Genetically Improved Yeast Strains with Lower Ethanol Yield for the Wine Industry Generated Through a Two-Round Breeding Program
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Populations and Breeding Program Design
2.2. Microscale Fermentations with Synthetic Must
2.3. SO2 Resistance Test
2.4. Pilot-Scale Fermentations with Natural Grape Must
2.5. Sensory Analysis
3. Results
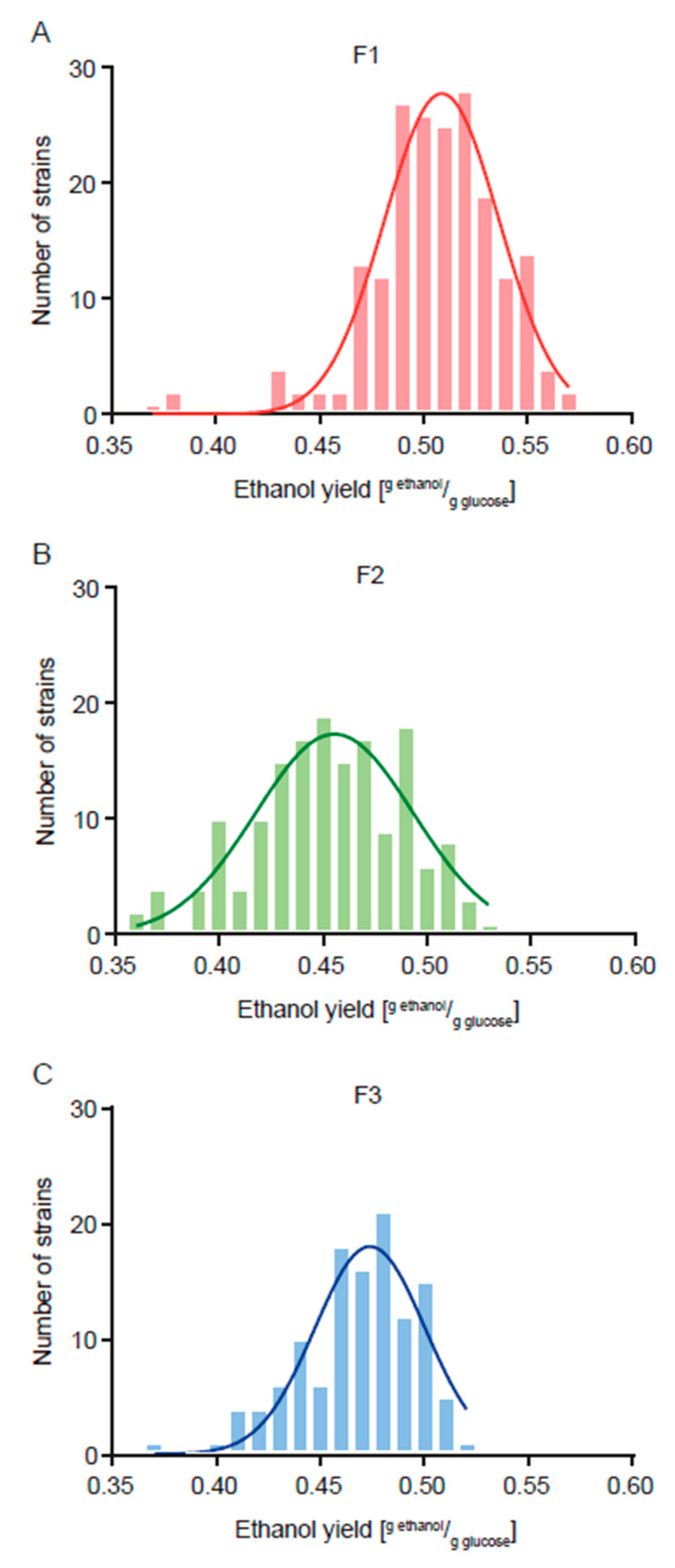
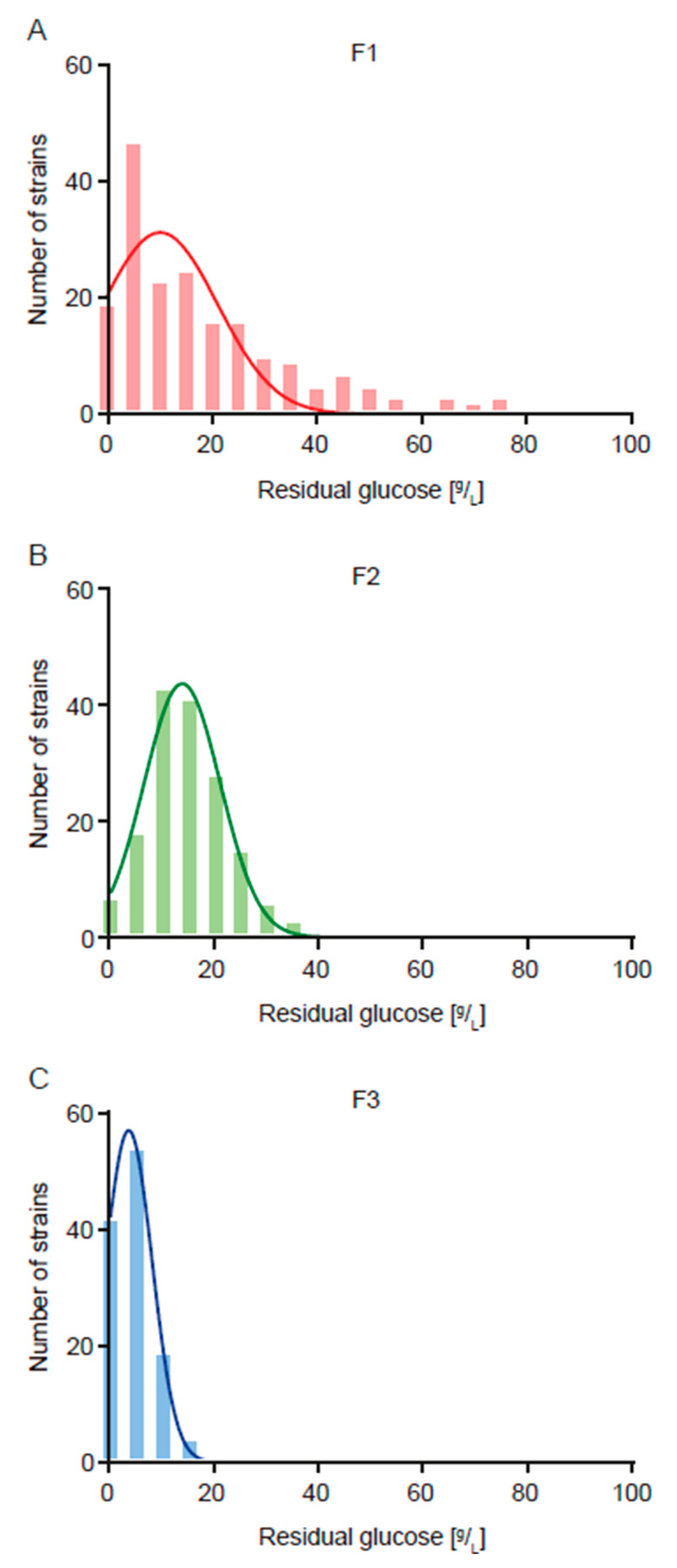
3.1. Two-Round Breeding Program to Obtain Strains with Less Ethanol Yield
3.2. Improved Strains with Applied Potential for the Wine Industry
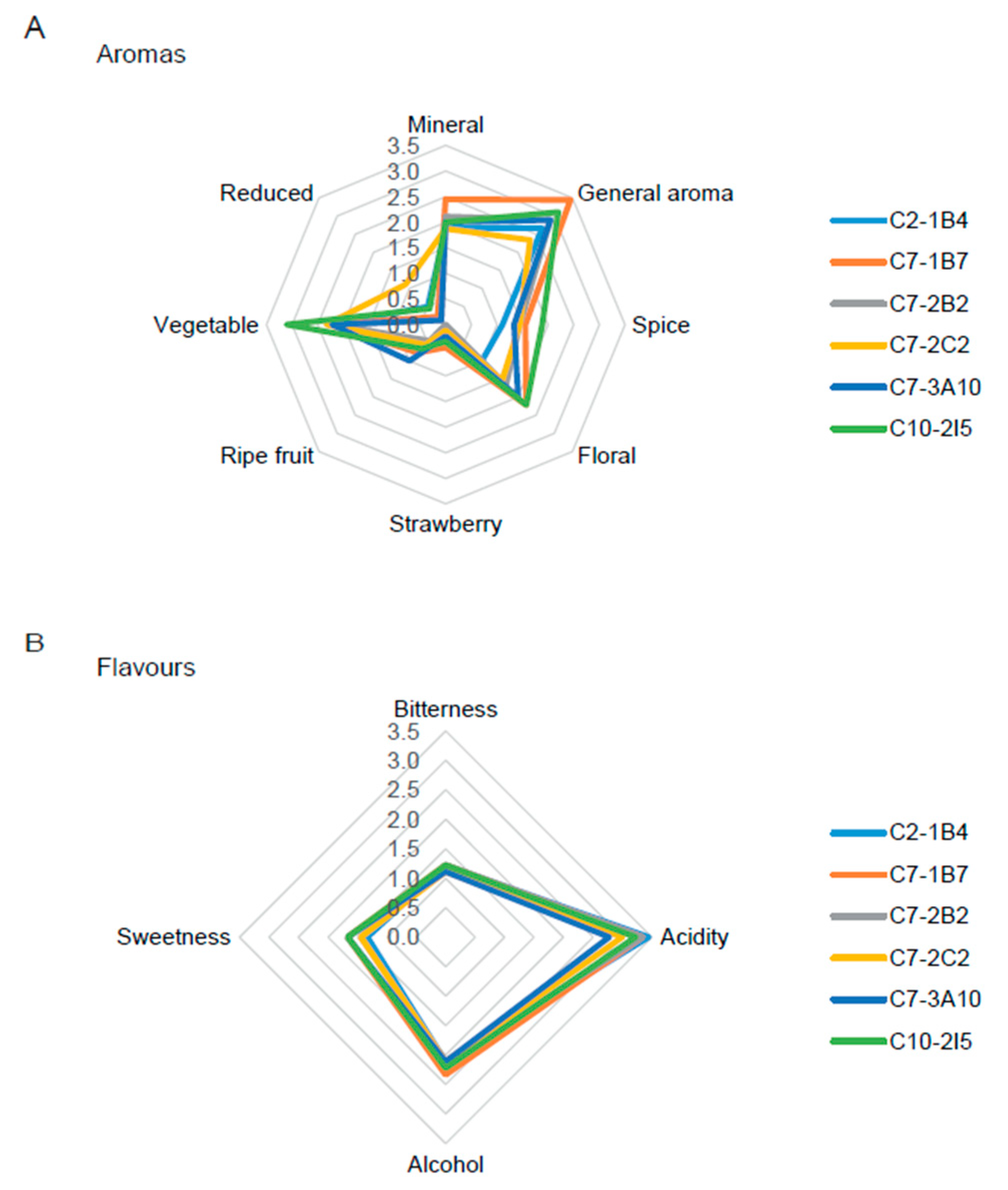
3.3. Improved Strains Able to Produce Wines with Commercial Potential
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pretorius, I.S. Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast 2000, 16, 675–729. [Google Scholar] [CrossRef]
- Marsit, S.; Dequin, S. Diversity and adaptive evolution of Saccharomyces wine yeast: A review. FEMS Yeast Res. 2015, 15, fov067. [Google Scholar] [CrossRef]
- Dequin, S.; Casaregola, S. The genomes of fermentative Saccharomyces. Comptes. Rendus Biol. 2011, 334, 687–693. [Google Scholar] [CrossRef]
- Querol, A. Adaptive evolution of wine yeast. Int. J. Food Microbiol. 2003, 86, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.V.; White, M.A.; Cooper, O.R.; Storchmann, K. Climate Change and Global Wine Quality. Clim. Change 2005, 73, 319–343. [Google Scholar] [CrossRef]
- Tilloy, V.; Ortiz-Julien, A.; Dequin, S. Reduction of ethanol yield and improvement of glycerol formation by adaptive evolution of the wine yeast Saccharomyces cerevisiae under hyperosmotic conditions. Appl. Environ. Microbiol. 2014, 80, 2623–2632. [Google Scholar] [CrossRef] [PubMed]
- Buescher, W.A.; Siler, C.E.; Morris, J.R.; Threlfall, R.T.; Main, G.L.; Cone, G.C. High Alcohol Wine Production from Grape Juice Concentrates. Am. J. Enol. Vitic. 2001, 52, 345–351. [Google Scholar] [CrossRef]
