Insights into the Gryllus bimaculatus Immune-Related Transcriptomic Profiling to Combat Naturally Invading Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection, Maintenance, and Tissue Extraction of Two-Spotted Field Crickets
2.2. Construction of cDNA Libraries of Two-Spotted Field Crickets
2.3. Sequence Assembly of the cDNA Libraries of Two-Spotted Field Crickets
2.4. Analysis of Differential Expression of Genes
2.5. Functional Annotation of the cDNA Libraries of Two-Spotted Field Crickets
3. Results
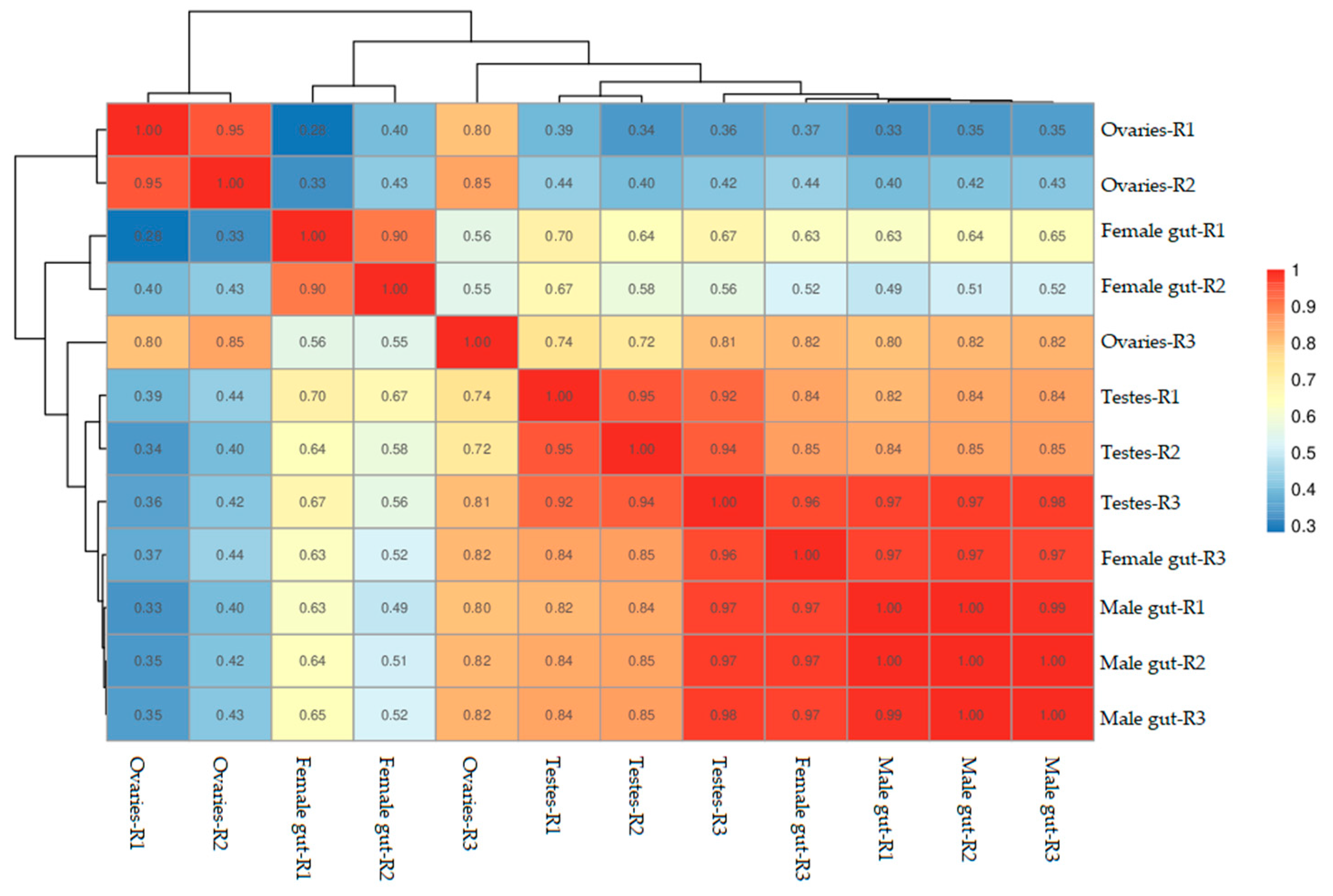
3.1. Samples Correlation Analysis
3.2. Sequence Assembly Characteristics of Two-Spotted Field Crickets
3.3. Classification and Functional Annotation of Two-Spotted Field Cricket Transcriptomes
3.4. Patterns of Differential Gene Expression Levels
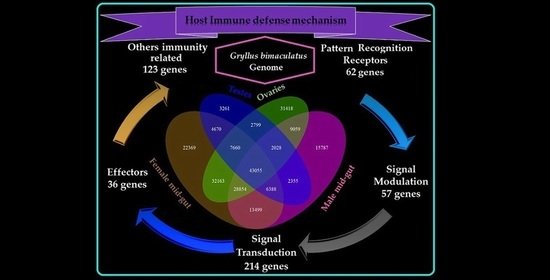
3.5. Immune-Related Transcriptome of Two-Spotted Field Crickets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Adamo, S.A.; Hoy, R.R. Agonistic behaviour in male and female field crickets, Gryllus bimaculatus, and how behavioural context influences its expression. Anim. Behav. 1995, 49, 1491–1501. [Google Scholar] [CrossRef]
- Haberkern, H.; Hedwig, B. Behavioural integration of auditory and antennal stimulation during phonotaxis in the field cricket Gryllus bimaculatus. J. Exp. Biol. 2016, 219, 3575–3586. [Google Scholar] [CrossRef] [PubMed]
- Rantala, M.J.; Roff, D.A. An analysis of trade-offs in immune function, body size and development time in the Mediterranean Field Cricket, Gryllus bimaculatus. Funct. Ecol. 2005, 19, 323–330. [Google Scholar] [CrossRef]
- Zorović, M.; Hedwig, B. Descending brain neurons in the cricket Gryllus bimaculatus (de Geer): Auditory responses and impact on walking. J. Comp. Physiol. A 2013, 199, 25–34. [Google Scholar] [CrossRef]
- Donoughe, S.; Extavour, C.G. Embryonic development of the cricket Gryllus bimaculatus. Dev. Biol. 2016, 411, 140–156. [Google Scholar] [CrossRef]
- Mito, T.; Noji, S. The Two-Spotted Cricket Gryllus bimaculatus: An emerging model for developmental and regeneration studies. Cold Spring Harb. Protoc. 2008, 2008, pdb-emo110. [Google Scholar] [CrossRef]
- Sorjonen, J.M.; Valtonen, A.; Hirvisalo, E.; Karhapää, M.; Lehtovaara, V.J.; Lindgren, J.; Marnila, P.; Mooney, P.; Mäki, M.; Siljander-Rasi, H.; et al. The plant-based by-product diets for the mass-rearing of Acheta domesticus and Gryllus bimaculatus. PLoS ONE 2019, 14, e0218830. [Google Scholar] [CrossRef]
- Hussain, A.; Rizwan-ul-Haq, M.; Al-Ayedh, H.; AlJabr, A. Susceptibility and immune defence mechanisms of Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae) against entomopathogenic fungal infections. Int. J. Mol. Sci. 2016, 17, 1518. [Google Scholar] [CrossRef]
- Hussain, A.; Tian, M.-Y.; Wen, S.-Y. Exploring the caste-specific multi-layer defense mechanism of Formosan Subterranean Termites, Coptotermes formosanus Shiraki. Int. J. Mol. Sci. 2017, 18, 2694. [Google Scholar] [CrossRef]
