Protective Efficacy of Lectin-Fc(IgG) Fusion Proteins In Vitro and in a Pulmonary Aspergillosis In Vivo Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Use and Ethics Statement
2.2. Organisms and Growth Conditions
2.3. Bone Marrow-Derived Macrophages Isolation
2.4. Construction of Lectin-Fc(IgG) Fusion Proteins
2.5. Lectin-Fc(IgG) Fusion Proteins Expression and Purification
2.6. Molecular Characterization of Lectin-Fc(IgG) Proteins
2.7. Binding of Lectin-Fc(IgG) Proteins to A. fumigatus Conidia
2.8. Conidia Germination and Lectin-Fc(IgG) Labeling by Immunofluorescence
2.9. Growth Inhibition Assay Using Lectin-Fc(IgG) Proteins
2.10. Biofilm
2.11. Lectin-Fc(IgG) Complement Activation and A. fumigatus Growth
2.12. Interference of Lectin-Fc(IgG) with the Interaction between Fungal Cells and Macrophages
2.13. Macrophage Killing/Fungal Growth Inhibition Assay
2.14. In Vivo Models
2.15. Statistical Analysis
3. Results
3.1. Construction, Expression and Molecular Characterization of Lectin-Fc(IgG) Proteins
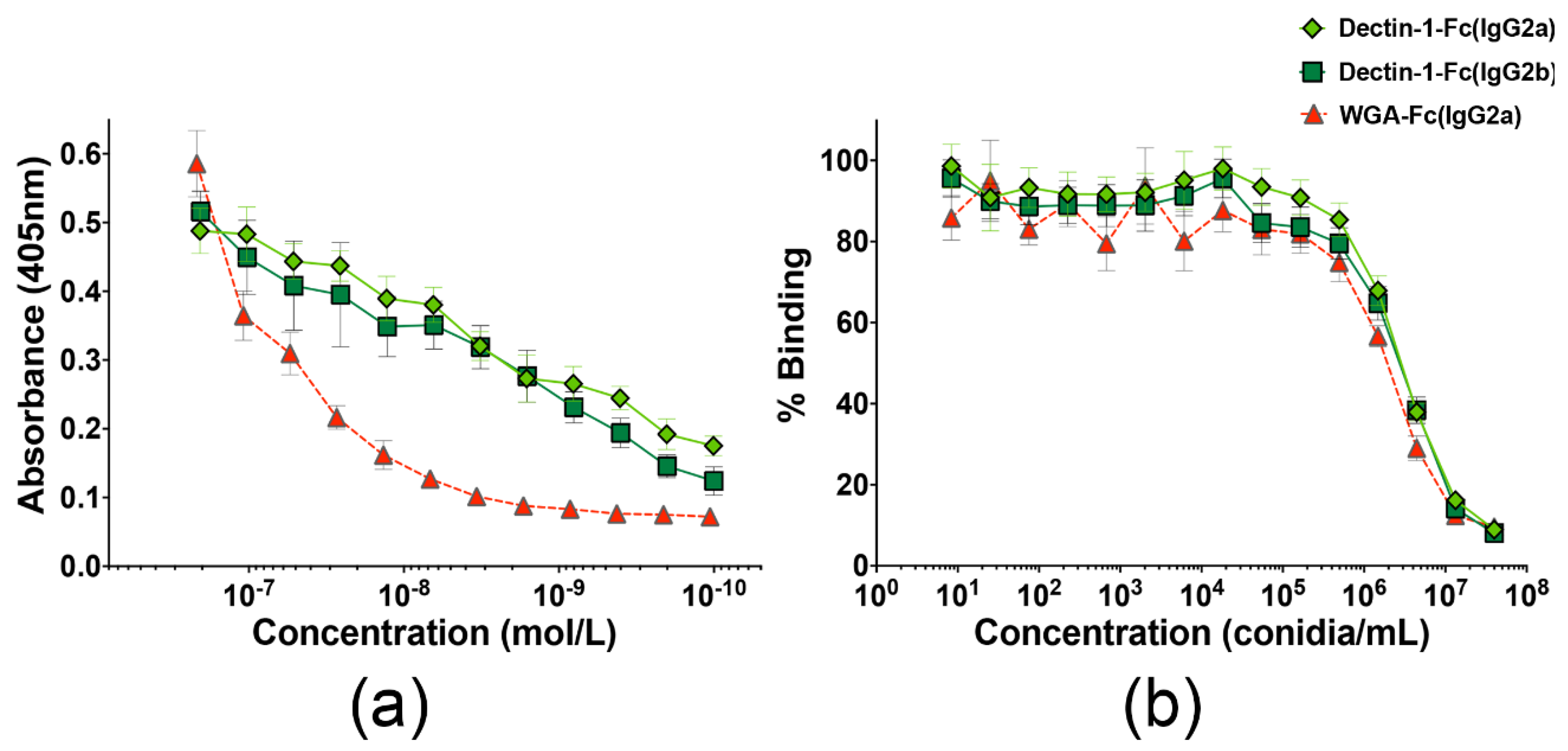
3.2. Lectin-Fc(IgG) Proteins Binding to A. fumigatus Germinating Conidia
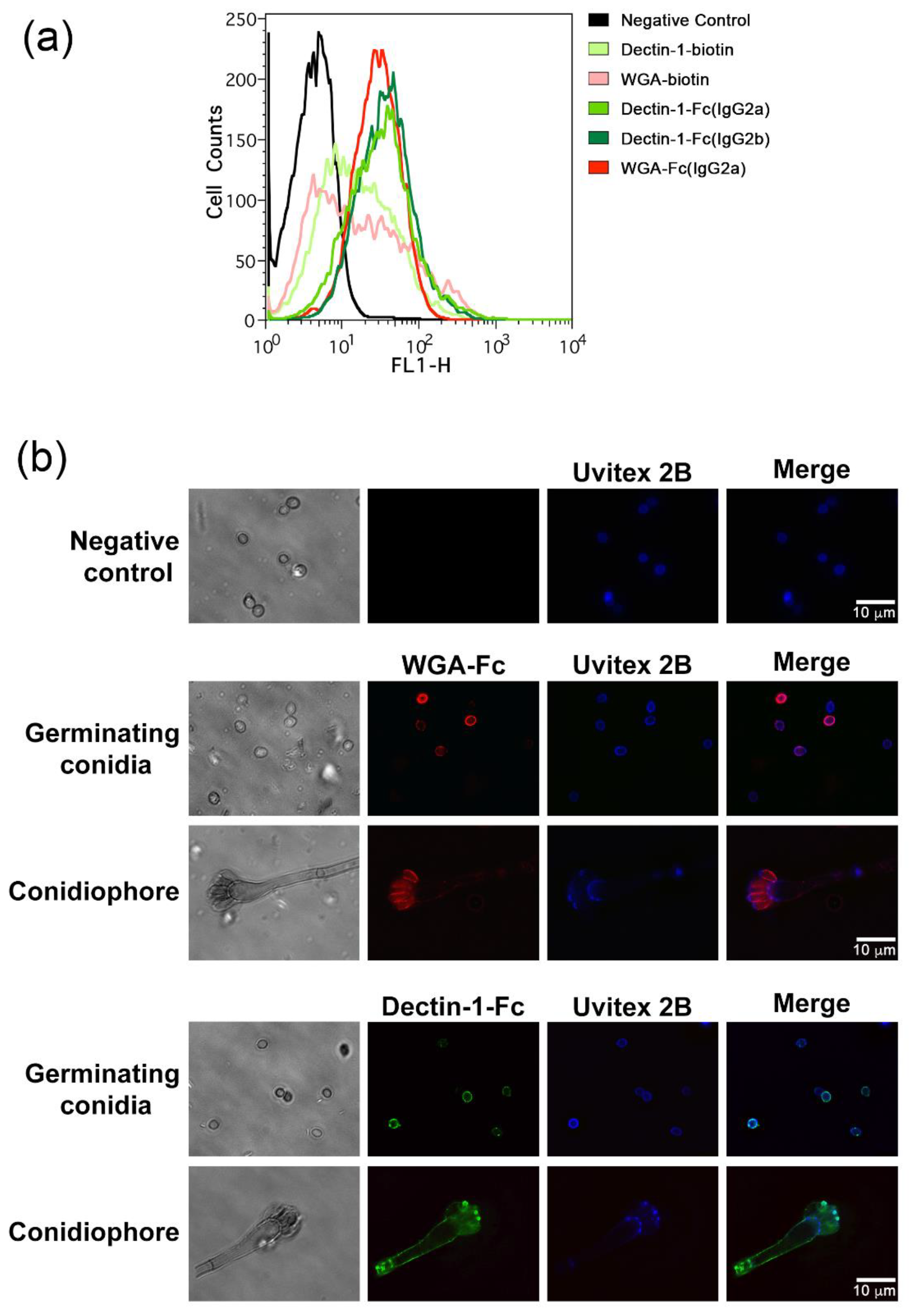
3.3. Lectin-Fc(IgG) Binding Pattern to A. fumigatus Germinating Conidia
3.4. Lectin-Fc(IgG) Proteins Show Enhanced Binding during Differentiation of A. fumigatus Conidia
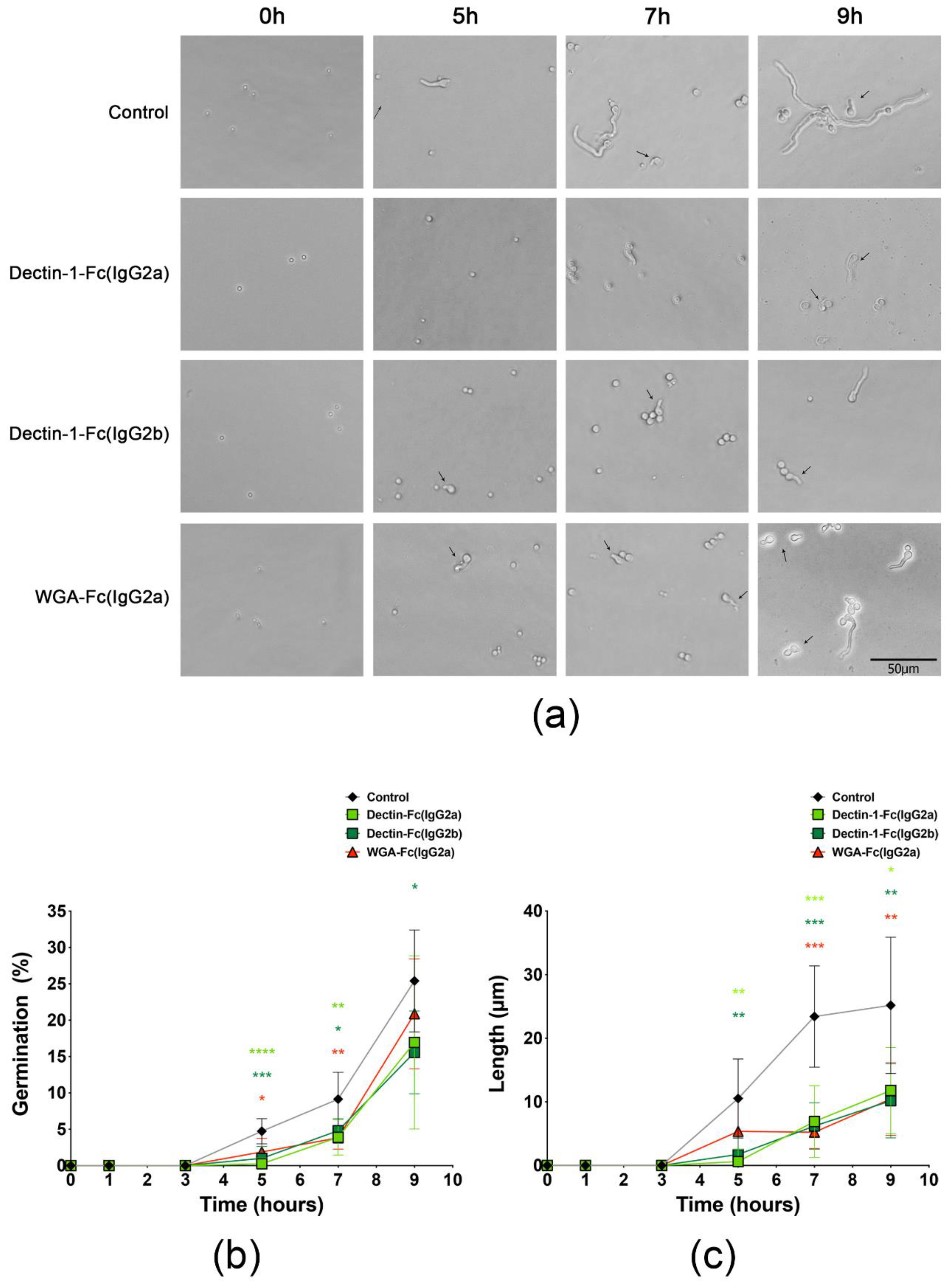
3.5. Lectin-Fc(IgG) Proteins Directly Inhibited A. fumigatus Germination
3.6. Influence of Lectin-Fc(IgG) Proteins on A. fumigatus Biofilm Formation
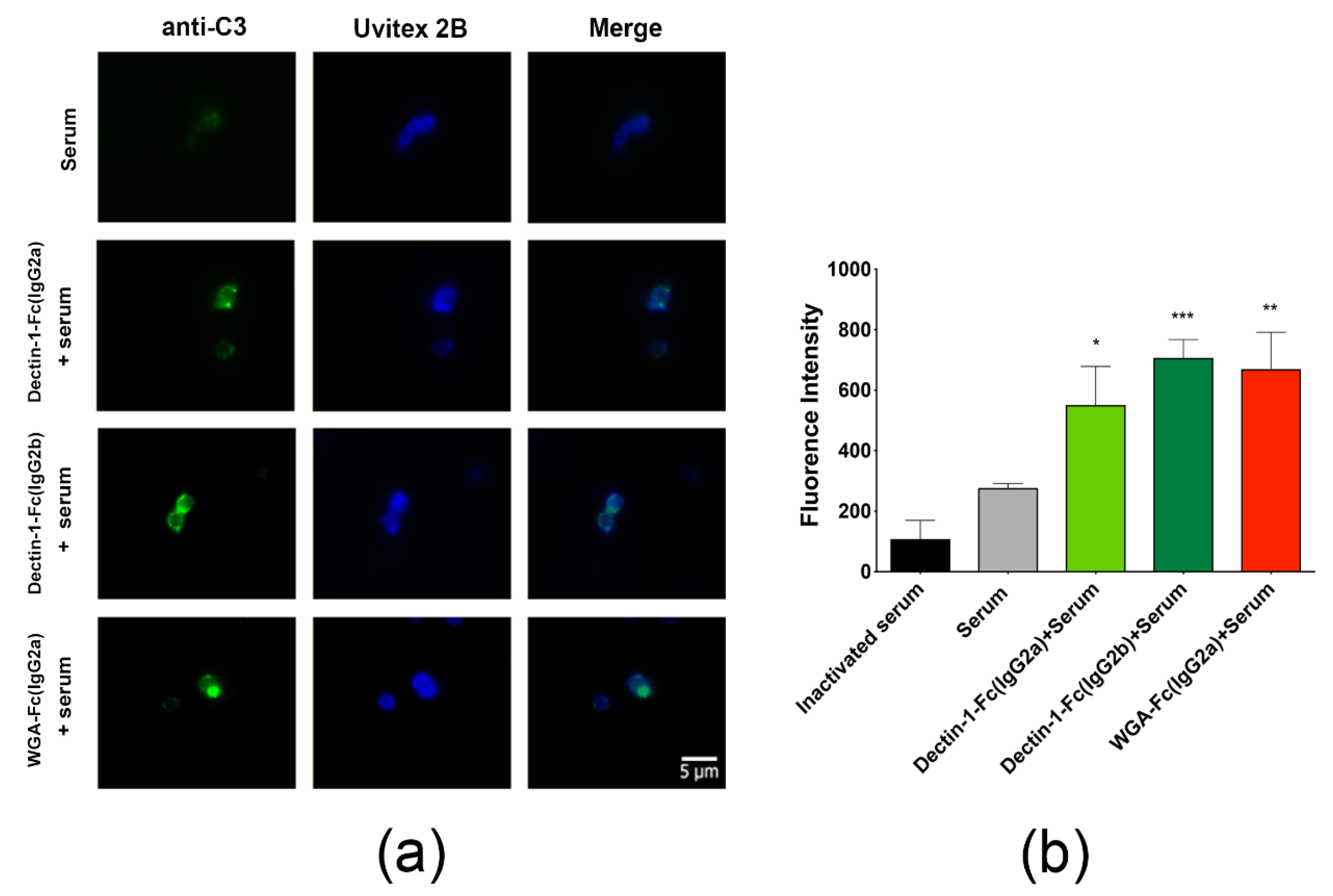
3.7. Lectin-Fc(IgG) Fusion Proteins Enhanced Complement Activation and A. fumigatus Growth Inhibition
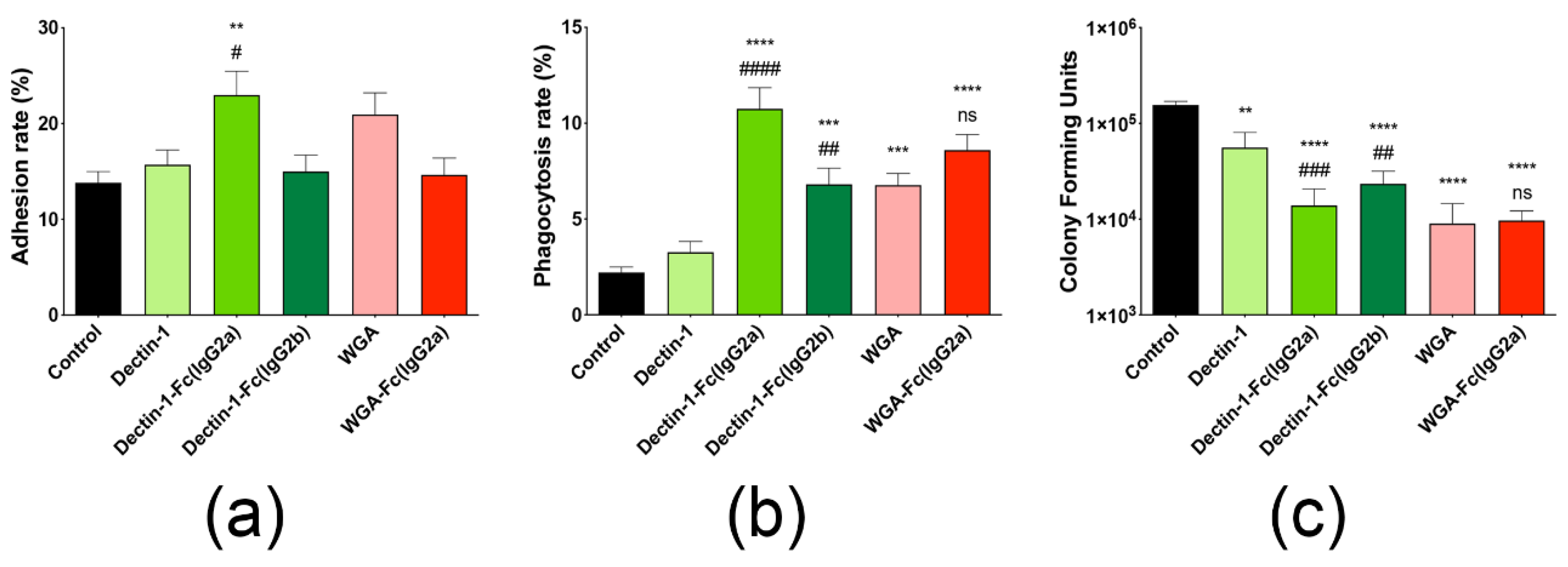
3.8. Opsonization with Lectin-Fc(IgG) Proteins Increased Phagocytosis of Germinating Conidia by Macrophages
