 2023, 13, e10559. [Google Scholar] [CrossRef]
- Ivajnšič, D.; Orlando-Bonaca, M.; Donša, D.; Grujić, V.J.; Trkov, D.; Mavrič, B.; Lipej, L. Evaluating seagrass meadow dynamics by integrating field-based and remote sensing techniques. Plants 2022, 11, 1196. [Google Scholar] [CrossRef]
- Zoffoli, M.L.; Gernez, P.; Oiry, S.; Godet, L.; Dalloyau, S.; Davies, B.F.R.; Barillé, L. Remote sensing in seagrass ecology: Coupled dynamics between migratory herbivorous birds and intertidal meadows observed by satellite during four decades. Remote Sens. Ecol. Conserv. 2023, 9, 420–433. [Google Scholar] [CrossRef]
- Picek, L.; Šulc, M.; Matas, J.; Heilmann-Clausen, J.; Jeppesen, T.S.; Lind, E. Automatic fungi recognition: Deep Learning meets Mycology. Sensors 2022, 22, 633. [Google Scholar] [CrossRef]
- Rahman, M.A.; Clinch, M.; Reynolds, J.; Dangott, B.; Meza Villegas, D.M.; Nassar, A.; Hata, D.J.; Akkus, Z. Classification of fungal genera from microscopic images using Artificial Intelligence. J. Pathol. Inform. 2023, 14, 100314. [Google Scholar] [CrossRef]
- Ghosh, S.; Dasgupta, R. Machine Learning in Biological Sciences; Springer Nature: Singapore, 2022; ISBN 978-981-16-8880-5. [Google Scholar]
- Basheer, M.A.; Abutaleb, K.; Abed, N.N.; Mekawey, A.A.I. Mycosynthesis of silver nanoparticles using marine fungi and their antimicrobial activity against pathogenic microorganisms. J. Genet. Eng. Biotechnol. 2023, 21, 127. [Google Scholar] [CrossRef]
- Rai, M.; Wypij, M.; Trzcińska-Wencel, J.; Yadav, A.; Ingel, I.P.; Avila-Quezada, G.D.; Golińska, P. Myconanotechnology: Opportunities and Challenges. In Myconanotechnology, 1st ed.; Rai, M., Golinska, P., Eds.; CRC Press: Boca Raton, FL, USA, 2023; p. 23. [Google Scholar]
- Suresh, J.I.; Lydia, N.J.; Atchayadana, U.; Mubina, J. Marine Endophytic Fungi Isolated from Gulf of Mannar—A Source for New Generation of Pharmaceutical Drugs and Biosynthesis of Silver Nanoparticles and Its Antibacterial Efficacy. In Fungi Bio-Prospects in Sustainable Agriculture, Environment and Nano-Technology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 175–185. [Google Scholar]
- Abdelrahman, S.E.S.A.H.; El Hawary, S.; Mohsen, E.; El Raey, M.A.; Selim, H.M.R.M.; Hamdan, A.M.E.; Ghareeb, M.A.; Hamed, A.A. Bio-fabricated zinc oxide nanoparticles mediated by endophytic fungus Aspergillus sp. SA17 with antimicrobial and anticancer activities: In vitro supported by in silico studies. Front. Microbiol. 2024, 15, 1366614. [Google Scholar] [CrossRef] [PubMed]
- Rina, D.Y.; Rozirwan, R.; Hendri, M. Karakteristik Jamur Endofit Pada Lamun Enhalus acoroides di Teluk Hurun Lampung Selatan. Ph.D. Thesis, Universitas Sriwijaya, Kota Palembang, Indonesia, 2018. [Google Scholar]
- Looney-Patterson, K. (University of Washington, Seattle, WA, USA). Diversity of Fungal Endophytes from Zostera marina eelgrass in False Bay Biological Preserve. 2021; (Unpublished Work). [Google Scholar]
- Kinamot, V.; Monotilla, A. Colonization and antagonistic activity of endophytic fungi in seagrasses: Understanding endophyte interactions. 2022; preprint. [Google Scholar] [CrossRef]
- Soperová, B. The Mycobiomes of Vegetative Organs of the Dominant Mediterranean Seagrass Posidonia oceanica (Posidoniaceae, Alismatales). Master’s Thesis, Univerzita Karlova, Staré Město, Czech Republic, 2022. [Google Scholar]
- Torta, L.; Burruano, S.; Giambra, S.; Conigliaro, G.; Piazza, G.; Mirabile, G.; Pirrotta, M.; Calvo, R.; Bellissimo, G.; Calvo, S.; et al. Cultivable fungal endophytes in roots, rhizomes and leaves of Posidonia oceanica (L.) Delile along the coast of Sicily, Italy. Plants 2022, 11, 1139. [Google Scholar] [CrossRef]