- Escudero, A.; Campo, E.; Farina, L.; Cacho, J.; Ferreira, V. Analytical characterization of the aroma of five premium red wines. Insights into the role of odor families and the concept of fruitiness of wines. J. Agric. Food Chem. 2007, 55, 4501–4510. [Google Scholar] [CrossRef]
- Gutierrez-Gamboa, G.; Zheng, W.; Martinez de Toda, F. Strategies in vineyard establishment to face global warming in viticulture: A mini review. J. Sci. Food Agric. 2021, 101, 1261–1269. [Google Scholar] [CrossRef] [PubMed]
- Schmidtke, L.M.; Blackman, J.W.; Agboola, S.O. Production technologies for reduced alcoholic wines. J. Food Sci. 2012, 77, R25–R41. [Google Scholar] [CrossRef] [PubMed]
- Varela, C.; Dry, P.R.; Kutyna, D.R.; Francis, I.L.; Henschke, P.A.; Curtin, C.D.; Chambers, P.J. Strategies for reducing alcohol concentration in wine. Aust. J. Grape Wine Res. 2015, 21, 670–679. [Google Scholar] [CrossRef]
- Zhu, X.; Torija, M.J.; Mas, A.; Beltran, G.; Navarro, Y. Effect of a Multistarter Yeast Inoculum on Ethanol Reduction and Population Dynamics in Wine Fermentation. Foods 2021, 10, 623. [Google Scholar] [CrossRef]
- Kessi-Perez, E.I.; Molinet, J.; Garcia, V.; Aguilera, O.; Cepeda, F.; Lopez, M.E.; Sari, S.; Cuello, R.; Ciklic, I.; Rojo, M.C.; et al. Generation of a Non-Transgenic Genetically Improved Yeast Strain for Wine Production from Nitrogen-Deficient Musts. Microorganisms 2020, 8, 1194. [Google Scholar] [CrossRef]
- Varela, J.; Varela, C. Microbiological strategies to produce beer and wine with reduced ethanol concentration. Curr. Opin. Biotechnol. 2019, 56, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Contreras, A.; Hidalgo, C.; Henschke, P.A.; Chambers, P.J.; Curtin, C.; Varela, C. Evaluation of non-Saccharomyces yeasts for the reduction of alcohol content in wine. Appl. Environ. Microbiol. 2014, 80, 1670–1678. [Google Scholar] [CrossRef] [PubMed]