3.9. Lectin-Fc(IgG) Proteins Enhance the Antifungal Efficacy of Macrophages
3.10. Administration of Dectin-1-Fc(IgG2b) or WGA-Fc(IgG2a) Protected Mice against A. fumigatus Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lamagni, T.L.; Evans, B.G.; Shigematsu, M.; Johnson, E.M. Emerging trends in the epidemiology of invasive mycoses in England and Wales (1990-9). Epidemiol. Infect. 2001, 126, 397–414. [Google Scholar] [CrossRef] [PubMed]
- van de Veerdonk, F.L.; Gresnigt, M.S.; Romani, L.; Netea, M.G.; Latge, J.P. Aspergillus fumigatus morphology and dynamic host interactions. Nat. Rev. Microbiol. 2017, 15, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Denis, B.; Guiguet, M.; de Castro, N.; Mechai, F.; Revest, M.; Melica, G.; Costagliola, D.; Lortholary, O. French Hospital Database on HIV National Agency for Research on AIDS and Viral Hepatitis, France CO4. Relevance of EORTC Criteria for the Diagnosis of Invasive Aspergillosis in HIV-Infected Patients, and Survival Trends Over a 20-Year Period in France. Clin. Infect. Dis. 2015, 61, 1273–1280. [Google Scholar] [CrossRef] [PubMed]
- Abad, A.; Fernandez-Molina, J.V.; Bikandi, J.; Ramirez, A.; Margareto, J.; Sendino, J.; Hernando, F.L.; Ponton, J.; Garaizar, J.; Rementeria, A. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev. Iberoam. Micol. 2010, 27, 155–182. [Google Scholar] [CrossRef] [PubMed]
- Low, C.Y.; Rotstein, C. Emerging fungal infections in immunocompromised patients. F1000 Med. Rep. 2011, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Jenks, J.D.; Spiess, B.; Buchheidt, D.; Hoenigl, M. (New) Methods for Detection of Aspergillus fumigatus Resistance in Clinical Samples. Curr. Fungal Infect. Rep. 2019, 13, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Mattila, P.E.; Metz, A.E.; Rapaka, R.R.; Bauer, L.D.; Steele, C. Dectin-1 Fc targeting of aspergillus fumigatus beta-glucans augments innate defense against invasive pulmonary aspergillosis. Antimicrob. Agents Chemother. 2008, 52, 1171–1172. [Google Scholar] [CrossRef] [PubMed]
