Clonal Dispersal of Cryptococcus gattii VGII in an Endemic Region of Cryptococcosis in Colombia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Isolates
2.2. Reference Strains
2.3. Molecular Type and Mating Type Determination
2.4. Multilocus Sequence Typing (MLST)
2.5. Phenotypic Characterization
2.6. Enzymatic Activities
2.7. Virulence Study
2.8. Statistical Analysis
3. Results
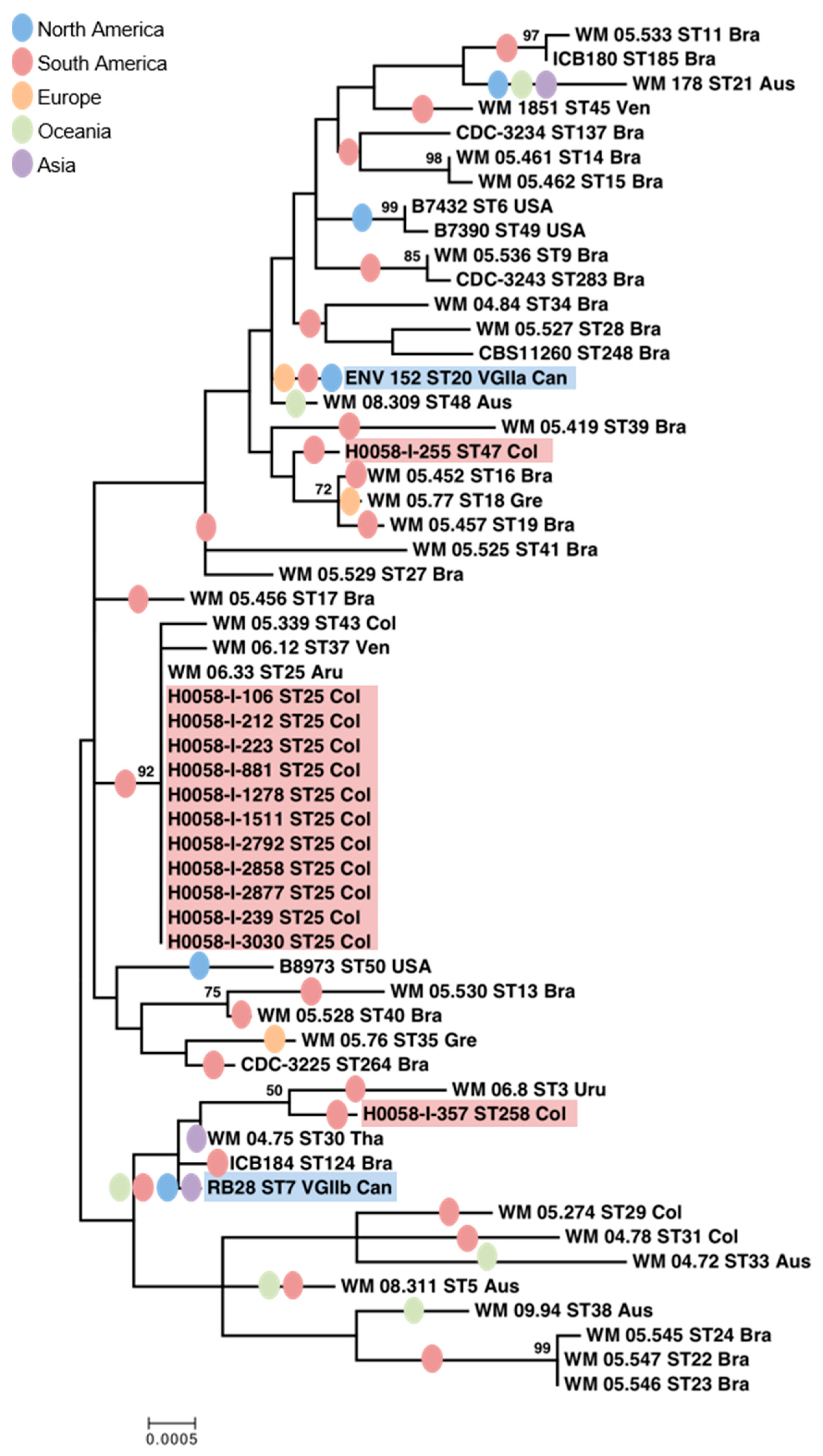
3.1. C. gattii VGII Isolates from Cucuta Were Highly Clonal as Established by MLST
3.2. Isolates Share Several Phenotypic Traits
3.3. Most Isolates Were Fertile and Mated with Opposite Mating Types
3.4. Enzymatic Activities Slightly Differ Among the Isolates
3.5. Isolates from Cucuta Are of Considerable Virulence
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kwon-Chung, K.J.; Fraser, J.A.; Doering, T.L.; Wang, Z.; Janbon, G.; Idnurm, A.; Bahn, Y.S. Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb. Perspect Med. 2014, 4, a019760. [Google Scholar] [CrossRef] [PubMed]
- Bovers, M.; Hagen, F.; Kuramae, E.E.; Diaz, M.R.; Spanjaard, L.; Dromer, F.; Hoogveld, H.L.; Boekhout, T. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 2006, 6, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Meyer, W.; Mitchell, T.G.; Freedman, E.Z.; Vilgalys, R. Hybridization probes for conventional DNA fingerprinting used as single primers in the polymerase chain reaction to distinguish strains of Cryptococcus neoformans. J. Clin. Microbiol. 1993, 31, 2274–2280. [Google Scholar] [PubMed]
- Meyer, W.; Castañeda, A.; Jackson, S.; Huynh, M.; Castañeda, E.; IberoAmerican Cryptococcal Study Group. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg. Infect. Dis. 2003, 9, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Meyer, W.; Aanensen, D.M.; Boekhout, T.; Cogliati, M.; Diaz, M.R.; Esposto, M.C.; Fisher, M.; Gilgado, F.; Hagen, F.; Kaocharoen, S.; et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 2009, 47, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Hagen, F.; Khayhan, K.; Theelen, B.; Kolecka, A.; Polacheck, I.; Sionov, E.; Falk, R.; Parnmen, S.; Lumbsch, H.T.; Boekhout, T. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet. Biol. 2015, 78, 16–48. [Google Scholar] [CrossRef] [PubMed]
- Kwon-Chung, K.J.; Bennett, J.E.; Wickes, B.L.; Meyer, W.; Cuomo, C.A.; Wollenburg, K.R.; Bicanic, T.A.; Castaneda, E.; Chang, Y.C.; Chen, J.; et al. The case for adopting the "Species Complex" nomenclature for the etiologic agents of cryptococcosis. mSphere 2017, 2. [Google Scholar] [CrossRef]