- Vohník, M. Bioerosion and fungal colonization of the invasive foraminiferan Amphistegina lobifera in a Mediterranean seagrass meadow. Biogeosciences 2021, 18, 2777–2790. [Google Scholar] [CrossRef]
- Suryanti, S.; Anggoro, S.; Sabdaningsih, A.; Febrianto, S.; Ayuningrum, D. Multi-temporal mapping and recent structures of seagrass community in Panjang Island. AACL Bioflux 2022, 15, 365–376. [Google Scholar]
- Htay, S.A.M.; Linn, K.T. The effects of pH and fermentation media on the antibacterial activity of secondary metabolite producing from marine derived fungi. J. Myanmar Acad. Arts Sci. 2021, 19, 4B. [Google Scholar]
- Newell, S. Fungi and bacteria in or on leaves of Eelgrass (Zostera marina L.) from Chesapeake Bay. Appl. Environ. Microbiol. 1981, 41, 1219–1224. [Google Scholar] [CrossRef]
- Hyde, K.D.; Jones, E.B.G. Ecological observations on marine fungi from the Seychelles. Bot. J. Linn. Soc. 1989, 100, 237–254. [Google Scholar] [CrossRef]
- Panno, L.; Voyron, S.; Anastasi, A.; Mussat Sartor, R.; Varese, G.C. Biodiversity of marine fungi associated with the seagrass Posidonia oceanica. Biol. Mar. Mediterr. 2011, 18, 85–88. [Google Scholar]
- Poli, A.; Bovio, E.; Ranieri, L.; Varese, G.C.; Prigione, V. Fungal diversity in the Neptune Forest: Comparison of the mycobiota of Posidonia oceanica, Flabellia petiolata, and Padina pavonica. Front. Microbiol. 2020, 11, 933. [Google Scholar] [CrossRef]
- Poli, A.; Vizzini, A.; Prigione, V.; Varese, G.C. Basidiomycota isolated from the Mediterranean Sea-Phylogeny and putative ecological roles. Fungal Ecol. 2018, 36, 51–62. [Google Scholar] [CrossRef]
- Atalla, M.M.; Zeinab, H.K.; Eman, R.H.; Amani, A.Y.; Abeer, A.A.E.A. Screening of some marine-derived fungal isolates for lignin degrading enzymes (LDEs) production. Agric. Biol. J. N. Am. 2010, 1, 591–599. [Google Scholar]
- Khatiwada, S.S. Prevalence and Distribution of Marine Phytophthora and Halophytophthora in Seagrass Habitats in Victoria, Australia. Master’s Thesis, Deakin University, Victoria, Australia, 2019. [Google Scholar]
- Man in ’t Veld, W.A.; Rosendahl, K.C.H.M.; van Rijswick, P.C.J.; Meffert, J.P.; Boer, E.; Westenberg, M.; van der Heide, T.; Govers, L.L. Multiple Halophytophthora spp. and Phytophthora spp. Including P. gemini, P. inundata and P. chesapeakensis sp. nov. isolated from the seagrass Zostera marina in the Northern Hemisphere. Eur. J. Plant Pathol. 2019, 153, 341–357. [Google Scholar] [CrossRef]
- Menning, D.M.; Gravley, H.A.; Cady, M.N.; Pepin, D.; Wyllie-Echeverria, S.; Ward, D.H.; Talbot, S.L. Metabarcoding of environmental samples suggest wide distribution of Eelgrass (Zostera marina) pathogens in the North Pacific. Metabarcoding Metagenom 2021, 5, 35–42. [Google Scholar] [CrossRef]
- Proctor, T.; Elliott, J. Undescribed oomycete pathogens on Zostera marina and Z. japonica in the Puget Sound. Summer Research, 353, University of Puget Sound. 2019. Available online: https://www.jstor.org/stable/community.36709228 (accessed on 21 August 2024).
- Wainwright, B.J.; Zahn, G.L.; Arlyza, I.S.; Amend, A.S. Seagrass-associated fungal communities follow Wallace’s Line, but host genotype does not structure fungal community. J. Biogeogr. 2018, 45, 762–770. [Google Scholar] [CrossRef]
- Pan, Y.; Li, G.; Su, L.; Zheng, P.; Wang, Y.; Shen, Z.; Chen, Z.; Han, Q.; Gong, J. Seagrass colonization alters diversity, abundance, taxonomic, and functional community structure of benthic microbial eukaryotes. Front. Microbiol. 2022, 13, 901741. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).