- Kontoyiannis, D.P.; Selleslag, D.; Mullane, K.; Cornely, O.A.; Hope, W.; Lortholary, O.; Croos-Dabrera, R.; Lademacher, C.; Engelhardt, M.; Patterson, T.F. Impact of unresolved neutropenia in patients with neutropenia and invasive aspergillosis: A post hoc analysis of the SECURE trial. J. Antimicrob. Chemother. 2018, 73, 757–763. [Google Scholar] [CrossRef]
- Procop, G.W.; Roberts, G.D. Emerging fungal diseases: The importance of the host. Clin. Lab. Med. 2004, 24, 691–719. [Google Scholar] [CrossRef]
- Goyal, S.; Castrillon-Betancur, J.C.; Klaile, E.; Slevogt, H. The Interaction of Human Pathogenic Fungi With C-Type Lectin Receptors. Front. Immunol 2018, 9, 1261. [Google Scholar] [CrossRef]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Shishodia, S.K.; Tiwari, S.; Shankar, J. Resistance mechanism and proteins in Aspergillus species against antifungal agents. Mycology 2019, 10, 151–165. [Google Scholar] [CrossRef]
- Desoubeaux, G.; Cray, C. Animal Models of Aspergillosis. Comp. Med. 2018, 68, 109–123. [Google Scholar] [PubMed]
- Taccone, F.S.; Van den Abeele, A.M.; Bulpa, P.; Misset, B.; Meersseman, W.; Cardoso, T.; Paiva, J.A.; Blasco-Navalpotro, M.; De Laere, E.; Dimopoulos, G.; et al. Epidemiology of invasive aspergillosis in critically ill patients: Clinical presentation, underlying conditions, and outcomes. Crit. Care 2015, 19, 7. [Google Scholar] [CrossRef] [PubMed]
- Leal, S.M., Jr.; Cowden, S.; Hsia, Y.C.; Ghannoum, M.A.; Momany, M.; Pearlman, E. Distinct roles for Dectin-1 and TLR4 in the pathogenesis of Aspergillus fumigatus keratitis. PLoS Pathog. 2010, 6, e1000976. [Google Scholar] [CrossRef]
