Abstract
Cryptococcus neoformans is a major fungal pathogen that infects immunocompromised people and causes life-threatening meningoencephalitis. C. neoformans does not occur in isolation either in the environment or in the human host, but is surrounded by other microorganisms. Bacteria are ubiquitously distributed in nature, including soil, and make up the dominant part of the human microbiota. Pioneering studies in the 1950s demonstrated antifungal activity of environmental bacteria against C. neoformans. However, the mechanisms and implications of these interactions remain largely unknown. Recently, interest in polymicrobial interaction studies has been reignited by the development of improved sequencing methodologies, and by the realization that such interactions may have a huge impact on ecology and human health. In this review, we summarize our current understanding of the interaction of bacteria with C. neoformans.
1. Introduction
Most microorganisms on earth do not occur in isolation, but interact with other microbes. This is true for interactions among members of the same kingdom, such as between different species of bacteria, as well as for mixed populations of multi-kingdom microbial consortia, e.g., interactions between bacteria and fungi [1,2]. Bacterial interactions with fungal pathogens are not only biologically interesting but may also provide new opportunities for antifungal therapy. Fortunately, the vast majority of fungi are harmless for humans [3]. However, a small number of fungi are human pathogens and can cause life-threatening infections [4]. Some of the predominant fungal pathogens include the human-associated fungus Candida albicans, and the environmental fungi Cryptococcus neoformans and Aspergillus fumigatus [5]. Fungal infections are usually very difficult to treat with current antifungal drugs, and the incidence of worldwide fungal resistance is on the rise [6].
C. neoformans is a common human fungal pathogen, and the yeast is responsible for almost a quarter million deaths annually [7]. The fungus primarily affects people with impaired immune systems, especially patients suffering from HIV/AIDS. Indeed, C. neoformans is estimated to be responsible for a staggering 15% of all AIDS-associated deaths worldwide [7]. Natural habitats for C. neoformans include soil, trees, and bird excreta [8,9,10]. C. neoformans cells are thought to be present in different morphologies in nature. Fungal cells can occur as desiccated yeast cells or as spores, which have the tendency to be distributed by wind and animals. For example, it has been shown that C. neoformans can be found attached to the feet and beaks of pigeons [11]. Interestingly, birds are not susceptible to the fungus. Humans, however, can inhale C. neoformans spores and yeast cells, leading to the development of pulmonary infections [12]. Such infections of the lung can further develop into life-threatening cryptococcal meningitis in a process involving the transition of the fungus from the lung to the brain via crossing the blood brain barrier.
So far, studies of fungal–bacterial interaction have mainly been performed with C. albicans, and it was found that these interkingdom interactions can dramatically influence human health and disease [13,14,15,16,17]. Relatively few studies have addressed interkingdom interactions between C. neoformans and bacteria. As mentioned, C. neoformans occurs both in the environment, for example in soil, and inside humans during infection, in comparison to C. albicans—which is obligately associated with a mammalian host. Therefore, C. neoformans is likely to come into contact and potentially interact with an enormous number of bacteria in its natural habitats and with the human microbiota during disease [18,19]. In this review, we provide background context and highlight new studies indicating that specific bacteria can have dramatic effects on cryptococcal growth and virulence factor expression. Understanding such interactions may lead to the discovery of novel antifungal drug targets or novel antifungal drugs.
2. Types of Fungal–Bacterial Interactions
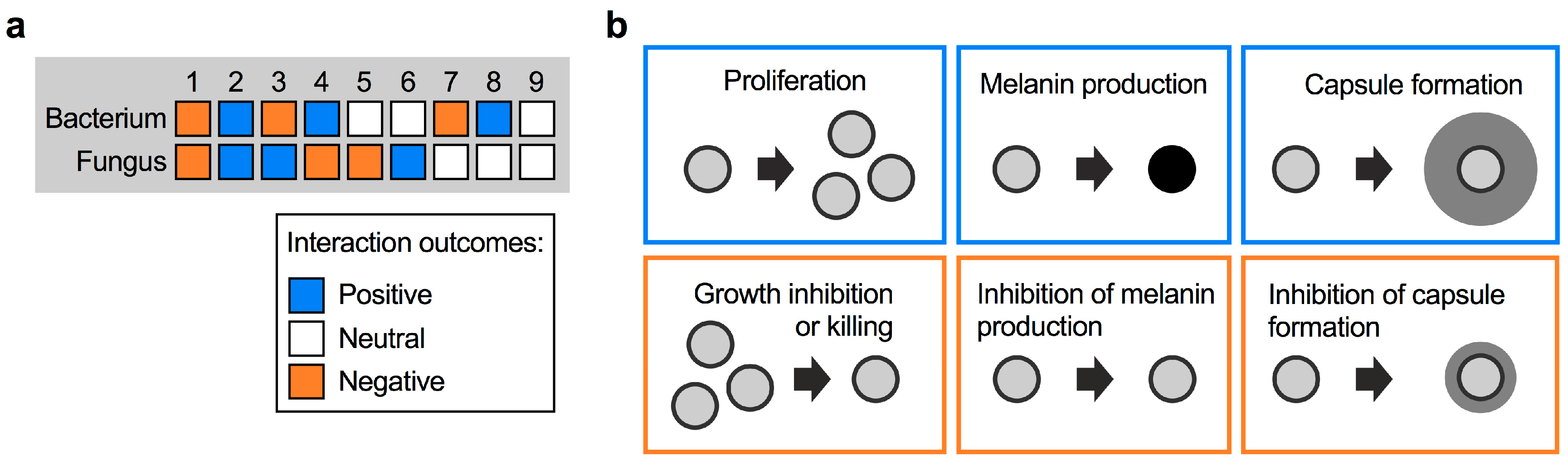
There are several ways that fungi and bacteria can interact with each other. Considering a bi-microbial interaction between a fungus and a bacterium, the outcome can be negative, positive, or neutral for both, or for either partner. In total, this results in nine possible interaction outcomes (Figure 1a). A useful illustrative example of positive and negative interactions comes from the bacterial pathogen Pseudomonas aeruginosa and C. albicans, which often co-infect patients with cystic fibrosis (CF). P. aeruginosa forms recalcitrant biofilms with increased antibiotic tolerance in the lungs, and it was shown that biofilm formation is promoted by C. albicans through ethanol production by the fungus [20]. P. aeruginosa itself stimulates fungal ethanol production by generating redox-active small molecules termed phenazines. This cyclic relationship was proposed to establish favorable conditions for both P. aeruginosa and C. albicans to co-infect CF patients [20]. Indeed, co-infection results in a significantly worse clinical outcome for CF patients compared with infection with P. aeruginosa alone [21]. The interaction between P. aeruginosa and C. albicans is more complex, however, and can have different outcomes than those mentioned in the context of CF. In general, pseudomonal phenazines are toxic to C. albicans at higher concentrations. At lower concentrations, phenazines were demonstrated to inhibit key C. albicans virulence factors, for example the yeast-to-hypha transition, adhesion to surfaces, and biofilm formation [22,23]. Conversely, in the context of gastrointestinal infections, C. albicans was recently shown to produce factors that inhibit P. aeruginosa virulence by suppressing production of the siderophores pyochelin and pyoverdine [24]. Notably, this process did not impact bacterial growth or gut colonization.
