3.1. Fungal Endophytic Diversity in Artemisia Species
3.1.1. Colonization Rate and Colonization Frequency of Endophytic Fungi in Artemisia thuscula
In this study, the employed analyses indicate that 37 fungal species and 25 fungal genera were isolated from 10 plants of Artemisia thuscula. Colonization rate (further CR) shows how much a plant can be colonized within predetermined conditions. It is valuable information as different plants showed distinct values of this index; therefore, low values could express plants poor in endophytic fungi culturable in the given conditions.
To calculate the colonization frequency (CF) of fungal endophytes in Artemisia species plants, we have considered same fungal endophytic species isolated from two or more plant fragments as being a distinct isolate belonging to the same species. Therefore, if the same species was isolated twice from the same plant fragment, it was considered only one time. This issue is to be expected at isolation moment, when no precise differentiation between the isolates can be defined, and only once purified and further analyzed then only the strain received a final identification. The CF% gives a hint over the distribution and abundance of a certain fungal species in a sample (i.e., plant/location/region). To know the “area” of the distribution and abundance of a certain endophytic fungal species, we have analyzed the data per plant individual or plant location, plant species, and plant region. Regions were grouped here as: La Palma Island and Tenerife Island. This way we can have an overview on where certain fungal species are more abundant or rare, as well as if there is a relation between their distribution and plant-specific parameters.
In
Artemisia thuscula, only one plant out of 10 had a colonization rate value over 90% (LP2). The lowest values (CR% = 25) were recorded for three plants (TF8, TF7 and TF3). Interestingly, as per variable geographical location there is a considerable variation between La Palma Island and Tenerife Island, with the former having the most colonized plant individuals (
Table 4).
Artemisia thuscula cannot escape of the “omnipresence” of
Alternaria alternata, this species was isolated from eight plants but with relevant differences in the frequency, CF% = 15–50%. A notable presence is remarked here,
Neofusicoccum australe, isolated from three plants at relatively high values (CF% = 25) when considering that the maximum value is 50. Moreover, the
Neofusicoccum genus, consisting here of three species was isolated from eight plants, one of which revealed a CF% of 34.21. Interestingly, around 70% of the fungal species in
Artemisia thuscula were isolated from only one plant each (
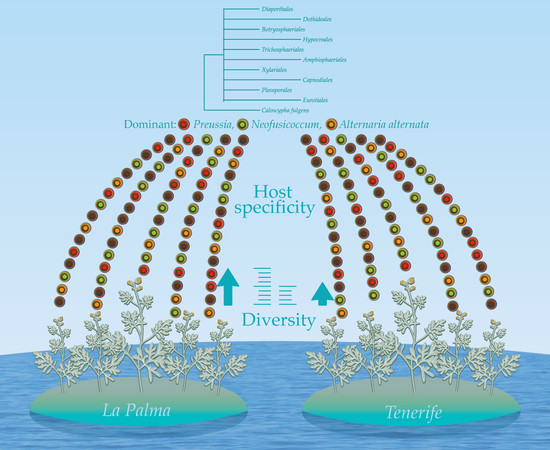
Table 5). This suggests a host specificity which was also exhibited by the low and moderate values of Sorensen–Dice coefficient when the similarity of the endophytic assemblages was analyzed (see further Diversity indices for endophytic fungi in
Artemisia thuscula).
34 endophytic fungal species were isolated from
Artemisia thuscula (
Table 6) and their frequency varied within a low range with two exceptions:
Alternaria alternata (CF% = 18.71) and
Neofusicoccum sp. 3 (CF% = 8.39).
Studies that are independent of fungal isolation and identification methods often revealed higher numbers of fungal species [
49]. We purposely chose the culture method to further select endophytic fungi of high interest according to their biological activities. Our goal was to yield a large number of endophytes, and not to produce a complete species list of fungal endophytes in these
Artemisia species. Nevertheless, the data obtained gave us an interesting fragment of knowledge about the communities of these microorganisms in their plant hosts.