- Tilloy, V.; Cadiere, A.; Ehsani, M.; Dequin, S. Reducing alcohol levels in wines through rational and evolutionary engineering of Saccharomyces cerevisiae. Int. J. Food Microbiol. 2015, 213, 49–58. [Google Scholar] [CrossRef]
- Rozas, P.; Kessi-Perez, E.I.; Martinez, C. Genetically modified organisms: Adapting regulatory frameworks for evolving genome editing technologies. Biol. Res. 2022, 55, 31. [Google Scholar] [CrossRef] [PubMed]
- Hart, R.S.; Jolly, N.P.; Ndimba, B.K. Characterisation of hybrid yeasts for the production of varietal Sauvignon blanc wine—A review. J. Microbiol. Methods 2019, 165, 105699. [Google Scholar] [CrossRef]
- Marullo, P.; Bely, M.; Masneuf-Pomarede, I.; Pons, M.; Aigle, M.; Dubourdieu, D. Breeding strategies for combining fermentative qualities and reducing off-flavor production in a wine yeast model. FEMS Yeast Res. 2006, 6, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Marullo, P.; Mansour, C.; Dufour, M.; Albertin, W.; Sicard, D.; Bely, M.; Dubourdieu, D. Genetic improvement of thermo-tolerance in wine Saccharomyces cerevisiae strains by a backcross approach. FEMS Yeast Res. 2009, 9, 1148–1160. [Google Scholar] [CrossRef]
- Perez-Torrado, R.; Barrio, E.; Querol, A. Alternative yeasts for winemaking: Saccharomyces non-cerevisiae and its hybrids. Crit. Rev. Food Sci. Nutr. 2018, 58, 1780–1790. [Google Scholar] [CrossRef] [PubMed]
- Peris, D.; Perez-Torrado, R.; Hittinger, C.T.; Barrio, E.; Querol, A. On the origins and industrial applications of Saccharomyces cerevisiae x Saccharomyces kudriavzevii hybrids. Yeast 2018, 35, 51–69. [Google Scholar] [CrossRef] [PubMed]