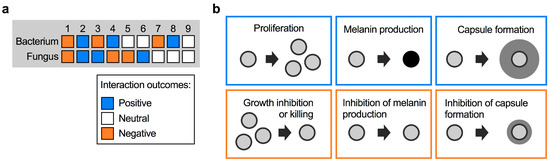
Figure 1.
Types of fungal–bacterial interactions. (a) Possible types of interactions between fungi and bacteria, and their respective outcomes. (b) Examples of positive and negative interaction outcomes for C. neoformans cells following exposure to bacteria. Bacteria may induce fungal proliferation, or kill fungal cells. Bacteria may also trigger the expression of fungal virulence factors (e.g., formation of melanin pigment or polysaccharide capsule), or repress formation of these factors. The different outcomes are color-coded depending on their impact on the fungus, i.e., outcomes likely to be beneficial to the fungus are boxed in blue, while outcomes likely to be unfavorable are boxed in orange.
For C. neoformans, interactions with bacteria could have a positive outcome for the fungus in the form of enhanced proliferation, or stimulation of protective fungal virulence factors such as melanin and capsule formation (Figure 1b). On the other hand, bacterial activities that result in inhibition of these processes may confer negative outcomes for C. neoformans (Figure 1b). Fungal proliferation is clearly important in competing for limited space and nutrient sources in a given niche. As virulence factors, melanin and capsule provide readily assayable readouts to observe the impact of bacteria. Melanin is a dark-brown/black, cell wall-anchored pigment that protects C. neoformans from multiple stresses in the environment and during host infection [25,26,27,28]. Melanin formation is one of the main cryptococcal virulence factors, and mutants with defects in melanization are usually attenuated for virulence in vivo [29,30,31]. The most important virulence factor in C. neoformans, however, is the polysaccharide capsule [32,33,34]. Polysaccharide fibers are anchored to the α-1,3-glucan layer of the cell wall and protect cells from phagocytosis by amoeba in the environment or macrophages in the human host [12,35]. Moreover, capsule polysaccharide can modulate the human immune response [36]. Capsule-deficient mutants are usually avirulent or strongly reduced for virulence in animal models [30,37,38,39]. Below we discuss the impact of specific bacterial species on cryptococcal proliferation and virulence factor production.
3. Interaction of C. neoformans with Bacteria
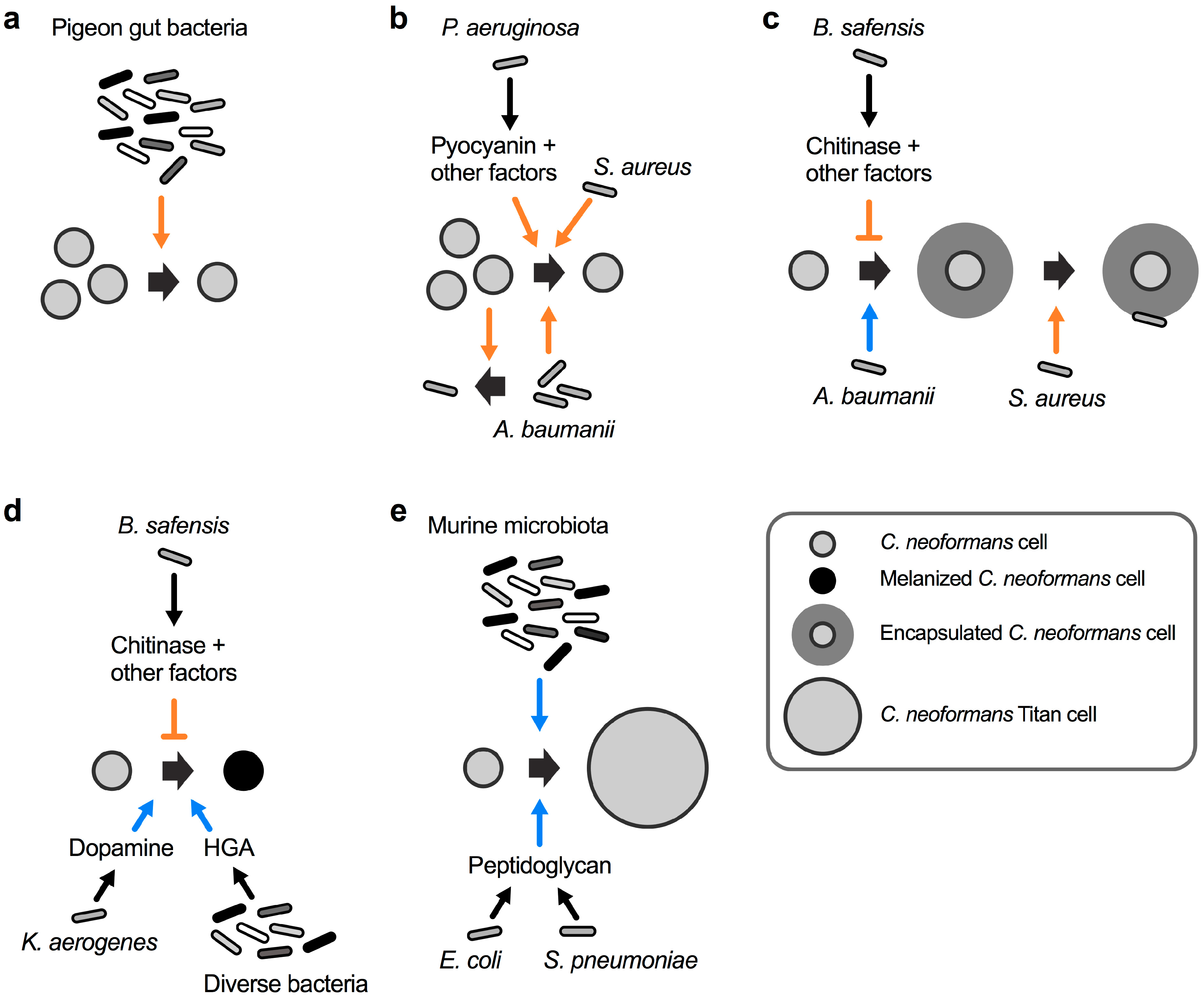
Cells of C. neoformans can be found in pigeon excreta, suggesting that the pigeon gastrointestinal tract is at least temporarily colonized with this fungus. Interestingly, pigeons as well as other birds are not susceptible to cryptococcal disease, and fungal cells are completely cleared from the excreta within 4 weeks [40]. This observation suggests that factors within the gastrointestinal tract may have fungicidal activity. In an attempt to examine the impact of the natural pigeon gut microbiota, researchers in the late 1970s isolated bacteria from the intestinal contents of healthy pigeons and studied their impact on C. neoformans viability [40]. Seven distinct bacterial species were isolated including Bacillus subtilis, Escherichia coli, Klebsiella aerogenes, Proteus mirabilis, Pseudomonas aeruginosa, Staphylococcus albus, and Streptococcus faecalis. Strikingly, a mixture of these seven bacteria completely inhibited growth of a suspension of C. neoformans cells (Figure 2a) [40]. These results indicated that the bacterial mixture had potent anticryptococcal activity. Therefore, it is tempting to speculate that, in addition to the elevated avian body temperature, a specialized avian microbiota may protect birds from infections by C. neoformans. Indeed, a study in the early 1980s re-analyzed most of the above mentioned bacteria in individual bacterium–cryptococcal co-cultures, and found that the growth-inhibiting activity was mainly exerted by P. aeruginosa and B. subtilis [41].