- Gupta, S.K.; Kupper, M.; Ratzka, C.; Feldhaar, H.; Vilcinskas, A.; Gross, R.; Dandekar, T.; Förster, F. Scrutinizing the immune defence inventory of Camponotus floridanus applying total transcriptome sequencing. BMC Genom. 2015, 16, 540. [Google Scholar] [CrossRef]
- Koch, S.I.; Groh, K.; Vogel, H.; Hannson, B.S.; Kleineidam, C.J.; Grosse-Wilde, E. Caste-specific expression patterns of immune response and chemosensory related genes in the leaf-cutting ant, Atta vollenweideri. PLoS ONE 2013, 8, e81518. [Google Scholar] [CrossRef]
- Sun, J.; Bai, Y. Predator-induced stress influences fall armyworm immune response to inoculating bacteria. J. Invertebr. Pathol. 2020, 172, 107352. [Google Scholar] [CrossRef] [PubMed]
- Nishide, Y.; Kageyama, D.; Yokoi, K.; Jouraku, A.; Tanaka, H.; Futahashi, R.; Fukatsu, T. Functional crosstalk across IMD and Toll pathways: Insight into the evolution of incomplete immune cascades. Proc. R. Soc. B Biol. Sci. 2019, 286, 20182207. [Google Scholar] [CrossRef] [PubMed]
- Sparks, M.E.; Shelby, K.S.; Kuhar, D.; Gundersen-Rindal, D.E. Transcriptome of the Invasive Brown Marmorated Stink Bug, Halyomorpha halys (Stål) (Heteroptera: Pentatomidae). PLoS ONE 2014, 9, e111646. [Google Scholar] [CrossRef]
- Gonella, E.; Mandrioli, M.; Tedeschi, R.; Crotti, E.; Pontini, M.; Alma, A. Activation of immune genes in leafhoppers by phytoplasmas and symbiotic bacteria. Front. Physiol. 2019, 10, 795. [Google Scholar] [CrossRef]
- Hussain, A.; Li, Y.F.; Cheng, Y.; Liu, Y.; Chen, C.C.; Wen, S.Y. Immune-related transcriptome of Coptotermes formosanus Shiraki workers: The defense mechanism. PLoS ONE 2013, 8, e69543. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Tian, M.-Y.; Wen, S.-Y. Proteomic analysis of Formosan Subterranean Termites during exposure to entomopathogenic fungi. Curr. Proteomics 2018, 15, 229–240. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- McIntyre, L.M.; Lopiano, K.K.; Morse, A.M.; Amin, V.; Oberg, A.L.; Young, L.J.; Nuzhdin, S. V RNA-seq: Technical variability and sampling. BMC Genom. 2011, 12, 293. [Google Scholar] [CrossRef]
- Wang, Y.; Sumathipala, N.; Rayaprolu, S.; Jiang, H. Recognition of microbial molecular patterns and stimulation of prophenoloxidase activation by a β-1,3-glucanase-related protein in Manduca sexta larval plasma. Insect Biochem. Mol. Biol. 2011, 41, 322–331. [Google Scholar] [CrossRef]
- De Gregorio, E. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002, 21, 2568–2579. [Google Scholar] [CrossRef] [PubMed]
- Dimopoulos, G. Insect immunity and its implication in mosquito-malaria interactions. Cell. Microbiol. 2003, 5, 3–14. [Google Scholar] [CrossRef]
- Hoffmann, J.A. The immune response of Drosophila. Nature 2003, 426, 33–38. [Google Scholar] [CrossRef]