- Lairon-Peris, M.; Perez-Traves, L.; Muniz-Calvo, S.; Guillamon, J.M.; Heras, J.M.; Barrio, E.; Querol, A. Differential Contribution of the Parental Genomes to a S. cerevisiae x S. uvarum Hybrid, Inferred by Phenomic, Genomic, and Transcriptomic Analyses, at Different Industrial Stress Conditions. Front. Bioeng. Biotechnol. 2020, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Contreras, A.; Garcia, V.; Salinas, F.; Urzua, U.; Ganga, M.A.; Martinez, C. Identification of genes related to nitrogen uptake in wine strains of Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2012, 28, 1107–1113. [Google Scholar] [CrossRef] [PubMed]
- Garcia, V.; Rivera, J.; Contreras, A.; Ganga, M.A.; Martinez, C. Development and characterization of hybrids from native wine yeasts. Braz. J. Microbiol. 2012, 43, 482–489. [Google Scholar] [CrossRef][Green Version]
- Bradbury, J.E.; Richards, K.D.; Niederer, H.A.; Lee, S.A.; Rod Dunbar, P.; Gardner, R.C. A homozygous diploid subset of commercial wine yeast strains. Antonie Van Leeuwenhoek 2006, 89, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Wills, C. Abundant microsatellite polymorphism in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc. Natl. Acad. Sci. USA 1998, 95, 1647–1652. [Google Scholar] [CrossRef]
- Rex, F.; Hirschler, A.; Scharfenberger-Schmeer, M. SSR-Marker Analysis—A Method for S. cerevisiae Strain Characterization and Its Application for Wineries. Fermentation 2020, 6, 101. [Google Scholar]
- Perez, M.A.; Gallego, F.J.; Martinez, I.; Hidalgo, P. Detection, distribution and selection of microsatellites (SSRs) in the genome of the yeast Saccharomyces cerevisiae as molecular markers. Lett. Appl. Microbiol. 2001, 33, 461–466. [Google Scholar] [CrossRef]
- Johnson, D.L.; Thompson, R. Restricted Maximum Likelihood Estimation of Variance Components for Univariate Animal Models Using Sparse Matrix Techniques and Average Information. J. Dairy. Sci. 1995, 78, 449–456. [Google Scholar] [CrossRef]
- Gilmour, A.; Gogel, B.; Cullisand, B.; Thompson, R. ASReml User Guide Release 3.0; VSN International Ltd.: Hemel Hempstead, UK, 2009. [Google Scholar]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics, 4th ed.; Longmans Green: Harlow, UK, 1996. [Google Scholar]