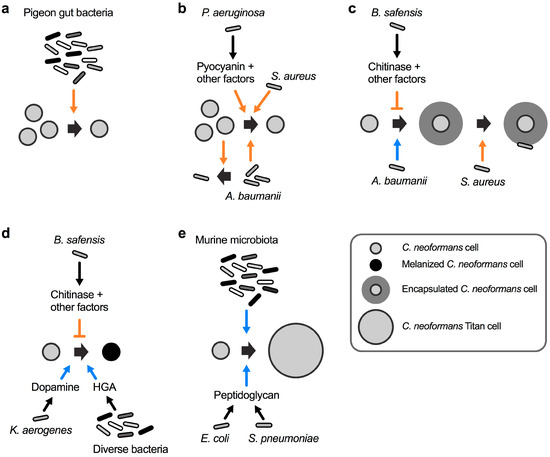
Figure 2.
The spectrum of interactions between C. neoformans and bacteria. (a) A mixture of bacteria isolated from the gastrointestinal tract of healthy pigeons kills C. neoformans. (b) Acinetobacter baumanii and C. neoformans reciprocally inhibit each other’s growth. Staphylococcus aureus kills C. neoformans by an unknown mechanism, and P. aeruginosa kills cryptococcal cells via production of pyocyanin and other factors. (c) A. baumanii induces C. neoformans capsule formation, and S. aureus preferentially attaches to and kills encapsulated C. neoformans cells. Bacillus safensis inhibits capsule formation via production of chitinase and other factors. (d) K. aerogenes produces dopamine, and diverse bacteria produce homogentisic acid (HGA), both of which serve as substrates for cryptococcal melanin biosynthesis. B. safensis inhibits fungal melanin production via chitinase activity and other factors. (e) Cell wall peptidoglycan from E. coli and Streptococcus pneumoniae induce C. neoformans titan cell formation. The murine microbiota induces fungal titanization by an unknown mechanism. The orange colored arrow-headed and blunt-ended lines indicate inducing and repressive processes, respectively, that have a negative impact on cryptococcal viability or virulence factor production. The blue colored arrow-headed lines indicate processes that have a positive influence on the formation of C. neoformans virulence factors.
Already in the mid 1950s, it was observed that cultures of the ubiquitous soil bacterium P. aeruginosa had the capacity to inhibit the growth of C. neoformans by an unknown mechanism [42]. A follow-up study in the mid 1970s analyzed 44 different P. aeruginosa clinical strains for their individual potential to inhibit 14 different clinical C. neoformans strains [43]. Strikingly, all pseudomonal strains inhibited growth of all 14 C. neoformans strains on solid media, although with varying efficiency. The authors noted that the more mucoid fungal strains had a tendency to display increased sensitivity towards P. aeruginosa-mediated inhibition [43]. Usually, mucoidy in C. neoformans is indicative of normal polysaccharide capsule formation, while strains with a dry colony appearance have reduced capsules [44]. This observation raises the possibility that P. aeruginosa may preferentially target encapsulated cryptococcal cells. Although unsuccessful in isolating the precise bacterial factor(s) responsible for the antifungal activity, the authors ruled out an involvement of the pseudomonal phenazine, pyocyanin [43]. Contrary to the findings in the study from 1975 [43], newer studies of the interaction between P. aeruginosa and C. neoformans indicate that direct bacterial–fungal cell-to-cell-contact triggers the production of pyocyanin and other factors to inhibit cryptococcal growth (Figure 2b) [45]. The differences in both studies regarding the role of pyocyanin may have been due to differences in the growth media used for cultivation. Indeed, it was shown that the inhibitory activity towards C. neoformans was fungicidal, and that it was dependent on the pseudomonal cell density and relative ratio of fungal and bacterial cells [45]. Since exogenously supplied pyocyanin only had fungistatic effects on C. neoformans cultures, it was concluded that additional bacterial factors, for example proteases and rhamnolipids, might also contribute to the antifungal activity [45]. Notably, C. neoformans did not impact the growth of P. aeruginosa, indicating that the bacteria have antagonistic activity, while the fungus remains neutral during this interaction [45].