- Theopold, U.; Schmidt, O.; Söderhäll, K.; Dushay, M.S. Coagulation in arthropods: Defence, wound closure and healing. Trends Immunol. 2004, 25, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, M.; Ashida, M. A pattern-recognition protein for beta-1,3-glucan. The binding domain and the cDNA cloning of beta-1,3-glucan recognition protein from the silkworm, Bombyx mori. J. Biol. Chem. 2000, 275, 4995–5002. [Google Scholar] [CrossRef]
- Rana, A.; Ahmed, M.; Rub, A.; Akhter, Y. A tug-of-war between the host and the pathogen generates strategic hotspots for the development of novel therapeutic interventions against infectious diseases. Virulence 2015, 6, 566–580. [Google Scholar] [CrossRef] [PubMed]
- Gotz, P.; Weise, C.; Kopacek, P.; Losen, S.; Wiesner, A. Isolated apolipophorin III from Galleria mellonella stimulates the immune reactions of this insect. J. Insect Physiol. 1997, 43, 383–391. [Google Scholar]
- Wang, C.; Cao, Y.; Wang, Z.; Yin, Y.; Peng, G.; Li, Z.; Zhao, H.; Xia, Y. Differentially-expressed glycoproteins in Locusta migratoria hemolymph infected with Metarhizium anisopliae. J. Invertebr. Pathol. 2007, 96, 230–236. [Google Scholar] [CrossRef]
- Whitten, M.M.A.; Tew, I.F.; Lee, B.L.; Ratcliffe, N.A. A novel role for an insect apolipoprotein (apolipophorin III) in beta-1,3-glucan pattern recognition and cellular encapsulation reactions. J. Immunol. 2004, 172, 2177–2185. [Google Scholar] [CrossRef] [PubMed]
- Zdybicka-Barabas, A.; Stączek, S.; Mak, P.; Piersiak, T.; Skrzypiec, K.; Cytryńska, M. The effect of Galleria mellonella apolipophorin III on yeasts and filamentous fungi. J. Insect Physiol. 2012, 58, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Zdybicka-Barabas, A.; Cytryńska, M. Involvement of apolipophorin III in antibacterial defense of Galleria mellonella larvae. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2011, 158, 90–98. [Google Scholar] [CrossRef]
- Weis, W.I.; Taylor, M.E.; Drickamer, K. The C-type lectin superfamily in the immune system. Immunol. Rev. 1998, 163, 19–34. [Google Scholar] [CrossRef]
- Zhang, D.; Lax, A.R.; Henrissat, B.; Coutinho, P.; Katiya, N.; Nierman, W.C.; Fedorova, N. Carbohydrate-active enzymes revealed in Coptotermes formosanus (Isoptera: Rhinotermitidae) transcriptome. Insect Mol. Biol. 2012, 21, 235–245. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen Recognition and Innate Immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Gorman, M.J.; Paskewitz, S.M. Serine proteases as mediators of mosquito immune responses. Insect Biochem. Mol. Biol. 2001, 31, 257–262. [Google Scholar] [CrossRef]
- Tang, H.; Kambris, Z.; Lemaitre, B.; Hashimoto, C. A serpin that regulates immune melanization in the respiratory system of Drosophila. Dev. Cell 2008, 15, 617–626. [Google Scholar] [CrossRef]
- Tanaka, H.; Ishibashi, J.; Fujita, K.; Nakajima, Y.; Sagisaka, A.; Tomimoto, K.; Suzuki, N.; Yoshiyama, M.; Kaneko, Y.; Iwasaki, T.; et al. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1087–1110. [Google Scholar] [CrossRef]