- Chen, S.C.; Meyer, W.; Sorrell, T.C. Cryptococcus gattii infections. Clin. Microbiol. Rev. 2014, 27, 980–1024. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Iqbal, N.; Harris, J.R.; Grossman, N.T.; DeBess, E.; Wohrle, R.; Marsden-Haug, N.; Vugia, D.J. Cryptococcus gattii in the United States: Genotypic diversity of human and veterinary isolates. PLoS ONE 2013, 8, e74737. [Google Scholar] [CrossRef]
- Kidd, S.E.; Hagen, F.; Tscharke, R.L.; Huynh, M.; Bartlett, K.H.; Fyfe, M.; Macdougall, L.; Boekhout, T.; Kwon-Chung, K.J.; Meyer, W. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc. Natl. Acad. Sci. USA 2004, 101, 17258–17263. [Google Scholar] [CrossRef] [PubMed]
- MacDougall, L.; Kidd, S.E.; Galanis, E.; Mak, S.; Leslie, M.J.; Cieslak, P.R.; Kronstad, J.W.; Morshed, M.G.; Bartlett, K.H. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg. Infect. Dis. 2007, 13, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.A.; Giles, S.S.; Wenink, E.C.; Geunes-Boyer, S.G.; Wright, J.R.; Diezmann, S.; Allen, A.; Stajich, J.E.; Dietrich, F.S.; Perfect, J.R.; et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 2005, 437, 1360–1364. [Google Scholar] [CrossRef] [PubMed]
- Gillece, J.D.; Schupp, J.M.; Balajee, S.A.; Harris, J.; Pearson, T.; Yan, Y.; Keim, P.; DeBess, E.; Marsden-Haug, N.; Wohrle, R.; et al. Whole genome sequence analysis of Cryptococcus gattii from the Pacific Northwest reveals unexpected diversity. PLoS ONE 2011, 6, e28550. [Google Scholar] [CrossRef] [PubMed]
- Ellis, D.H. Cryptococcus neoformans var. gattii in Australia. J. Clin. Microbiol. 1987, 25, 430–431. [Google Scholar] [PubMed]
- Laurenson, I.F.; Lalloo, D.G.; Naraqi, S.; Seaton, R.A.; Trevett, A.J.; Matuka, A.; Kevau, I.H. Cryptococcus neoformans in Papua New Guinea: A common pathogen but an elusive source. J. Med. Vet. Mycol. 1997, 35, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Firacative, C.; Lizarazo, J.; Illnait-Zaragozi, M.T.; Castaneda, E.; Latin American Cryptococcal Study Group. The status of cryptococcosis in Latin America. Mem. Inst. Oswaldo Cruz 2018, 113, e170554. [Google Scholar] [CrossRef] [PubMed]
- Colom, M.F.; Hagen, F.; Gonzalez, A.; Mellado, A.; Morera, N.; Linares, C.; Garcia, D.F.; Penataro, J.S.; Boekhout, T.; Sanchez, M. Ceratonia siliqua (carob) trees as natural habitat and source of infection by Cryptococcus gattii in the Mediterranean environment. Med. Mycol. 2012, 50, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Kohl, K.H.; Hof, H.; Schrettenbrunner, A.; Seeliger, H.P.; Kwon-Chung, K.J. Cryptococcus neoformans var. gattii in Europe. Lancet 1985, 1, 1515. [Google Scholar] [CrossRef]