- Kruuk, L.E. Estimating genetic parameters in natural populations using the “animal model”. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004, 359, 873–890. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Walsh, B. Genetics and Analysis of Quantitative Traits; Sinauer Associates: Sunderland, MA, USA, 1998. [Google Scholar]
- Kessi-Perez, E.I.; Araos, S.; Garcia, V.; Salinas, F.; Abarca, V.; Larrondo, L.F.; Martinez, C.; Cubillos, F.A. RIM15 antagonistic pleiotropy is responsible for differences in fermentation and stress response kinetics in budding yeast. FEMS Yeast Res. 2016, 16, fow021. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kessi-Perez, E.I.; Salinas, F.; Gonzalez, A.; Su, Y.; Guillamon, J.M.; Hall, M.N.; Larrondo, L.F.; Martinez, C. KAE1 Allelic Variants Affect TORC1 Activation and Fermentation Kinetics in Saccharomyces cerevisiae. Front. Microbiol. 2019, 10, 1686. [Google Scholar] [CrossRef]
- Bely, M.; Sablayrolles, J.M.; Barre, P. Description of Alcoholic Fermentation Kinetics: Its Variability and Significance. Am. J. Enol. Vitic. 1990, 41, 319–324. [Google Scholar] [CrossRef]
- Rossignol, T.; Dulau, L.; Julien, A.; Blondin, B. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast 2003, 20, 1369–1385. [Google Scholar] [CrossRef]
- Nissen, T.L.; Schulze, U.; Nielsen, J.; Villadsen, J. Flux distributions in anaerobic, glucose-limited continuous cultures of Saccharomyces cerevisiae. Microbiology 1997, 143 Pt 1, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Kessi-Perez, E.I.; Acuna, E.; Bastias, C.; Fundora, L.; Villalobos-Cid, M.; Romero, A.; Khaiwal, S.; De Chiara, M.; Liti, G.; Salinas, F.; et al. Single nucleotide polymorphisms associated with wine fermentation and adaptation to nitrogen limitation in wild and domesticated yeast strains. Biol. Res. 2023, 56, 43. [Google Scholar] [CrossRef] [PubMed]