- Buil, J.B.; Hare, R.K.; Zwaan, B.J.; Arendrup, M.C.; Melchers, W.J.G.; Verweij, P.E. The fading boundaries between patient and environmental routes of triazole resistance selection in Aspergillus fumigatus. PLoS Pathog. 2019, 15, e1007858. [Google Scholar] [CrossRef] [PubMed]
- Nami, S.; Mohammadi, R.; Vakili, M.; Khezripour, K.; Mirzaei, H.; Morovati, H. Fungal vaccines, mechanism of actions and immunology: A comprehensive review. Biomed. Pharmacother. 2019, 109, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Patterson, T.F.; Thompson, G.R., 3rd; Denning, D.W.; Fishman, J.A.; Hadley, S.; Herbrecht, R.; Kontoyiannis, D.P.; Marr, K.A.; Morrison, V.A.; Nguyen, M.H.; et al. Practice Guidelines for the Diagnosis and Management of Aspergillosis: 2016 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016, 63, e1–e60. [Google Scholar] [CrossRef]
- Verweij, P.E.; Chowdhary, A.; Melchers, W.J.; Meis, J.F. Azole Resistance in Aspergillus fumigatus: Can We Retain the Clinical Use of Mold-Active Antifungal Azoles? Clin. Infect. Dis. 2016, 62, 362–368. [Google Scholar] [CrossRef]
- Saylor, C.; Dadachova, E.; Casadevall, A. Monoclonal antibody-based therapies for microbial diseases. Vaccine 2009, 27 (Suppl. 6), G38–G46. [Google Scholar] [CrossRef]
- Levitz, S.M. Aspergillus vaccines: Hardly worth studying or worthy of hard study? Med. Mycol. 2017, 55, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Vargas, G.; Honorato, L.; Guimaraes, A.J.; Rodrigues, M.L.; Reis, F.C.G.; Vale, A.M.; Ray, A.; Nosanchuk, J.D.; Nimrichter, L. Protective effect of fungal extracellular vesicles against murine candidiasis. Cell Microbiol. 2020, 22, e13238. [Google Scholar] [CrossRef]
- Bryan, R.A.; Guimaraes, A.J.; Hopcraft, S.; Jiang, Z.; Bonilla, K.; Morgenstern, A.; Bruchertseifer, F.; Del Poeta, M.; Torosantucci, A.; Cassone, A.; et al. Toward developing a universal treatment for fungal disease using radioimmunotherapy targeting common fungal antigens. Mycopathologia 2012, 173, 463–471. [Google Scholar] [CrossRef]