- Romeo, O.; Scordino, F.; Criseo, G. Environmental isolation of Cryptococcus gattii serotype B, VGI/MATalpha strains in southern Italy. Mycopathologia 2011, 171, 423–430. [Google Scholar] [CrossRef]
- Escandón, P.; Lizarazo, J.; Agudelo, C.I.; Castañeda, E. Cryptococcosis in Colombia: Compilation and analysis of data from laboratory-based surveillance. J. Fungi (Basel) 2018, 4, 32. [Google Scholar] [CrossRef]
- Escandón, P.; Sánchez, A.; Martinez, M.; Meyer, W.; Castaneda, E. Molecular epidemiology of clinical and environmental isolates of the Cryptococcus neoformans species complex reveals a high genetic diversity and the presence of the molecular type VGII mating type a in Colombia. FEMS Yeast Res. 2006, 6, 625–635. [Google Scholar] [CrossRef]
- Lizarazo, J.; Escandon, P.; Agudelo, C.I.; Firacative, C.; Meyer, W.; Castaneda, E. Retrospective study of the epidemiology and clinical manifestations of Cryptococcus gattii infections in Colombia from 1997–2011. PLoS Negl. Trop. Dis. 2014, 8, e3272. [Google Scholar] [CrossRef]
- Engelthaler, D.M.; Hicks, N.D.; Gillece, J.D.; Roe, C.C.; Schupp, J.M.; Driebe, E.M.; Gilgado, F.; Carriconde, F.; Trilles, L.; Firacative, C.; et al. Cryptococcus gattii in North American Pacific Northwest: Whole-population genome analysis provides insights into species evolution and dispersal. MBio 2014, 5, e01464-14. [Google Scholar] [CrossRef] [PubMed]
- Lizarazo, J.; Linares, M.; de Bedout, C.; Restrepo, A.; Agudelo, C.I.; Castañeda, E. Estudio clínico y epidemiológico de la criptococosis en Colombia: Resultados de nueve años de la encuesta nacional, 1997–2005. Biomedica 2007, 27, 94–109. [Google Scholar] [CrossRef] [PubMed]
- Firacative, C.; Torres, G.; Rodriguez, M.C.; Escandon, P. First environmental isolation of Cryptococcus gattii serotype B, from Cucuta, Colombia. Biomedica 2011, 31, 118–123. [Google Scholar] [CrossRef]
- Meyer, W.; Marszewska, K.; Amirmostofian, M.; Igreja, R.P.; Hardtke, C.; Methling, K.; Viviani, M.A.; Chindamporn, A.; Sukroongreung, S.; John, M.A.; et al. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA-a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis 1999, 20, 1790–1799. [Google Scholar] [CrossRef]
- Yan, Z.; Li, X.; Xu, J. Geographic distribution of mating type alleles of Cryptococcus neoformans in four areas of the United States. J. Clin. Microbiol. 2002, 40, 965–972. [Google Scholar] [CrossRef]