- Zhao, P.; Dong, Z.; Duan, J.; Wang, G.; Wang, L.; Li, Y.; Xiang, Z.; Xia, Q. Genome-wide identification and immune response analysis of serine protease inhibitor genes in the silkworm, Bombyx mori. PLoS ONE 2012, 7, e31168. [Google Scholar] [CrossRef]
- Park, D.S.; Shin, S.W.; Hong, S.D.; Park, H.Y. Immunological detection of serpin in the fall webworm, Hyphantria cunea and its inhibitory activity on the prophenoloxidase system. Mol. Cells 2000, 10, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Zettervall, C.J.; Anderl, I.; Williams, M.J.; Palmer, R.; Kurucz, E.; Ando, I.; Hultmark, D. A directed screen for genes involved in Drosophila blood cell activation. Proc. Natl. Acad. Sci. USA 2004, 101, 14192–14197. [Google Scholar] [CrossRef]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef]
- Rojas, A.M.; Fuentes, G.; Rausell, A.; Valencia, A. The Ras protein superfamily: Evolutionary tree and role of conserved amino acids. J. Cell Biol. 2012, 196, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Shrestha, S.; Prasad, S.V.; Kim, Y. Role of a small G protein Ras in cellular immune response of the beet armyworm, Spodoptera exigua. J. Insect Physiol. 2011, 57, 356–362. [Google Scholar] [CrossRef]
- Wixler, V.; Hirner, S.; Müller, J.M.; Gullotti, L.; Will, C.; Kirfel, J.; Günther, T.; Schneider, H.; Bosserhoff, A.; Schorle, H.; et al. Deficiency in the LIM-only protein Fhl2 impairs skin wound healing. J. Cell Biol. 2007, 177, 163–172. [Google Scholar] [CrossRef]
- Martin, G.B.; Brommonschenkel, S.H.; Chunwongse, J.; Frary, A.; Ganal, M.W.; Spivey, R.; Wu, T.; Earle, E.D.; Tanksley, S.D. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 1993, 262, 1432–1436. [Google Scholar] [CrossRef] [PubMed]
- Belvin, M.P.; Anderson, K.V. A conserved signaling pathway: The Drosophila toll-dorsal pathway. Annu. Rev. Cell Dev. Biol. 1996, 12, 393–416. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, P.; Bao, Y.; Han, Y.; Wang, Y.; Zhang, Q.; Zhan, Z.; Meng, J.; Li, Y.; Li, N.; et al. Zinc finger protein ZBTB20 promotes Toll-like receptor-triggered innate immune responses by repressing IκBα gene transcription. Proc. Natl. Acad. Sci. USA 2013, 110, 11097–11102. [Google Scholar] [CrossRef] [PubMed]
- Tsuruma, R.; Ohbayashi, N.; Kamitani, S.; Ikeda, O.; Sato, N.; Muromoto, R.; Sekine, Y.; Oritani, K.; Matsuda, T. Physical and functional interactions between STAT3 and KAP1. Oncogene 2008, 27, 3054–3059. [Google Scholar] [CrossRef] [PubMed]
- Ulvila, J.; Vanha-aho, L.-M.; Kleino, A.; Vaha-Makila, M.; Vuoksio, M.; Eskelinen, S.; Hultmark, D.; Kocks, C.; Hallman, M.; Parikka, M.; et al. Cofilin regulator 14-3-3 is an evolutionarily conserved protein required for phagocytosis and microbial resistance. J. Leukoc. Biol. 2011, 89, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Harari-Steinberg, O.; Cantera, R.; Denti, S.; Bianchi, E.; Oron, E.; Segal, D.; Chamovitz, D.A. COP9 signalosome subunit 5 (CSN5/Jab1) regulates the development of the Drosophila immune system: Effects on Cactus, Dorsal and hematopoiesis. Genes Cells 2007, 12, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Nikapitiya, C.; De Zoysa, M.; Whang, I.; Kim, S.