In terms of endophytic fungal species CF%, the most isolated species was
Alternaria alternata (CF = 18.71; eight of ten plants), as expected. It is a common saprobe found on various plants and other substrata worldwide [
50,
51] and has often been isolated as endophyte in previous studies [
52,
53,
54,
55]. Qian et al., 2014 [
27] isolated endophytic fungi from
Artemisia argy and found Pleosporales to be the most represented group, with three species of
Alternaria present. It was found as the most predominant species in grasses [
56] and various plants families, also [
57]. Among dominant endophytic fungal species, we observed taxa like
Neofusicoccum and
Preussia. These genera of endophytic fungi were previously isolated from a wide range of host plants including
Artemisia spp. [
24,
57,
58,
59,
60].
Interestingly, it was observed a tendency on host specificity of most endophytic fungal species. In
Cirsium arvense similarity in endophytic communities decreased with increasing intersite distance [
61] while in
Holcus lanatus the similarity between leaf and root myco-assemblages at the same location was lower than that observed in leaves at different locations [
58]. Further, in leaf fungal communities the average number of species shared by any pair of location was 3.13 and in root assemblages was 1.73 out of an average of 12.2 species identified at each location [
58].
Despite the dominant species, the rest of the endophytic fungal species reflect an unequal distribution of a certain endophytic species among plant individuals. This same issue was previously observed [
58] but no definitive answer has been found. Some hypotheses were proposed like ubiquitous taxa with spatial dominance or selection of certain dependent on culture conditions [
58]. In the case study on
Artemisia thuscula (see
Section 3.1.3) taxa such as
Preussia,
Pestalotiopsis,
Aplosporella,
Chaetomium and
Cladosporium were isolated from only one nutrient medium out of the three media tested. Nevertheless, this is not a unique parameter, which should account for the determination of an endophytic taxa preference for a nutrient medium. One of the major variables which we consider is the rest of the community involved and their role in the interaction when the isolation performed. That is, which are the other taxa living in the same “space” (i.e., plated plant fragment) and we must consider if there are (i) fast-growing taxa versus slow growing taxa; (ii) nutrient deficiency or promoting medium for certain taxa, as well as (iii) the interaction between the taxa (i.e. antagonism).
3.1.2. Diversity Indices for Endophytic Fungi in Artemisia thuscula
In the La Palma results of diversity, Margalef index revealed the highest value for species richness in San Bartolo (Margalef = 4.24) followed by El Granel (Margalef = 3.69). The Brillouin index agrees that the highest diversity is found in San Bartolo (Brillouin = 1.8) but Fisher’s alpha index shows a higher abundance of rare species in El Granel (Fisher’s alpha = 18.6) than in San Bartolo (Fisher’s alpha = 13.9). Diversity regarded as evenness was found to be similar in both localities (Simpson’s index: El Granel = 0.88 and San Bartolo = 0.87). In La Palma Island, San Bartolo locality was revealed as having the highest value for species richness and diversity. Yet, El Granel was shown as having a higher abundance of rare species (Fisher’s alpha: El Granel = 18.6 and San Bartolo = 13.9) and a higher value of evenness than San Bartolo (Simpson’s index: El Granel = 0.88 and San Bartolo = 0.87). In Tenerife, the locality San Andres showed by far the highest diversity in all previously mentioned terms and all the indices confirm it (
Table 7).
Sorensen–Dice index revealed that of 45 cases in the matrix, 27 of them were of zero similarity. Further, only one case was found to have 57% similarity (TF2 versus TF7) and one case with 50% similarity (TF1 versus TF4). The rest of the cases had values ranging between 11% and 40% similarity. These different similarity values may be due to distance among hosts, soil composition and/or climatic conditions. When the distance was plotted (UPGMA), the Sorensen–Dice coefficient clustered plants LP1 and LP2 with maximum bootstrap support (BPP = 100), although these plants had only 38% similarity in between. Nevertheless, this is to be considered a high value of similarity in the given matrix and one of the reasons for obtaining it might be the proximity of the collection places (approx. 5 km) between the host plants and similar altitudes and climate. Further clusters were formed like LP4 and TF1; TF2 and TF3; TF4, TF5 and TF7 (
Figure 1). As we expected (from CF and CR values) TF8 is the most different host plant, the backbone of the dendrogram divides into this branch and the other branches which form various clusters of similarity. Also, cluster LP1 and LP2 is a sister cluster of the other clusters which were exhibited as more related in terms of similarity.