- Kessi-Perez, E.I.; Ponce, B.; Li, J.; Molinet, J.; Baeza, C.; Figueroa, D.; Bastias, C.; Gaete, M.; Liti, G.; Diaz-Barrera, A.; et al. Differential Gene Expression and Allele Frequency Changes Favour Adaptation of a Heterogeneous Yeast Population to Nitrogen-Limited Fermentations. Front. Microbiol. 2020, 11, 1204. [Google Scholar] [CrossRef] [PubMed]
- Martinez, C.; Cosgaya, P.; Vasquez, C.; Gac, S.; Ganga, A. High degree of correlation between molecular polymorphism and geographic origin of wine yeast strains. J. Appl. Microbiol. 2007, 103, 2185–2195. [Google Scholar] [CrossRef] [PubMed]
- Martinez, C.; Gac, S.; Lavin, A.; Ganga, M. Genomic characterization of Saccharomyces cerevisiae strains isolated from wine-producing areas in South America. J. Appl. Microbiol. 2004, 96, 1161–1168. [Google Scholar] [CrossRef] [PubMed]
- Querol, A.; Barrio, E.; Huerta, T.; Ramon, D. Molecular monitoring of wine fermentations conducted by active dry yeast strains. Appl. Environ. Microbiol. 1992, 58, 2948–2953. [Google Scholar] [CrossRef]
- Schwartz, D.C.; Cantor, C.R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell 1984, 37, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Lawless, H.T.; Heymann, H. Sensory Evaluation of Food: Principles and Practices; Springer Science & Business Media: New York, NY, USA, 2010. [Google Scholar]
- Noble, A.C.; Arnold, R.A.; Buechsenstein, J.; Leach, E.J.; Schmidt, J.O.; Stern, P.M. Modification of a Standarized System of Wine Aroma Terminology. Am. J. Enol. Vitic. 1987, 38, 143–146. [Google Scholar] [CrossRef]
- Colonna, A.E.; Adams, D.O.; Noble, A.C. Comparison of procedures for reducing astringency carry-over effects in evaluation of red wines. Aust. J. Grape Wine Res. 2004, 10, 26–31. [Google Scholar] [CrossRef]
- R-Core-Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: http://www.R-project.org/ (accessed on 17 January 2025).