-J.; Choi, C.Y.; Lee, J.-S.; Lee, J. Characterization and expression analysis of EF hand domain-containing calcium-regulatory gene from disk abalone: Calcium homeostasis and its role in immunity. Fish Shellfish. Immunol. 2010, 29, 334–342. [Google Scholar] [CrossRef]
- Ghosh, R.; Chakrabarti, C. Crystal structure analysis of NP24-I: A thaumatin-like protein. Planta 2008, 228, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Altincicek, B.; Gross, J.; Vilcinskas, A. Wounding-mediated gene expression and accelerated viviparous reproduction of the pea aphid Acyrthosiphon pisum. Insect Mol. Biol. 2008, 17, 711–716. [Google Scholar] [CrossRef]
- Liu, J.-J.; Sturrock, R.; Ekramoddoullah, A.K.M. The superfamily of thaumatin-like proteins: Its origin, evolution, and expression towards biological function. Plant Cell Rep. 2010, 29, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Wel, H.; Loeve, K. Isolation and Characterization of Thaumatin I and II, the Sweet-Tasting Proteins from Thaumatococcus daniellii Benth. Eur. J. Biochem. 1972, 31, 221–225. [Google Scholar] [CrossRef]
- Brandazza, A.; Angeli, S.; Tegoni, M.; Cambillau, C.; Pelosi, P. Plant stress proteins of the thaumatin-like family discovered in animals. FEBS Lett. 2004, 572, 3–7. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Watanabe, H.; Nagai, M.; Nakade, K.; Takahashi, M.; Sato, T. Lentinula edode s tlg1 encodes a Thaumatin-Like Protein that is involved in lentinan degradation and fruiting body senescence. Plant Physiol. 2006, 141, 793–801. [Google Scholar] [CrossRef]
- Gerardo, N.M.; Altincicek, B.; Anselme, C.; Atamian, H.; Barribeau, S.M.; de Vos, M.; Duncan, E.J.; Evans, J.D.; Gabaldón, T.; Ghanim, M.; et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010, 11, R21. [Google Scholar] [CrossRef]
- Daffre, S.; Kylsten, P.; Samakovlis, C.; Hultmark, D. The lysozyme locus in Drosophila melanogaster: An expanded gene family adapted for expression in the digestive tract. Mol. Gen. Genet. MGG 1994, 242, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Fujita, A. Lysozymes in insects: What role do they play in nitrogen metabolism? Physiol. Entomol. 2004, 29, 305–310. [Google Scholar] [CrossRef]
- Trevijano-Contador, N.; Zaragoza, O. Immune Response of Galleria mellonella against human fungal pathogens. J. Fungi 2018, 5, 3. [Google Scholar] [CrossRef]
- Matsuura, K.; Tamura, T.; Kobayashi, N.; Yashiro, T.; Tatsumi, S. The antibacterial protein lysozyme identified as the termite egg recognition pheromone. PLoS ONE 2007, 2, e813. [Google Scholar] [CrossRef] [PubMed]