- Cassone, A. Fungal vaccines: Real progress from real challenges. Lancet Infect. Dis. 2008, 8, 114–124. [Google Scholar] [CrossRef]
- Sheppard, D.C.; Edwards, J.E., Jr. Development of a vaccine for invasive aspergillosis. Clin. Infect. Dis. 2004, 38, 1137–1138. [Google Scholar] [CrossRef]
- Pachl, J.; Svoboda, P.; Jacobs, F.; Vandewoude, K.; van der Hoven, B.; Spronk, P.; Masterson, G.; Malbrain, M.; Aoun, M.; Garbino, J.; et al. A randomized, blinded, multicenter trial of lipid-associated amphotericin B alone versus in combination with an antibody-based inhibitor of heat shock protein 90 in patients with invasive candidiasis. Clin. Infect. Dis. 2006, 42, 1404–1413. [Google Scholar] [CrossRef] [PubMed]
- Larsen, R.A.; Pappas, P.G.; Perfect, J.; Aberg, J.A.; Casadevall, A.; Cloud, G.A.; James, R.; Filler, S.; Dismukes, W.E. Phase I evaluation of the safety and pharmacokinetics of murine-derived anticryptococcal antibody 18B7 in subjects with treated cryptococcal meningitis. Antimicrob. Agents Chemother. 2005, 49, 952–958. [Google Scholar] [CrossRef]
- Moragues, M.D.; Omaetxebarria, M.J.; Elguezabal, N.; Sevilla, M.J.; Conti, S.; Polonelli, L.; Ponton, J. A monoclonal antibody directed against a Candida albicans cell wall mannoprotein exerts three anti-C. albicans activities. Infect. Immun. 2003, 71, 5273–5279. [Google Scholar] [CrossRef] [PubMed]
- Torosantucci, A.; Bromuro, C.; Chiani, P.; De Bernardis, F.; Berti, F.; Galli, C.; Norelli, F.; Bellucci, C.; Polonelli, L.; Costantino, P.; et al. A novel glyco-conjugate vaccine against fungal pathogens. J. Exp. Med. 2005, 202, 597–606. [Google Scholar] [CrossRef]