The bacterial pathogen Staphylococcus aureus can cause life-threatening infections in humans. Similar to P. aeruginosa, S. aureus also displays fungicidal activity towards C. neoformans during fungal–bacterial co-culture (Figure 2b) [46]. Again, bacterial growth was not affected by C. neoformans, indicating a mono-directional interaction. Interestingly, a control experiment also revealed that C. albicans growth and survival was not affected by S. aureus [46]. This points to the possibility of Cryptococcus-specific proteins or factors targeted by the bacterium. Indeed, S. aureus cells were observed to preferentially attach to C. neoformans cells that have the capacity to form capsule (Figure 2c). Bacteria did not attach to an acapsular mutant of C. neoformans [46]. However, since the experiments were not performed under robust capsule-inducing conditions, it would be interesting to study the interaction of S. aureus and C. neoformans under conditions that promote capsule biosynthesis. Nevertheless, the likelihood that S. aureus attaches to capsule polysaccharide is quite high because exogenously added capsular polysaccharides reduced bacterial binding to and killing of C. neoformans [46].
Recently, the nosocomial bacterial pathogen Acinetobacter baumanii was demonstrated to induce cryptococcal capsule and biofilm formation during co-cultivation (Figure 2c) [47]. The exact molecular mechanism of this interaction remains to be determined, however, the authors established that physical contact was not required, at least for the biofilm-inducing activity. This indicates that A. baumanii likely secretes specific factors that affect the fungus either at the cell surface or inside the cell. The interaction between both organisms also resulted in reciprocal killing. Co-incubation experiments revealed that 40–75% of cryptococcal cells were killed by A. baumanii, while ~65% of bacterial cells were killed by C. neoformans [47]. As noted by other researchers however, A. baumanii is not a common soil bacterium and it is unclear whether the observed effects have clinical or biological significance [48].
We recently showed that the ubiquitous soil bacterium Bacillus safensis has potent anti-capsular activity, in part via the action of chitinase activity upon cell-to-cell contact (Figure 2c) [49]. B. safensis is a Gram-positive, spore-forming bacterium that was first isolated from a Spacecraft Assembly Facility at the Jet Propulsion Laboratory, USA, and it obtained its name from this location (SAFensis) [50]. B. safensis was also demonstrated to inhibit melanin formation by C. neoformans in a process that relied in part on chitinase activity (Figure 2d) [49]. Physical contact was required for the anti-virulence factor activities, and we hypothesized that the bacterial cell may produce cell surface-associated chitinase(s) upon contact with the fungus, or that contact may trigger close-range secretion of the enzyme [51]. Bacterial proteases and lipopeptides could be other factors involved during the interaction of B. safensis with C. neoformans. B. safensis specifically inhibited cryptococcal virulence factor production without significantly affecting overall fungal growth, thus, it is tempting to speculate that this bacterium or similar Bacilli may have the potential of being developed into antifungal probiotics that exclusively target virulence factor production by C. neoformans [52,53]. Encouragingly, some Bacillus spp., including B. subtilis and Bacillus pumilus, the latter being closely related to B. safensis, have recently been demonstrated to have potent anti-pathogen activities and are already being used as probiotics in certain countries [54,55,56,57].
While B. safensis inhibits cryptococcal melanin production, the opportunistic bacterial pathogen K. aerogenes was shown to promote melanization of C. neoformans cells during co-cultivation (Figure 2d) [58]. The basis for the activity was the bacterial production of dopamine that can serve as a precursor for cryptococcal melanin biosynthesis [58]. Another study established that bacterial homogentisic acid, which is an intermediate product of tyrosine and phenylalanine catabolism, can serve as a precursor for melanin formation by bacteria and C. neoformans (Figure 2d) [59].
It was recently shown that the murine microbiota has the capacity to induce titan cell formation by C. neoformans (Figure 2e) [60]. Titan cells are cryptococcal cells with enormous dimensions and clinical relevance due to being refractory to phagocytosis by human immune cells [61,62]. The in vivo significance of the microbiota in promoting titan cell formation was established by the finding that mice pre-treated with antibiotics prior to infection with C. neoformans had significantly less fungal cells with the titan morphology compared to antibiotic-free mice [60]. Further analysis of the titan cell-inducing mechanisms revealed that bacteria such as E. coli, and Streptococcus pneumoniae trigger cryptococcal titanization via shedding of peptidoglycan, a component of the bacterial cell wall (Figure 2e) [60].
In summary, these studies demonstrate that different bacteria can have disparate effects on C. neoformans, either promoting or preventing growth and survival, and either enhancing or blocking production of virulence factors.
4. Direct Cell-To-Cell Contact during Fungal–Bacterial Encounters
Direct cell-to-cell contact is known to be important for interactions among bacteria. For example, E. coli has a contact-dependent inhibition system to prevent the growth of competing bacteria [63]. Contact-dependent interactions between bacteria and fungi are far less well understood. Studies on C. albicans–bacteria co-incubations revealed that several bacteria have the capacity to attach to the fungal cells via interaction with the fungal surface-localized, hypha-specific and agglutinin-like protein Als3 [64,65]. Accordingly, P. aeruginosa only attaches to and kills C. albicans hyphal cells. Yeast cells of C. albicans are not affected by the bacterium [66].