3.1.3. Case Study: Artemisia thuscula of Tenerife, Endophytic Fungi Isolated from Two Types of Stems on Three Media: Colonization Frequency and Colonization Rate
In this study, we can observe throughout various individual plants from the same species (i.e., Artemisia thuscula) the relevance of nutrient media and the age of the stem as the selected organ to yield endophytic fungi. When averaged the colonization rates of the three nutrient media selected (PDA, LCA, and V8) do not show relevant differences (CR% = 33.93. 33.93 and 37.50, respectively). Neither do the differences of age in stems; stems with the age < 1 year have CR% = 30.95 and stems with age > 1 year have CR% = 36.90.
Differences may be observed (
Table 8) when comparing different plants, as for instance TF3 and TF4 had the lowest colonization rates (CR% = 16.67) and no endophytic fungi was isolated from V8 or stems with age of more than 1 year for TF4 and TF3, respectively. In addition, there is no higher value than 58.33 of colonization rate, as observed in other individuals of
Artemisia.
If colonization frequency data is segregated into plants sampled (
Table 9), we observe that
Alternaria alternata is the major colonizer in three out of seven plants, namely TF2, TF4, and TF5. Plants had different yields considering number of endophytic fungal species, ranging between three (TF3) and eight (TF7).
Among the major colonizers we observed
Neofusicoccum austral and
Neofusicoccum parvum in TF3 (CF% = 8.33),
Chaetomium sp. 1 and
Phoma sp. 1 in TF7 and
Phoma with two different species in TF8 (CF% = 16.67;
Table 8). Myrchiang et al., 2014 [
28] investigated the endophytic fungi associated with
Artemisia nilagirica and comparing the colonization of three organs (i.e., root, stem and leaf), the authors obtained the highest diversity in the roots (i.e., 14 species), less in stem (i.e., 10 species) and the smallest number in the leaves (i.e., 6 species). Similarly, in
Artemisia thuscula Cosoveanu et al., 2012 [
62] isolated 29 distinct morphotypes: 20 from roots, 7 from stem and 2 from leaves. In addition, Myrchiang et al., 2014 [
28] observed that from all fungal endophytic species, only
Phoma eupyrena was found to be a common occurrence in all plants sample, the other species having a certain preference for one or maximum two organs.
Comparing different plant individuals of the same species and observing the distribution of fungal endophytes provides insights to determine the occurrence of a certain species. For instance, in TF2 four fungal species were isolated only from one nutrient medium, namely
Biscogniauxia mediterranea in PDA,
Alternaria sp. on LCA,
Phoma sp. on PDA and
Pestalotiopsis sp. on LCA (
Table 9). Furthermore, we may observe that the same species of
Phoma sp. 1 was also isolated from TF8 on PDA, similar to
Pestalotiopsis isolated from TF3 on LCA while
Biscogniauxia mediterranea was isolated on V8 from TF8.
When the distribution of endophytic fungi species is observed in terms of colonization frequency per total number of the studied plants (
Figure 2), data showed several species like
Aplosporella prunicola,
Camarosporium sp.,
Chaetomium sp.,
Cladosporium sp,
Nectria mauritiicola and others with certain “preference” for nutrient medium. It is well known that fungi have specific carbon and nitrogen requirements for sporulation [
63,
64,
65]. However, the requirements for fungal growth are less stringent but not less important when isolation is pursued. Nutrient - rich media result in selective isolation for fast-growing fungi, overlooking slow growing species if present [
66]. Osono and Takeda [
29] stated that LCA due to its low glucose content suppresses the overgrowth of fast-growing species. 22 species of fungal endophytes were isolated from all
Artemisia thuscula plants in this case study and 14 species (63%) were isolated only from one nutrient medium. Additionally, 12 fungal species were isolated from stems older than 1 year and seven were isolated from stems younger than one year. Seven fungal species are to be considered rare, as their colonization frequency value is the lowest one, throughout the data set (CF% = 0.60;
Figure 2).