- Matveev, A.L.; Krylov, V.B.; Khlusevich, Y.A.; Baykov, I.K.; Yashunsky, D.V.; Emelyanova, L.A.; Tsvetkov, Y.E.; Karelin, A.A.; Bardashova, A.V.; Wong, S.S.W.; et al. Novel mouse monoclonal antibodies specifically recognizing beta-(1-->3)-D-glucan antigen. PLoS ONE 2019, 14, e0215535. [Google Scholar] [CrossRef]
- Guimaraes, A.J.; Frases, S.; Gomez, F.J.; Zancope-Oliveira, R.M.; Nosanchuk, J.D. Monoclonal antibodies to heat shock protein 60 alter the pathogenesis of Histoplasma capsulatum. Infect. Immun. 2009, 77, 1357–1367. [Google Scholar] [CrossRef]
- Guimaraes, A.J.; Frases, S.; Pontes, B.; de Cerqueira, M.D.; Rodrigues, M.L.; Viana, N.B.; Nimrichter, L.; Nosanchuk, J.D. Agglutination of Histoplasma capsulatum by IgG monoclonal antibodies against Hsp60 impacts macrophage effector functions. Infect. Immun. 2011, 79, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Nosanchuk, J.D.; Steenbergen, J.N.; Shi, L.; Deepe, G.S., Jr.; Casadevall, A. Antibodies to a cell surface histone-like protein protect against Histoplasma capsulatum. J. Clin. Investig. 2003, 112, 1164–1175. [Google Scholar] [CrossRef] [PubMed]
- Boniche, C.; Rossi, S.A.; Kischkel, B.; Barbalho, F.V.; Moura, A.N.D.; Nosanchuk, J.D.; Travassos, L.R.; Taborda, C.P. Immunotherapy against Systemic Fungal Infections Based on Monoclonal Antibodies. J. Fungi 2020, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Pirofski, L.A. Antibody-mediated protection through cross-reactivity introduces a fungal heresy into immunological dogma. Infect. Immun. 2007, 75, 5074–5078. [Google Scholar] [CrossRef][Green Version]
- Guimaraes, A.J.; de Cerqueira, M.D.; Nosanchuk, J.D. Surface architecture of histoplasma capsulatum. Front. Microbiol. 2011, 2, 225. [Google Scholar] [CrossRef]
- Guimaraes, A.J.; Nakayasu, E.S.; Sobreira, T.J.; Cordero, R.J.; Nimrichter, L.; Almeida, I.C.; Nosanchuk, J.D. Histoplasma capsulatum heat-shock 60 orchestrates the adaptation of the fungus to temperature stress. PLoS ONE 2011, 6, e14660. [Google Scholar] [CrossRef]