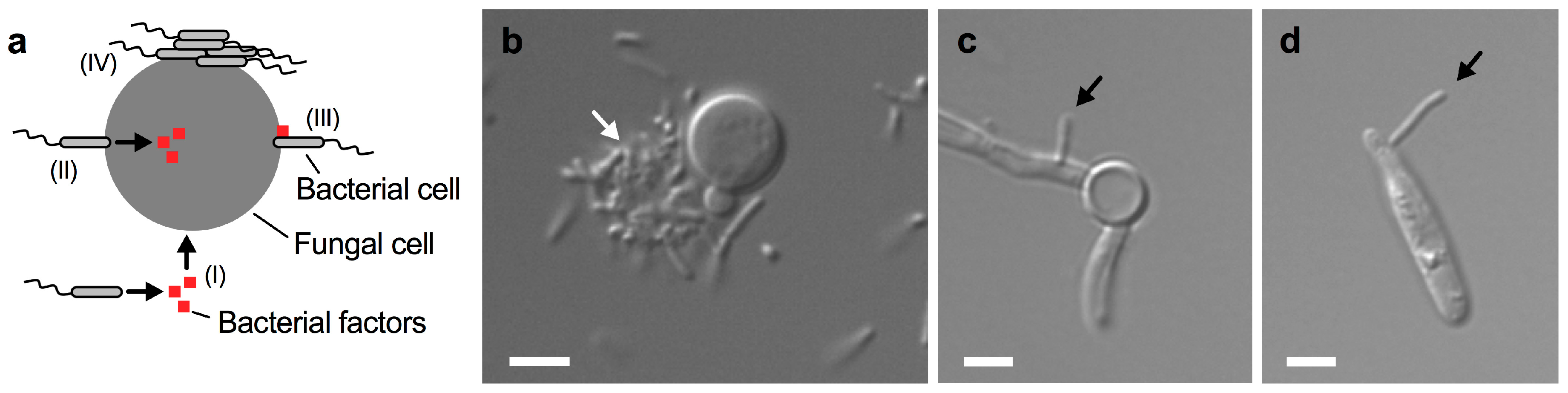
Bacterium-driven interactions that do not require direct cell contact are usually based on the secretion of specific bacterial molecules that can enter the fungal cell on their own (Figure 3a I). In contrast, there are several ways that direct cell-to-cell contact may trigger an interaction. First, bacterial attachment to the fungus may trigger the injection of factors into the fungus (Figure 3a II). Second, bacterial factors may be produced and impact the fungus at its surface following attachment of bacterial cells (Figure 3a III). Finally, bacteria may attach to the fungal cell surface and form aggregates and biofilms (Figure 3a IV). We have recently shown that the soil bacterium B. safensis forms aggregates on some C. neoformans yeast cells during co-cultivation (Figure 3b) [49]. Moreover, we found that B. safensis can attach to C. albicans hypha (Figure 3c) [49]. Interestingly, our unpublished results indicate that B. safensis can also attach to cells of the plant-pathogenic fungus Ustilago maydis (Figure 3d). Therefore, B. safensis has the capacity to attach to diverse fungi. These results also point to the possibility that the fungal molecule(s) that the bacteria use for docking may be conserved among these different fungi. Direct cell contact between bacteria and pathogenic fungi was also observed for the interaction of C. neoformans with P. aeruginosa and S. aureus [45,46].
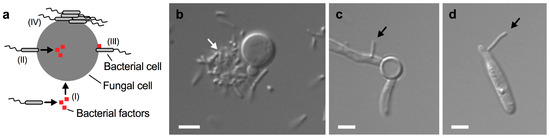
Figure 3.
Close-contact interactions of B. safensis with diverse fungal pathogens. (a) Schematic representation of some fungal-bacterial interactions. (I) Bacteria may secrete certain factors that enter the fungal cell; (II) bacteria may attach to the fungal cell surface and inject factors into the fungus; (III) bacteria may attach to the fungal cell and express cell-surface associated factors; and (IV) bacteria may attach to the fungal cell surface and form cell aggregates and biofilms. Additional mechanisms may exist. (b) Differential interference contrast (DIC) microscopy image of C. neoformans cells grown with B. safensis in yeast peptone dextrose medium for 24 h. Note that B. safensis appears to form a cluster of cells (indicated by a white arrow) on one side of the C. neoformans cell. (c) DIC microscopy image of C. albicans cells grown with B. safensis under fungal hypha-inducing conditions for 4 h. A bacterial cell (black arrow) can be seen attached to the fungal filament. (d) DIC microscopy image of U. maydis cells grown with B. safensis in potato dextrose broth for 24 h. A bacterial cell (black arrow) has attached to the fungal cell. Scale bars, 2 µm.
In summary, it appears that direct cell-to-cell contact between bacteria and fungi is common during interactions, and understanding mechanisms of attachment and bacterial factors that are delivered could potentially lead to the identification of novel antifungal activities.
5. Open Questions for Further Study
Despite recent insights into some specific C. neoformans–bacterial interactions, many questions remain to be investigated. Specifically, it will be crucial to identify the molecular mechanisms underlying the different types of interactions between C. neoformans and bacteria. Some of the open questions are as follows:
- Which types of bacteria interact with C. neoformans in the environment and in the host?
- How prevalent are cryptococcal–bacterial interactions in nature and in the human host?
- Do polymicrobial interactions between C. neoformans and bacteria lead to emergent properties?
- Do bacteria impact cryptococcal gene expression?
- What effect does C. neoformans have on bacterial interaction partners?
- What are the immunological implications of C. neoformans–bacteria interactions?
- Do interactions, e.g., with the microbiota, impact the clinical outcome of cryptococcosis?
- What are the detailed molecular mechanisms in play during polymicrobial interactions?
- How may the bacterial factors mediating interaction with C. neoformans be identified?
- How do C. neoformans-bacteria interactions evolve?
- Will it be possible to use certain bacteria as probiotics to prevent or treat cryptococcosis?