- Ngamskulrungroj, P.; Sorrell, T.C.; Chindamporn, A.; Chaiprasert, A.; Poonwan, N.; Meyer, W. Association between fertility and molecular sub-type of global isolates of Cryptococcus gattii molecular type VGII. Med. Mycol. 2008, 46, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Casali, A.K.; Goulart, L.; Rosa e Silva, L.K.; Ribeiro, A.M.; Amaral, A.A.; Alves, S.H.; Schrank, A.; Meyer, W.; Vainstein, M.H. Molecular typing of clinical and environmental Cryptococcus neoformans isolates in the Brazilian state Rio Grande do Sul. FEMS Yeast Res. 2003, 3, 405–415. [Google Scholar] [CrossRef]
- Halliday, C.L.; Bui, T.; Krockenberger, M.; Malik, R.; Ellis, D.H.; Carter, D.A. Presence of alpha and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia. J. Clin. Microbiol. 1999, 37, 2920–2926. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Carriconde, F.; Gilgado, F.; Arthur, I.; Ellis, D.; Malik, R.; van de Wiele, N.; Robert, V.; Currie, B.J.; Meyer, W. Clonality and alpha-a recombination in the Australian Cryptococcus gattii VGII population-an emerging outbreak in Australia. PLoS ONE 2011, 6, e16936. [Google Scholar] [CrossRef] [PubMed]
- Hagen, F.; Colom, M.F.; Swinne, D.; Tintelnot, K.; Iatta, R.; Montagna, M.T.; Torres-Rodriguez, J.M.; Cogliati, M.; Velegraki, A.; Burggraaf, A.; et al. Autochthonous and dormant Cryptococcus gattii infections in Europe. Emerg. Infect. Dis. 2012, 18, 1618–1624. [Google Scholar] [CrossRef] [PubMed]
- Kaocharoen, S.; Ngamskulrungroj, P.; Firacative, C.; Trilles, L.; Piyabongkarn, D.; Banlunara, W.; Poonwan, N.; Chaiprasert, A.; Meyer, W.; Chindamporn, A. Molecular epidemiology reveals genetic diversity amongst isolates of the Cryptococcus neoformans/C. gattii species complex in Thailand. PLoS Negl. Trop. Dis. 2013, 7, e2297. [Google Scholar] [CrossRef] [PubMed]
- Franzot, S.P.; Mukherjee, J.; Cherniak, R.; Chen, L.C.; Hamdan, J.S.; Casadevall, A. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect. Immun. 1998, 66, 89–97. [Google Scholar]
- Fries, B.C.; Casadevall, A. Serial isolates of Cryptococcus neoformans from patients with AIDS differ in virulence for mice. J. Infect. Dis. 1998, 178, 1761–1766. [Google Scholar] [CrossRef]
- Mackenzie, D.W. Effect of relative humidity on survival of Candida albicans and other yeasts. Appl. Microbiol. 1971, 22, 678–682. [Google Scholar] [PubMed]
- Rosa e Silva, L.K.; Staats, C.C.; Goulart, L.S.; Morello, L.G.; Pelegrinelli Fungaro, M.H.; Schrank, A.; Vainstein, M.H. Identification of novel temperature-regulated genes in the human pathogen Cryptococcus neoformans using representational difference analysis. Res. Microbiol. 2008, 159, 221–229. [Google Scholar] [CrossRef]