- Bewley, M.A.; Marriott, H.M.; Tulone, C.; Francis, S.E.; Mitchell, T.J.; Read, R.C.; Chain, B.; Kroemer, G.; Whyte, M.K.B.; Dockrell, D.H. A cardinal role for Cathepsin D in co-ordinating the host-mediated apoptosis of macrophages and killing of Pneumococci. PLoS Pathog. 2011, 7, e1001262. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.; Zhang, H.; Liu, H.; Zhou, Z.; Niu, D.; Wong, L.; Kucuktas, H.; Liu, X.; Peatman, E.; Liu, Z. Molecular characterization and expression analysis of the channel catfish cathepsin D genes. Fish Shellfish. Immunol. 2011, 31, 164–169. [Google Scholar] [CrossRef]
- Hultmark, D.; Engström, A.; Andersson, K.; Steiner, H.; Bennich, H.; Boman, H.G. Insect immunity. Attacins, a family of antibacterial proteins from Hyalophora cecropia. EMBO J. 1983, 2, 571–576. [Google Scholar] [CrossRef]
- Wu, Q.; Patočka, J.; Kuča, K. Insect Antimicrobial Peptides, a Mini Review. Toxins 2018, 10, 461. [Google Scholar] [CrossRef]


| Sample | NCBI SRA Accession | Biosample Accession | Clean Reads Number | No of Bases | Q20 Content (%) | Q30 Content (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| Mid-Gut of Male Two-Spotted Field Crickets | |||||||
| R1 | SRX8826426 | SAMN15594732 (SRS7091132) | 42,341,558 | 6.7 G | 93 | 83 | 38.1 |
| R2 | SRX8826427 | SAMN15594733 (SRS7091133) | 52,441,310 | 8.3 G | 93 | 84 | 39.0 |
| R3 | SRX8826430 | SAMN15594734 (SRS7091136) | 55,325,614 | 8.7 G | 93 | 84 | 39.2 |
| Mid-Gut of Female Two-Spotted Field Crickets | |||||||
| R1 | SRX8826431 | SAMN15594735 (SRS7091137) | 88,936,348 | 14.0 G | 94 | 84 | 49.9 |
| R2 | SRX8826432 | SAMN15594736 (SRS7091138) | 80,383,866 | 12.6 G | 94 | 84 | 47.2 |
| R3 | SRX8826433 | SAMN15594737 (SRS7091139) | 55,325,614 | 8.7 G | 94 | 85 | 44.3 |
| Testes of Male Two-Spotted Field Crickets | |||||||
| R1 | SRX8826434 | SAMN15594738 (SRS7091140) | 67,447,566 | 10.6 G | 94 | 84 | 49.3 |
| R2 | SRX8826435 | SAMN15594739 (SRS7091141) | 59,198,274 | 9.4 G | 93 | 84 | 49.0 |
| R3 | SRX8826436 | SAMN15594740 (SRS7091142) | 65,058,620 | 10.2 G | 94 | 85 | 45.2 |
| Ovaries of Female Two-Spotted Field Crickets | |||||||
| R1 | SRX8826437 | SAMN15594741(SRS7091143) | 70,968,356 | 11.1 G | 95 | 86 | 43.6 |
| R2 | SRX8826428 | SAMN15594742 (SRS7091134) | 58,530,318 | 9.1 G | 95 | 87 | 41.9 |
| R3 | SRX8826429 | SAMN15594743 (SRS7091135) | 64,386,128 | 10.0 G | 95 | 87 | 42.5 |
| Assembled Reads Information | Male Mid-Gut | Female Mid-Gut | Ovaries | Testes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R1 (%) | R2 (%) | R3 (%) | R1 (%) | R2 (%) | R3 (%) | R1 (%) | R2 (%) | R3 (%) | R1 (%) | R2 (%) | R3 (%) | |
| Unidentified Reads | 68.68 | 79.08 | 84.68 | 95.38 | 91.87 | 81.92 | 96.3 | 97.28 | 95.9 | 87.76 | 89.21 | 85.83 |
| Identified Reads | 31.32 | 20.92 | 15.32 | 4.62 | 8.13 | 18.08 | 3.7 | 2.72 | 4.10 | 12.24 | 10.79 | 14.17 |
| Matched with Arthropoda | 23.71 | 15.32 | 11.69 | 2.76 | 5.61 | 11.47 | 1.56 | 1.13 | 2.05 | 5.96 | 5.24 | 8.84 |
| Functional Categories | Characterized Annotations |
|---|---|