- Gow, N.A.R.; Latge, J.P.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Hopke, A.; Brown, A.J.P.; Hall, R.A.; Wheeler, R.T. Dynamic Fungal Cell Wall Architecture in Stress Adaptation and Immune Evasion. Trends Microbiol. 2018, 26, 284–295. [Google Scholar] [CrossRef]
- Allen, A.K.; Neuberger, A.; Sharon, N. The purification, composition and specificity of wheat-germ agglutinin. Biochem. J. 1973, 131, 155–162. [Google Scholar] [CrossRef]
- Herre, J.; Gordon, S.; Brown, G.D. Dectin-1 and its role in the recognition of beta-glucans by macrophages. Mol. Immunol. 2004, 40, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Czajkowsky, D.M.; Hu, J.; Shao, Z.; Pleass, R.J. Fc-fusion proteins: New developments and future perspectives. EMBO Mol. Med. 2012, 4, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Liedke, S.C. Proteínas Fc de Fusão Contra Glicanas da Parede Celular Fúngica: Construção e Avaliação das suas Propriedades Antifúngicas. Ph.D. Thesis, Microbiologia, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil, 2018. [Google Scholar]
- Liedke, S.C.; Miranda, D.Z.; Gomes, K.X.; Goncalves, J.L.S.; Frases, S.; Nosanchuk, J.D.; Rodrigues, M.L.; Nimrichter, L.; Peralta, J.M.; Guimaraes, A.J. Characterization of the antifungal functions of a WGA-Fc (IgG2a) fusion protein binding to cell wall chitin oligomers. Sci. Rep. 2017, 7, 12187. [Google Scholar] [CrossRef]
- Luther, K.; Torosantucci, A.; Brakhage, A.A.; Heesemann, J.; Ebel, F. Phagocytosis of Aspergillus fumigatus conidia by murine macrophages involves recognition by the dectin-1 beta-glucan receptor and Toll-like receptor 2. Cell Microbiol. 2007, 9, 368–381. [Google Scholar] [CrossRef]