- Marullo, P.; Bely, M.; Masneuf-Pomarede, I.; Aigle, M.; Dubourdieu, D. Inheritable nature of enological quantitative traits is demonstrated by meiotic segregation of industrial wine yeast strains. FEMS Yeast Res. 2004, 4, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Ambroset, C.; Petit, M.; Brion, C.; Sanchez, I.; Delobel, P.; Guerin, C.; Chiapello, H.; Nicolas, P.; Bigey, F.; Dequin, S.; et al. Deciphering the molecular basis of wine yeast fermentation traits using a combined genetic and genomic approach. G3 2011, 1, 263–281. [Google Scholar] [CrossRef]
- Salinas, F.; Cubillos, F.A.; Soto, D.; Garcia, V.; Bergstrom, A.; Warringer, J.; Ganga, M.A.; Louis, E.J.; Liti, G.; Martinez, C. The genetic basis of natural variation in oenological traits in Saccharomyces cerevisiae. PLoS ONE 2012, 7, e49640. [Google Scholar] [CrossRef] [PubMed]
- Capece, A.; Pietrafesa, R.; Siesto, G.; Romano, P. Biotechnological Approach Based on Selected Saccharomyces cerevisiae Starters for Reducing the Use of Sulfur Dioxide in Wine. Microorganisms 2020, 8, 738. [Google Scholar] [CrossRef]
- AWRI. Predicting alcohol levels. Grapegrow. Winemak. 2016, 626, 68. [Google Scholar]
- Hidalgo Togores, J. Tratado de Enología, 2nd ed.; Ediciones Mundi-Prensa: Madrid, Spain, 2011. [Google Scholar]
- European Union. Publication of a communication of approval of a standard amendment to the product specification for a name in the wine sector referred to in Article 17(2) and (3) of Commission Delegated Regulation (EU) 2019/33. Off. J. Eur. Union. 2022, 381, 25–35. [Google Scholar]
- Gagneur, J.; Stegle, O.; Zhu, C.; Jakob, P.; Tekkedil, M.M.; Aiyar, R.S.; Schuon, A.K.; Pe’er, D.; Steinmetz, L.M. Genotype-environment interactions reveal causal pathways that mediate genetic effects on phenotype. PLoS Genet. 2013, 9, e1003803. [Google Scholar] [CrossRef] [PubMed]
- Granek, J.A.; Magwene, P.M. Environmental and genetic determinants of colony morphology in yeast. PLoS Genet. 2010, 6, e1000823. [Google Scholar] [CrossRef] [PubMed]

| First Round of Breeding (Ethanol Yield) | Second Round of Breeding (Residual Glucose) | |
|---|---|---|
| Phenotypic variance (VP) | 0.00107 ± 0.00013 | 59.691 ± 11.055 |
| Additive genetic variance (VA) | 0.00066 ± 0.00024 | 56.345 ± 21.304 |
| Residual variance (VR) | 0.00041 ± 0.00016 | 3.346 ± 11.544 |
| Heritability (h2) | 0.619 ± 0.173 * | 0.944 ± 0.202 * |
| Expected response to selection (G) | 0.030 | 12.62 |
| Percentage of response to selection (%G) | 5.8% | 86.4% |
| Population | Ethanol Yield ± SD (g Ethanol/g Sugar) | Residual Glucose ± SD (g/L) |
|---|---|---|
| F1 | 0.506 ± 0.032 a | 19.7 ± 18.4 a |
| F2 | 0.452 ± 0.036 c | 14.6 ± 7.7 b |
| F3 | 0.468 ± 0.028 b | 4.3 ± 3.9 c |
| Strain | Ethanol Yield (g Ethanol/g Sugar) | Residual Glucose (g/L) | Relative SO2 Resistance | |
|---|---|---|---|---|