- Fries, B.C.; Taborda, C.P.; Serfass, E.; Casadevall, A. Phenotypic switching of Cryptococcus neoformans occurs in vivo and influences the outcome of infection. J. Clin. Investig. 2001, 108, 1639–1648. [Google Scholar] [CrossRef]
- Hicks, J.K.; Bahn, Y.S.; Heitman, J. Pde1 phosphodiesterase modulates cyclic AMP levels through a protein kinase A-mediated negative feedback loop in Cryptococcus neoformans. Eukaryot. Cell 2005, 4, 1971–1981. [Google Scholar] [CrossRef]
- Leone, R.; Cabeli, P.; Sinicco, A.; Ito-Kuwa, S.; Aoki, S.; Vidotto, V. Relationship between protease production and capsule size in Cryptococcus neoformans. J. Med. Mycol. 1999, 9, 42–44. [Google Scholar]
- Price, M.F.; Wilkinson, I.D.; Gentry, L.O. Plate method for detection of phospholipase activity in Candida albicans. Sabouraudia 1982, 20, 7–14. [Google Scholar] [CrossRef]
- Torres-Rodriguez, J.M.; Alvarado-Ramirez, E.; Gutierrez-Gallego, R. Urease activity in Cryptococcus neoformans and Cryptococcus gattii. Rev. Iberoam. Micol. 2008, 25, 27–31. [Google Scholar] [CrossRef]
- Krockenberger, M.B.; Malik, R.; Ngamskulrungroj, P.; Trilles, L.; Escandon, P.; Dowd, S.; Allen, C.; Himmelreich, U.; Canfield, P.J.; Sorrell, T.C.; et al. Pathogenesis of pulmonary Cryptococcus gattii infection: A rat model. Mycopathologia 2010, 170, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Lizarazo, J.; Escandón, P.; Agudelo, C.I.; Castañeda, E. Cryptococcosis in Colombian children and literature review. Mem. Inst. Oswaldo Cruz 2014, 109, 797–804. [Google Scholar] [CrossRef] [Green Version]
- Severo, C.B.; Xavier, M.O.; Gazzoni, A.F.; Severo, L.C. Cryptococcosis in children. Paediatr. Respir. Rev. 2009, 10, 166–171. [Google Scholar] [CrossRef]
- Billmyre, R.B.; Croll, D.; Li, W.; Mieczkowski, P.; Carter, D.A.; Cuomo, C.A.; Kronstad, J.W.; Heitman, J. Highly recombinant VGII Cryptococcus gattii population develops clonal outbreak clusters through both sexual macroevolution and asexual microevolution. MBio 2014, 5, e01494-14. [Google Scholar] [CrossRef]
- Aksenov, S.I.; Babyeva, I.P.; Golubev, V.I. On the mechanism of adaptation of micro-organisms to conditions of extreme low humidity. Life Sci. Space Res. 1973, 11, 55–61. [Google Scholar]
- Ruiz, A.; Neilson, J.B.; Bulmer, G.S. A one year study on the viability of Cryptococcus neoformans in nature. Mycopathologia 1982, 77, 117–122. [Google Scholar] [CrossRef]
- Granados, D.P.; Castaneda, E. Influence of climatic conditions on the isolation of members of the Cryptococcus neoformans species complex from trees in Colombia from 1992–2004. FEMS Yeast Res. 2006, 6, 636–644. [Google Scholar] [CrossRef]