| Pattern Recognition Receptors | Apolipophorin, Ataxin, Beta-1,3-glucan-binding protein, C-type lectin (CTL), Down syndrome cell adhesion molecule-like protein (DSCAM), Galectin (GALE), Gram negative binding protein (GNBP), Hemolymph lipopolysaccharide-binding protein, Hemolymph juvenile hormone binding protein, Immulectin, Lectin, Peptidoglycan-recognition protein (PRP), Regenectin, Septin, Spondin, Techylectin |
| Signal Modulators | Allatotropin (ATs), Allatostatin (Ast), Angiopoietin (Ang), Chymotrypsin, Kazal domain-containing peptide, Kunitz-type protease inhibitor, Porin, proPO, Prostaglandin, Serine protease (SPs), Serpin, Tetraspanin |
| Signal Transductors | Adiponectin receptor protein, Allatostatin A prohormone, Ankyrin repeat and fibronectin type-III domain-containing protein 1, Angiomotin, Beta-arrestin2, Bursicon-beta, C2 domain-containing protein, CSN5 cop9 signalosome subunit 5, C-Jun-amino-terminal kinase-interacting protein 3, Calmodulin, Cactin, Calpain, Chimaerin, Contactin, COP9 signalosome complex, EF-hand domain-containing protein, Folliculin, Four-and-a-half LIM domain protein 1 isoform B, Frizzled, Hippo, Hippocampus abundant transcript 1 protein, IMD-like protein, JNK-interacting protein, Kruppel-like protein, Leucine-rich repeat-containing protein, Malectin-B, NACHT and WD repeat domain-containing protein 1, NACHT and Ankyrin domain protein, Nesprin, Notch, Octopamine, Pelle, Rab, Ras, Rho, Serine/threonine-protein kinase, Spaetzle, Striatin, TATA-box binding protein, Toll-like receptors, Transgelin, Transducin beta-like protein, Target of rapamycin complex 2 subunit MAPKAP1, Tubulin beta chain, WD repeat-containing protein, Wnt, WW domin protein, Zinc finger protein, 14-3-3 family protein |
| Effectors | Attacin, Bacteriocin, Carboxypeptidase, Cathepsin, Caspase, Lysozyme, Pyocin, C-type lysozyme, I-type lysozyme, Pyocin, Thaumatin-like protein 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, A.; Ali, M.W.; AlJabr, A.M.; AL-Kahtani, S.N. Insights into the Gryllus bimaculatus Immune-Related Transcriptomic Profiling to Combat Naturally Invading Pathogens. J. Fungi 2020, 6, 232. https://doi.org/10.3390/jof6040232
Hussain A, Ali MW, AlJabr AM, AL-Kahtani SN. Insights into the Gryllus bimaculatus Immune-Related Transcriptomic Profiling to Combat Naturally Invading Pathogens. Journal of Fungi. 2020; 6(4):232. https://doi.org/10.3390/jof6040232
Chicago/Turabian StyleHussain, Abid, Muhammad Waqar Ali, Ahmed Mohammed AlJabr, and Saad Naser AL-Kahtani. 2020. "Insights into the Gryllus bimaculatus Immune-Related Transcriptomic Profiling to Combat Naturally Invading Pathogens" Journal of Fungi 6, no. 4: 232. https://doi.org/10.3390/jof6040232
APA StyleHussain, A., Ali, M. W., AlJabr, A. M., & AL-Kahtani, S. N. (2020). Insights into the Gryllus bimaculatus Immune-Related Transcriptomic Profiling to Combat Naturally Invading Pathogens. Journal of Fungi, 6(4), 232. https://doi.org/10.3390/jof6040232