Further, Sorensen–Dice similarity coefficient reveals proximate values among the similarities of the endophytic communities isolated on the three tested media (
Table 10). Yet, none of them overpassed 52% similarity (i.e., LCA versus V8). As for the stem ages, the index showed a value of 43% similarity. Evidence for tissue specificity was previously demonstrated for phloem and xylem tissue, where the value of endophytic similarity reached 36% in roots of
Sophora tonkinensis [
67]. This suggests the necessity to broad both culture media and diversity of tissues to obtain a higher richness of endophytic fungal taxa.
Among the singleton species that occur only in the Artemisia thuscula plant individuals selected for this case study (i.e., limited to Tenerife) we have: Aplosporella prunicola, Camarosporium sp. 1, Macrophomina phaseolina, Chaetomium sp. 1, Nectria mauritiicola, Neofusicoccum australe, Pestalotiopsis sp., Phoma sp. 1 and Stachybotrys longispora. Except Phoma sp., all endophytic fungal species previously mentioned were isolated from only one nutrient medium. In addition, except Camarosporium sp. (isolated also from Artemisia thuscula in Palma Island), all endophytic fungal species previously named were isolated only from Artemisia thuscula in Tenerife Island.
3.2. Phylogenetic Relations
54 endophytic fungal ITS sequences and the associated GenBank sequences were used for the phylogenetic analysis (
Table 3; sequences of strains HLP16, HLP22, HLP28, HLP33, HLP48A, HLP4, HTF29, HTF33, HTF43, and HTF61 are not listed and are only available at request). The dataset consists of 603 characters after alignment, 43 characters are conserved, and 447 characters are parsimony informative, while 557 are variable characters. Bayesian Posterior Probabilities (BPPs) given below each node are shown on the upper branches.
Ten orders (Diaporthales, Dothideales, Botryosphaeriales, Hypocreales, Trichosphaeriales, Amphisphaeriales, Xylariales, Capnodiales, Pleosporales and Eurotiales) are recognized (
Figure 3). The phylogenetic tree divides the taxa in five main clades, leaving
Diaporthe sequences unclustered. Clade 1 consists of Dothideales and Botryosphaeriales (BPP = 0.98), Clade 2 groups Hypocreales, Trichosphaeriales, Amphisphaeriales and Xylariales (BPP =0.88), Clade 3 and Clade 4 contain Capnodiales (BPP = 0.79) and Pleosporales (BPP = 0.63), respectively while Clade 5 accommodates Eurotiales (BPP = 0.62).
Interestingly,
Diaporthe sequences are not clustered but several show different branch lengths. Yet, taxa
D. novem and
D. phaseolorum do not differentiate. Endophytic fungi were basically identified using morphology; therefore, HLP15 and HLP23 were considered
D. phaseolorum and
D. novem, respectively while structures of HLP37 did not allow an accurate species level identification. Apparently, the ITS region in
Diaporthe is evolving at higher rates than TEF1 or MAT genes [
68], therefore presenting a wider variation than advisable for species boundaries. Thus, a slowly evolving gene region should be used in order to establish species limits [
69]. Nevertheless, ITS sequence data can be used for reliable identification of phylogenetic relationships as long as they are interpreted with care [
69]. Several arrangements of genera draw the attention, like
Aureobasidium (Dothideales) and
Aplosporella (Botryosphaeriales) which are shown with an immediate common ancestor (BPP = 0.97).
Aplosporella has over 300 species and appears to be heterogenous; therefore not all species are likely to belong in Botryosphaeriaceae [
70]. The ascomycete genus
Aureobasidium is a member of the family Aureobasidiaceae within the class of the Dothideomycetes [
71]. Dothideomycetidae subclass was emended by Schoch et al., 2006 [
72] and a new subclass was proposed, Pleosporomycetidae, with an additional order, the Botryosphariales.