- Ophir, T.; Gutnick, D.L. A role for exopolysaccharides in the protection of microorganisms from desiccation. Appl. Environ. Microbiol. 1994, 60, 740–745. [Google Scholar]
- Bulmer, G.S.; Sans, M.D. Cryptococcus neoformans. II. Phagocytosis by human leukocytes. J. Bacteriol. 1967, 94, 1480–1483. [Google Scholar] [PubMed]
- Jain, N.; Fries, B.C. Phenotypic switching of Cryptococcus neoformans and Cryptococcus gattii. Mycopathologia 2008, 166, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Huérfano, S.; Cepero, M.C.; Castañeda, E. Phenotype characterization of environmental Cryptococcus neoformans isolates. Biomedica 2003, 23, 328–340. [Google Scholar] [CrossRef] [PubMed]



| Strain Number | Other Number | Year of Isolation | Patient’s Age (Years) | Patient’s Gender 1 | Clinical Presentation | Outcome | Ref. |
|---|---|---|---|---|---|---|---|
| H0058-I-106 | WM 08.290 | 1999 | 43 | M | Meningitis | Living | - |
| H0058-I-212 | WM 08.288 | 1993 | 54 | M | Meningitis | Deceased | - |
| H0058-I-223 | WM 08.289 | 1993 | 41 | M | Meningitis | Living | - |
| H0058-I-239 | WM 08.291 | 1993 | 8 | F | Meningitis | Deceased | - |
| H0058-I-255 | WM 08.292 | 1999 | 25 | M 2 | Meningitis | Living | - |
| H0058-I-357 | WM 08.293 | 1995 | 11 | M | Meningitis | ND | - |
| H0058-I-881 | WM 08.295 | 1999 | 34 | M | Meningitis | Living | [22] |
| H0058-I-1278 | WM 05.275 | 2001 | 39 | M | Meningitis | Living | [22,23] |
| H0058-I-1511 | WM 05.399 | 2002 | 56 | M | Meningitis | Living | [22] |
| H0058-I-2792 | WM 08.297 | 2007 | 51 | M | Meningitis | ND | [22] |
| H0058-I-2858 | WM 08.298 | 2007 | 60 | M | Meningitis | ND | [22] |
| H0058-I-2877 | WM 08.299 | 2008 | 46 | F 3 | Meningitis | Deceased | [22] |
| H0058-I-3030 | WM 08.305 | 2008 | 31 | M | Meningitis | Living | - |
| Strain Number | Mating Type | CAP59 | GPD1 | IGS1 | LAC1 | PLB1 | SOD1 | URA5 | ST |
|---|---|---|---|---|---|---|---|---|---|
| H0058-I-106 | a | 2 | 6 | 25 | 4 | 18 | 12 | 10 | 25 |
| H0058-I-212 | a | 2 | 6 | 25 | 4 | 18 | 12 | 10 | 25 |
| H0058-I-223 | a | 2 | 6 | 25 | 4 | 18 | 12 | 10 | 25 |
| H0058-I-239 | a | 2 | 6 | 25 | 4 | 18 | 12 | 10 | 25 |
| H0058-I-881 | a | 2 | 6 | 25 | 4 | 18 | 12 | 10 | 25 |
| H0058-I-1278 | a | 2 | 6 | 25 | 4 | 18 | 12 | 10 | 25 |
| H0058-I-1511 | a | 2 | 6 | 25 | 4 | 18 | 12 | 10 | 25 |
| H0058-I-2792 | a | 2 | 6 | 25 | 4 | 18 | 12 | 10 | 25 |
| H0058-I-2858 | a | 2 | 6 | 25 | 4 | 18 | 12 | 10 | 25 |
| H0058-I-2877 | a | 2 | 6 | 25 | 4 | 18 | 12 | 10 | 25 |
| H0058-I-3030 | a | 2 | 6 | 25 | 4 | 18 | 12 | 10 | 25 |