In order to answer these questions, it will be important to carefully increase the level of sophistication of fungal-bacterial interaction studies. First, these interactions should be studied in vitro to potentially uncover mechanisms-of-action, and then the complexity could be increased by including interactions in the presence of human cell lines (e.g., lung epithelial cells). For a long time, the human lung has been thought to be a sterile organ. Recent studies however suggest that the lungs have a distinct microbiota [67,68]. In this context, it is intriguing to consider the possibility that the lung microbiota may influence the initial pulmonary infection with C. neoformans. Furthermore, fungal–bacterial interactions may be studied in vivo in animal models leading potentially to the discovery of novel probiotic bacteria that antagonize the growth or virulence of C. neoformans. Clinical trials with new probiotics may then represent an important translational outcome. Finally, it should be kept in mind that, while bi-microbial interaction studies provide a phenomenal opportunity to uncover potentially new antifungal treatment strategies, ultimately the challenge will be understanding fungal virulence within the complete consortia of thousands of niche-specific microbes. The establishment of model polymicrobial communities may help in studying such interactions [69,70,71].
6. Conclusions
The era of multi-species interaction studies has just begun. New discoveries involving the human microbiota are made on an almost daily basis, and many of them have potentially huge clinical implications. Most current research efforts are focused on the impact of the bacteriome on the host, and it will therefore be important to include fungi into future analyses. Fungal pathogens such as C. neoformans can cause debilitating infections in humans. Therefore, finding new approaches to tackle these neglected infections is extremely important, and the study of fungal–bacterial interactions may open up the way to discover novel antifungal drug targets and new antifungal compounds.
Author Contributions
Conceptualization, F.L.M. and J.W.K.; Writing—Original Draft preparation, F.L.M.; Writing—Review and Editing, F.L.M. and J.W.K.; visualization, F.L.M.; funding acquisition, F.L.M. and J.W.K.
Funding
This research was funded by the Deutsche Forschungsgemeinschaft, grant number MA6248/1-1 (to F.L.M.), and the Canadian Institutes of Health Research, grant number MOP 13234 (to J.W.K.). J.W.K. is a Burroughs Wellcome Fund Scholar in Molecular Pathogenic Mycology.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wargo, M.J.; Hogan, D.A. Fungal-bacterial interactions: A mixed bag of mingling microbes. Curr. Opin. Microbiol. 2006, 9, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Deveau, A.; Bonito, G.; Uehling, J.; Paoletti, M.; Becker, M.; Bindschedler, S.; Hacquard, S.; Herve, V.; Labbe, J.; Lastovetsky, O.A.; et al. Bacterial-fungal interactions: Ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018, 42, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, M. The fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Denning, D.W.; Levitz, S.M. Tackling human fungal infections. Science 2012, 336, 647. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Fisher, M.C.; Hawkins, N.J.; Sanglard, D.; Gurr, S.J. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 2018, 360, 739–742. [Google Scholar] [CrossRef]
- Rajasingham, R.; Smith, R.M.; Park, B.J.; Jarvis, J.N.; Govender, N.P.; Chiller, T.M.; Denning, D.W.; Loyse, A.; Boulware, D.R. Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis 2017, 17, 873–881. [Google Scholar] [CrossRef]
- May, R.C.; Stone, N.R.; Wiesner, D.L.; Bicanic, T.; Nielsen, K. Cryptococcus: From environmental saprophyte to global pathogen. Nat. Rev. Microbiol. 2016, 14, 106–117. [Google Scholar] [CrossRef]
- Emmons, C.W. Isolation of Cryptococcus neoformans from soil. J. Bacteriol. 1951, 62, 685–690. [Google Scholar]
- Emmons, C.W. Saprophytic sources of Cryptococcus neoformans associated with the pigeon (Columba livia). Am. J. Hyg. 1955, 62, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Littman, M.L.; Borok, R. Relation of the pigeon to cryptococcosis: Natural carrier state, heat resistance and survival of Cryptococcus neoformans. Mycopathol. Mycol. Appl. 1968, 35, 329–345. [Google Scholar] [CrossRef]
- Kronstad, J.W.; Attarian, R.; Cadieux, B.; Choi, J.; D’Souza, C.A.; Griffiths, E.J.; Geddes, J.M.; Hu, G.; Jung, W.H.; Kretschmer, M.; et al. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat. Rev. Microbiol. 2011, 9, 193–203. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Hogan, D.A.; Mylonakis, E. Medically important bacterial-fungal interactions. Nat. Rev. Microbiol. 2010, 8, 340–349. [Google Scholar] [CrossRef]
- Allison, D.L.; Willems, H.M.; Jayatilake, J.A.; Bruno, V.M.; Peters, B.M.; Shirtliff, M.E. Candida-bacteria interactions: Their impact on human disease. Microbiol. Spectr. 2016, 4. [Google Scholar]
- Arvanitis, M.; Mylonakis, E. Fungal-bacterial interactions and their relevance in health. Cell Microbiol. 2015, 17, 1442–1446. [Google Scholar] [CrossRef]