| 40 ppm | 60 ppm | |||
| C1-1C5 | 0.458 | 0.9 | 11% | 3% |
| C2-1B4 | 0.421 | 0.9 | 56% | 17% |
| C3-1B7 | 0.452 | 0.7 | 33% | 5% |
| C3-2A10 | 0.457 | 0.2 | 34% | 3% |
| C6-1B3 | 0.400 | 0.6 | 15% | 1% |
| C6-2C6 | 0.460 | 1.3 | 17% | 8% |
| C7-1B5 | 0.419 | 0.4 | 34% | 24% |
| C7-1B7 | 0.369 | 2.0 | 56% | 32% |
| C7-2A2 | 0.411 | 0.1 | 42% | 2% |
| C7-2B2 | 0.438 | 0.0 | 70% | 34% |
| C7-2C2 | 0.438 | 0.0 | 32% | 27% |
| C7-3A10 | 0.429 | 0.1 | 75% | 56% |
| C7-3A6 | 0.469 | 0.8 | 33% | 4% |
| C7-3B4 | 0.447 | 0.0 | 42% | 18% |
| C10-2I5 | 0.462 | 2.5 | 59% | 80% |
| C12-1A5 | 0.432 | 0.8 | 18% | 11% |
| C12-1A9 | 0.461 | 0.2 | 1% | 0% |
| EC1118 | 0.482 | 3.9 | 69% | 56% |
| Strain | Implantation Percentage | |
|---|---|---|
| Sauvignon Blanc | Carmenere | |
| C2-1B4 | 90.9% | 81.8% |
| C7-1B7 | 100% | 100% |
| C7-2B2 | 100% | 90.9% |
| C7-2C2 | 100% | 100% |
| C7-3A10 | 100% | 100% |
| C10-2I5 | 100% | 100% |
| Strain | Sauvignon Blanc | Carmenere | ||
|---|---|---|---|---|
| Reducing Sugars (g/L) | Alcohol (% ABV) | Reducing Sugars (g/L) | Alcohol (% ABV) | |
| C2-1B4 | 0.96 | 13.6 | 2.89 | 12.9 |
| C7-1B7 | 1.00 | 13.0 | 2.77 | 13.6 |
| C7-2B2 | 1.08 | 13.3 | 2.74 | 13.7 |
| C7-2C2 | 1.77 | 13.4 | 2.92 | 13.5 |
| C7-3A10 | 1.00 | 13.5 | 2.98 | 13.5 |
| C10-2I5 | 0.82 | 13.4 | 2.91 | 13.4 |
| Strain | Sauvignon Blanc | Carmenere |
|---|---|---|
| C2-1B4 | 3.3 ± 0.9 | 3.7 ± 1.0 |
| C7-1B7 | 4.2 ± 0.7 | 3.0 ± 1.1 |
| C7-2B2 | 3.9 ± 0.6 | 3.6 ± 1.1 |
| C7-2C2 | 3.4 ± 1.1 | 3.1 ± 1.1 |
| C7-3A10 | 3.6 ± 0.9 | 3.9 ± 0.6 |
| C10-2I5 | 3.2 ± 1.1 | 3.7 ± 0.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kessi-Pérez, E.I.; Gómez, M.; Farías, W.; García, V.; Ganga, M.A.; Querol, A.; Martínez, C. Genetically Improved Yeast Strains with Lower Ethanol Yield for the Wine Industry Generated Through a Two-Round Breeding Program. J. Fungi 2025, 11, 137. https://doi.org/10.3390/jof11020137
Kessi-Pérez EI, Gómez M, Farías W, García V, Ganga MA, Querol A, Martínez C. Genetically Improved Yeast Strains with Lower Ethanol Yield for the Wine Industry Generated Through a Two-Round Breeding Program. Journal of Fungi. 2025; 11(2):137. https://doi.org/10.3390/jof11020137
Chicago/Turabian StyleKessi-Pérez, Eduardo I., Melissa Gómez, William Farías, Verónica García, María Angélica Ganga, Amparo Querol, and Claudio Martínez. 2025. "Genetically Improved Yeast Strains with Lower Ethanol Yield for the Wine Industry Generated Through a Two-Round Breeding Program" Journal of Fungi 11, no. 2: 137. https://doi.org/10.3390/jof11020137
APA StyleKessi-Pérez, E. I., Gómez, M., Farías, W., García, V., Ganga, M. A., Querol, A., & Martínez, C. (2025). Genetically Improved Yeast Strains with Lower Ethanol Yield for the Wine Industry Generated Through a Two-Round Breeding Program. Journal of Fungi, 11(2), 137. https://doi.org/10.3390/jof11020137