| H0058-I-255 | alpha | 2 | 6 | 15 | 4 | 1 | 42 | 7 | 47 |
| H0058-I-357 | a | 7 | 2 | 32 | 7 | 25 | 15 | 2 | 258 |
| RB28 (VGIIb) | alpha | 2 | 6 | 10 | 4 | 2 | 15 | 2 | 7 |
| ENV152 (VGIIa) | alpha | 1 | 1 | 4 | 4 | 1 | 14 | 7 | 20 |
| Strain Number | Switching | Colony Morphology | Phenol-Oxydase (μg/mL) | Proteases | Phospholipases | |||
|---|---|---|---|---|---|---|---|---|
| Diameter (mm) | Texture | Mean (Pz) | Activity | Mean (Pz) | Activity | |||
| Isolates from Cucuta | ||||||||
| H0058-I-106 | + | 7.5 | Smooth | 196.3 | 1 | None | 0.60 | Medium |
| H0058-I-212 | + | 9.5 | Mucoid | 35.3 | 1 | None | 0.57 | Medium |
| H0058-I-223 | - | 12.0 | Mucoid | 216.2 | 1 | None | 0.67 | Medium |
| H0058-I-239 | + | 7.6 | Mucoid | 153.9 | 1 | None | 0.63 | Medium |
| H0058-I-255 | + | 7.0 | Mucoid | 1.2 | 1 | None | 0.60 | Medium |
| H0058-I-357 | + | 10.0 | Mucoid | 193.1 | 1 | None | 0.68 | Medium |
| H0058-I-881 | + | 10.3 | Mucoid | 185.4 | 1 | None | 0.63 | Medium |
| H0058-I-1278 | + | 9.1 | Mucoid | 201.6 | 1 | None | 0.68 | Medium |
| H0058-I-1511 | - | 7.1 | Smooth | 168.7 | 1 | None | 0.66 | Medium |
| H0058-I-2792 | + | 8.4 | Smooth | 203.9 | 1 | None | 0.60 | Medium |
| H0058-I-2858 | + | 8.0 | Smooth | 239.8 | 1 | None | 0.61 | Medium |
| H0058-I-2877 | + | 9.3 | Smooth | 119.1 | 1 | None | 0.57 | Medium |
| H0058-I-3030 | + | 7.8 | Smooth | 159.5 | 1 | None | 0.57 | Medium |
| Vancouver Island Isolates | ||||||||
| ENV152 (VGIIa) | + | 8.6 | Smooth | 150.82 | 0.82 | Low | 0.70 | Low |
| RB28 (VGIIb) | + | 8.3 | Smooth | 116.68 | 0.78 | Low | 0.68 | Medium |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Firacative, C.; Torres, G.; Meyer, W.; Escandón, P. Clonal Dispersal of Cryptococcus gattii VGII in an Endemic Region of Cryptococcosis in Colombia. J. Fungi 2019, 5, 32. https://doi.org/10.3390/jof5020032
Firacative C, Torres G, Meyer W, Escandón P. Clonal Dispersal of Cryptococcus gattii VGII in an Endemic Region of Cryptococcosis in Colombia. Journal of Fungi. 2019; 5(2):32. https://doi.org/10.3390/jof5020032
Chicago/Turabian StyleFiracative, Carolina, Germán Torres, Wieland Meyer, and Patricia Escandón. 2019. "Clonal Dispersal of Cryptococcus gattii VGII in an Endemic Region of Cryptococcosis in Colombia" Journal of Fungi 5, no. 2: 32. https://doi.org/10.3390/jof5020032
APA StyleFiracative, C., Torres, G., Meyer, W., & Escandón, P. (2019). Clonal Dispersal of Cryptococcus gattii VGII in an Endemic Region of Cryptococcosis in Colombia. Journal of Fungi, 5(2), 32. https://doi.org/10.3390/jof5020032