- Harriott, M.M.; Noverr, M.C. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 2011, 19, 557–563. [Google Scholar] [CrossRef]
- Morales, D.K.; Hogan, D.A. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 2010, 6, e1000886. [Google Scholar] [CrossRef]
- Whitman, W.B.; Coleman, D.C.; Wiebe, W.J. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Chen, A.I.; Dolben, E.F.; Okegbe, C.; Harty, C.E.; Golub, Y.; Thao, S.; Ha, D.G.; Willger, S.D.; O’Toole, G.A.; Harwood, C.S.; et al. Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog. 2014, 10, e1004480. [Google Scholar]
- Chotirmall, S.H.; O’Donoghue, E.; Bennett, K.; Gunaratnam, C.; O’Neill, S.J.; McElvaney, N.G. Sputum Candida albicans presages FEV(1) decline and hospital-treated exacerbations in cystic fibrosis. Chest 2010, 138, 1186–1195. [Google Scholar] [CrossRef]
- Morales, D.K.; Grahl, N.; Okegbe, C.; Dietrich, L.E.; Jacobs, N.J.; Hogan, D.A. Control of Candida albicans metabolism and biofilm formation by Pseudomonas aeruginosa phenazines. MBio 2013, 4, e00526-12. [Google Scholar] [CrossRef]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef]
- Lopez-Medina, E.; Fan, D.; Coughlin, L.A.; Ho, E.X.; Lamont, I.L.; Reimmann, C.; Hooper, L.V.; Koh, A.Y. Candida albicans Inhibits Pseudomonas aeruginosa Virulence through Suppression of Pyochelin and Pyoverdine Biosynthesis. PLoS Pathog. 2015, 11, e1005129. [Google Scholar] [CrossRef]
- Rosas, A.L.; Nosanchuk, J.D.; Feldmesser, M.; Cox, G.M.; McDade, H.C.; Casadevall, A. Synthesis of polymerized melanin by Cryptococcus neoformans in infected rodents. Infect. Immun 2000, 68, 2845–2853. [Google Scholar] [CrossRef]
- Wang, Y.; Casadevall, A. Susceptibility of melanized and nonmelanized Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infect. Immun. 1994, 62, 3004–3007. [Google Scholar]
- Wang, Y.; Casadevall, A. Decreased susceptibility of melanized Cryptococcus neoformans to UV light. Appl. Environ. Microbiol. 1994, 60, 3864–3866. [Google Scholar]
- Rosas, A.L.; Casadevall, A. Melanization affects susceptibility of Cryptococcus neoformans to heat and cold. FEMS Microbiol. Lett. 1997, 153, 265–272. [Google Scholar] [CrossRef]
- Kwon-Chung, K.J.; Polacheck, I.; Popkin, T.J. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J. Bacteriol. 1982, 150, 1414–1421. [Google Scholar]
- Kwon-Chung, K.J.; Rhodes, J.C. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect. Immun. 1986, 51, 218–223. [Google Scholar]
- Salas, S.D.; Bennett, J.E.; Kwon-Chung, K.J.; Perfect, J.R.; Williamson, P.R. Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans. J. Exp. Med. 1996, 184, 377–386. [Google Scholar] [CrossRef]
- McClelland, E.E.; Bernhardt, P.; Casadevall, A. Estimating the relative contributions of virulence factors for pathogenic microbes. Infect. Immun. 2006, 74, 1500–1504. [Google Scholar] [CrossRef]
- Casadevall, A.; Coelho, C.; Cordero, R.J.B.; Dragotakes, Q.; Jung, E.; Vij, R.; Wear, M.P. The capsule of Cryptococcus neoformans. Virulence 2018, 1–10. [Google Scholar] [CrossRef]
- Ding, H.; Mayer, F.L.; Sanchez-Leon, E.; de S. Araújo, G.R.; Frases, S.; Kronstad, J.W. Networks of fibers and factors: Regulation of capsule formation in Cryptococcus neoformans. F1000Res 2016, 5. [Google Scholar] [CrossRef]
- Reese, A.J.; Doering, T.L. Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol. Microbiol. 2003, 50, 1401–1409. [Google Scholar] [CrossRef]
- Kozel, T.R.; Gulley, W.F.; Cazin, J., Jr. Immune response to Cryptococcus neoformans soluble polysaccharide: Immunological unresponsiveness. Infect. Immun. 1977, 18, 701–707. [Google Scholar]
- Chang, Y.C.; Kwon-Chung, K.J. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell Biol. 1994, 14, 4912–4919. [Google Scholar] [CrossRef]
- Chang, Y.C.; Penoyer, L.A.; Kwon-Chung, K.J. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 1996, 64, 1977–1983. [Google Scholar]
- Chang, Y.C.; Kwon-Chung, K.J. Isolation of the third capsule-associated gene, CAP60, required for virulence in Cryptococcus neoformans. Infect. Immun. 1998, 66, 2230–2236. [Google Scholar]
- Abou-Gabal, M.; Atia, M. Study of the role of pigeons in the dissemination of Cryptococcus neoformans in nature. Sabouraudia 1978, 16, 63–68. [Google Scholar] [CrossRef]
- Ruiz, A.; Neilson, J.B.; Bulmer, G.S. Control of Cryptococcus neoformans in nature by biotic factors. Sabouraudia 1982, 20, 21–29. [Google Scholar] [CrossRef]
- Fisher, A.M. Inhibition of growth of Cryptococcus neoformans by cultures of Pseudomonas aeruginosa. Bull. Johns Hopkins Hosp. 1954, 95, 157–161. [Google Scholar]
- Teoh-Chan, H.; Chau, P.Y.; Ng, M.H.; Wong, P.C. Inhibition of Cryptococcus neoformans by Pseudomonas aeruginosa. J. Med. Microbiol. 1975, 8, 77–81. [Google Scholar] [CrossRef][Green Version]
- Liu, O.W.; Chun, C.D.; Chow, E.D.; Chen, C.; Madhani, H.D.; Noble, S.M. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 2008, 135, 174–188. [Google Scholar] [CrossRef]
- Rella, A.; Yang, M.W.; Gruber, J.; Montagna, M.T.; Luberto, C.; Zhang, Y.M.; Del Poeta, M. Pseudomonas aeruginosa inhibits the growth of Cryptococcus species. Mycopathologia 2012, 173, 451–461. [Google Scholar] [CrossRef]
- Saito, F.; Ikeda, R. Killing of Cryptococcus neoformans by Staphylococcus aureus: The role of cryptococcal capsular polysaccharide in the fungal-bacteria interaction. Med. Mycol. 2005, 43, 603–612. [Google Scholar] [CrossRef]