Penicillium and
Aspergillus sequences form two sister clades as expected (BPP = 0.60). Three species of
Neofusicoccum are clustered with relevant support (BPP = 0.89) while
N. parvum is drawn outside. Hypocreales taxa are split in two sister clusters along with
Stachybotrys,
Grandibotrys,
Melanopsamma and
Sirastachys in one sister clade although with no relevant support (BPP = 0.55) and
Nectria,
Sarocladium and
Corallomycetela as another sister clade (BPP = 0.87). Also, internal clustering is revealed between several taxa of the mentioned genera. Trichosphaeriales and Amphisphaeriales are shown having a common recent ancestor (BPP = 0.98). Hypocreales is recognized as monophyletic [
73]. The order Hypocreales incorporates Nectriaceae and Stachybotriaceae beyond other six families [
74]. Maharachchikumbura et al., 2014 [
75] found using a combined LSU, SSU, TEF and RPB2 sequences data that
Stachybotrys and related taxa (Stachybotriaceae) form a sister cluster of
Nectria and related taxa (Nectriaceae). The results obtained with the ITS region are in accordance with the combined inference obtained by Maharachchikumbura et al., [
75]. The Nectriaceae group (BPP = 0.87) comprises
Nectria (Nectriaceae),
Sarocladium (Hypocreomycetidae) and
Corallomycetella—shown to comprise two distinct clades in Nectriaceae [
76]. The second cluster joints
Stachybotrys,
Grandibotrys,
Sirastachys,
Stachybotrys (Stachybotriaceae, Hypocreomycetidae) and
Melanopsamma (Chaetosphaeriaceae, Sordariomycetidae).
Melanopsamma pomiformis was recently excluded from the genus [
77] and it was linked to the asexual morph
Stachybotrys albipes [
78]. Strains of Sordariomycetes clustered into six subclasses among which Diaporthomycetidae, Xylariomycetidae and Hypocreomycetidae [
75]. Our Bayesian analysis resulted in a monophyletic clade (Clade 2) which accommodates Hypocreales (Hypocreomycetidae), Trichosphaeriales (Diaporthomycetidae), Amphisphaeriales (Xylariomycetidae) and Xylariales (Xylariomycetidae). Yet, Diaporthales taxa (
Diaporthe spp.) were left outside this clade. A resulting parsimonious tree of multi-locus based (LSU, ITS, and
TEF1) sequences shows that the genus
Diaporthe has paraphyletic origins [
79]. Xylariales and Amphisphaeriales were found as sister clusters in Xylariomycetidae sharing a common ancestor [
80]. Yet, the clade which accommodates Xylariomycetidae is a sister clade of Diaporthomycetidae (Diaporthales) and Hypocreomycetidae (Hypocreales).
Cladosporium sequences are clustered (BPP = 0.79) and different branch lengths between species are revealed, grouping C. ossifragi, C. antarcticum and C. iridis (BPP = 0.78). Conversely, Aplosporella sequences do not differentiate in between, showing all species with same branch lengths.
Stemphylium sequences are grouped but support does not avail this grouping (BPP = 0.53).
Phoma-like sequences are clustered as expected (BPP = 0.93) showing higher differences between
Phoma,
Didymella,
Dothiorella and
Notophoma on one side (BPP = 0.93) and
Paraphoma chrysantemicola on the other side (BPP = 0.88). It is curious that several sequences of endophytes are grouped in a sister clade of
Alternaria clade,
Phoma-like clade and
Stemphylium clade with high probability (BPP = 1), indicating different branch lengths.
Alternaria sequences are not grouped in a single cluster but different branch lengths are drawn among the species. Similarly,
Preussia and
Sporormiella taxa are spread.
Coniothyrium-like sequences are clustered, but support has an average value, BPP = 0.78. Coniothyriaceae and Camarosporiaceae grouping as well as the
Coniothyrium-like sequences cluster and its sister cluster of Pleosporaceae is supported by the findings of Wijayawardene et al., 2014 [
81]. Mainly the sequences obtained from the endophytic strains are grouped with the external sequences as expected (i.e., morphological identification) but several are left unclustered. For instance, inside the group of Pleosporales three endophytic sequences (HLP16, HLP22 and HLP48A) appear as more related, forming a strong-supported cluster (BPP = 1). This apparently new lineage should be confirmed with another phylogenetic study based on large subunit and small subunit nuclear rDNA regions, where only Pleosporales taxa would be included. In the present study none of the methods used like the morphology (absence of the sporulating structures), BLAST alignment (values of similarity with GenBank provided sequences did not exceed 86%, 88% and 84% for HLP16, HLP22, and HLP48A, respectively) and the ITS inference, could provide their proper identification or genetic stronger alliances inside Pleosporales.