- Abdulkareem, A.F.; Lee, H.H.; Ahmadi, M.; Martinez, L.R. Fungal serotype-specific differences in bacterial-yeast interactions. Virulence 2015, 6, 652–657. [Google Scholar] [CrossRef]
- Dhamgaye, S.; Murray, G.L.; Peleg, A.Y. The influence of bacterial interaction on the virulence of Cryptococcus neoformans. Virulence 2015, 6, 677–678. [Google Scholar] [CrossRef][Green Version]
- Mayer, F.L.; Kronstad, J.W. Disarming Fungal Pathogens: Bacillus safensis Inhibits Virulence Factor Production and Biofilm Formation by Cryptococcus neoformans and Candida albicans. MBio 2017, 8. [Google Scholar] [CrossRef]
- Satomi, M.; La Duc, M.T.; Venkateswaran, K. Bacillus safensis sp. nov., isolated from spacecraft and assembly-facility surfaces. Int. J. Syst. Evol. Microbiol. 2006, 56, 1735–1740. [Google Scholar] [CrossRef]
- Mayer, F.L.; Kronstad, J.W. Breaking the bad: Bacillus blocks fungal virulence factors. Microb. Cell 2017, 4, 384–386. [Google Scholar] [CrossRef]
- Clatworthy, A.E.; Pierson, E.; Hung, D.T. Targeting virulence: A new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007, 3, 541–548. [Google Scholar] [CrossRef]
- Azevedo, R.; Rizzo, J.; Rodrigues, M.L. Virulence Factors as Targets for Anticryptococcal Therapy. J. Fungi 2016, 2, 29. [Google Scholar] [CrossRef]
- Duc le, H.; Hong, H.A.; Barbosa, T.M.; Henriques, A.O.; Cutting, S.M. Characterization of Bacillus probiotics available for human use. Appl. Environ. Microbiol. 2004, 70, 2161–2171. [Google Scholar] [CrossRef]
- O’Toole, P.W.; Marchesi, J.R.; Hill, C. Next-generation probiotics: The spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2017, 2, 17057. [Google Scholar] [CrossRef]
- Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Joo, H.S.; Villaruz, A.E.; Glose, K.A.; Fisher, E.L.; Hunt, R.L.; Li, B.; et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 2018, 562, 532–537. [Google Scholar] [CrossRef]
- Hong, H.A.; Duc le, H.; Cutting, S.M. The use of bacterial spore formers as probiotics. FEMS Microbiol. Rev. 2005, 29, 813–835. [Google Scholar] [CrossRef]
- Frases, S.; Chaskes, S.; Dadachova, E.; Casadevall, A. Induction by Klebsiella aerogenes of a melanin-like pigment in Cryptococcus neoformans. Appl Environ. Microbiol. 2006, 72, 1542–1550. [Google Scholar] [CrossRef]
- Frases, S.; Salazar, A.; Dadachova, E.; Casadevall, A. Cryptococcus neoformans can utilize the bacterial melanin precursor homogentisic acid for fungal melanogenesis. Appl. Environ. Microbiol. 2007, 73, 615–621. [Google Scholar] [CrossRef]
- Dambuza, I.M.; Drake, T.; Chapuis, A.; Zhou, X.; Correia, J.; Taylor-Smith, L.; LeGrave, N.; Rasmussen, T.; Fisher, M.C.; Bicanic, T.; et al. The Cryptococcus neoformans Titan cell is an inducible and regulated morphotype underlying pathogenesis. PLoS Pathog. 2018, 14, e1006978. [Google Scholar] [CrossRef]
- Okagaki, L.H.; Strain, A.K.; Nielsen, J.N.; Charlier, C.; Baltes, N.J.; Chretien, F.; Heitman, J.; Dromer, F.; Nielsen, K. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 2010, 6, e1000953. [Google Scholar] [CrossRef]
- Zaragoza, O.; Garcia-Rodas, R.; Nosanchuk, J.D.; Cuenca-Estrella, M.; Rodriguez-Tudela, J.L.; Casadevall, A. Fungal cell gigantism during mammalian infection. PLoS Pathog. 2010, 6, e1000945. [Google Scholar] [CrossRef]
- Aoki, S.K.; Pamma, R.; Hernday, A.D.; Bickham, J.E.; Braaten, B.A.; Low, D.A. Contact-dependent inhibition of growth in Escherichia coli. Science 2005, 309, 1245–1248. [Google Scholar] [CrossRef]
- Liu, Y.; Filler, S.G. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot. Cell 2011, 10, 168–173. [Google Scholar] [CrossRef]
- Bamford, C.V.; Nobbs, A.H.; Barbour, M.E.; Lamont, R.J.; Jenkinson, H.F. Functional regions of Candida albicans hyphal cell wall protein Als3 that determine interaction with the oral bacterium Streptococcus gordonii. Microbiology 2015, 161, 18–29. [Google Scholar] [CrossRef]
- Hogan, D.A.; Kolter, R. Pseudomonas-Candida interactions: An ecological role for virulence factors. Science 2002, 296, 2229–2232. [Google Scholar] [CrossRef]
- Dickson, R.P.; Huffnagle, G.B. The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease. PLoS Pathog. 2015, 11, e1004923. [Google Scholar] [CrossRef]
- Bassis, C.M.; Erb-Downward, J.R.; Dickson, R.P.; Freeman, C.M.; Schmidt, T.M.; Young, V.B.; Beck, J.M.; Curtis, J.L.; Huffnagle, G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 2015, 6, e00037. [Google Scholar] [CrossRef]
- Niu, B.; Paulson, J.N.; Zheng, X.; Kolter, R. Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. USA 2017, 114, E2450–E2459. [Google Scholar] [CrossRef]
- Lozano, G.L.; Bravo, J.I.; Garavito Diago, M.F.; Park, H.B.; Hurley, A.; Peterson, S.B.; Stabb, E.V.; Crawford, J.M.; Broderick, N.A.; Handelsman, J. Introducing THOR, a Model Microbiome for Genetic Dissection of Community Behavior. MBio 2019, 10. [Google Scholar] [CrossRef]
- Goldford, J.E.; Lu, N.; Bajic, D.; Estrela, S.; Tikhonov, M.; Sanchez-Gorostiaga, A.; Segre, D.; Mehta, P.; Sanchez, A. Emergent simplicity in microbial community assembly. Science 2018, 361, 469–474. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).