- Steele, C.; Rapaka, R.R.; Metz, A.; Pop, S.M.; Williams, D.L.; Gordon, S.; Kolls, J.K.; Brown, G.D. The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLoS Pathog. 2005, 1, e42. [Google Scholar] [CrossRef] [PubMed]
- Torosantucci, A.; Chiani, P.; Bromuro, C.; De Bernardis, F.; Palma, A.S.; Liu, Y.; Mignogna, G.; Maras, B.; Colone, M.; Stringaro, A.; et al. Protection by anti-beta-glucan antibodies is associated with restricted beta-1,3 glucan binding specificity and inhibition of fungal growth and adherence. PLoS ONE 2009, 4, e5392. [Google Scholar] [CrossRef] [PubMed]
- Englen, M.D.; Valdez, Y.E.; Lehnert, N.M.; Lehnert, B.E. Granulocyte/macrophage colony-stimulating factor is expressed and secreted in cultures of murine L929 cells. J. Immunol. Methods 1995, 184, 281–283. [Google Scholar] [CrossRef]
- Trouplin, V.; Boucherit, N.; Gorvel, L.; Conti, F.; Mottola, G.; Ghigo, E. Bone marrow-derived macrophage production. J. Vis. Exp. 2013, 81, e50966. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Guimarães, A.J.; Frases, S.; Cordero, R.J.; Nimrichter, L.; Casadevall, A.; Nosanchuk, J.D. Cryptococcus neoformans responds to mannitol by increasing capsule size in vitro and in vivo. Cell Microbiol. 2010, 12, 740–753. [Google Scholar] [CrossRef]
- Kaur, S.; Singh, S. Biofilm formation by Aspergillus fumigatus. Med. Mycol. 2014, 52, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Mowat, E.; Butcher, J.; Lang, S.; Williams, C.; Ramage, G. Development of a simple model for studying the effects of antifungal agents on multicellular communities of Aspergillus fumigatus. J. Med. Microbiol. 2007, 56, 1205–1212. [Google Scholar] [CrossRef]
- Pierce, C.G.; Uppuluri, P.; Tristan, A.R.; Wormley, F.L., Jr.; Mowat, E.; Ramage, G.; Lopez-Ribot, J.L. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat. Protoc. 2008, 3, 1494–1500. [Google Scholar] [CrossRef] [PubMed]
- van de Sande, W.W.; Tavakol, M.; van Vianen, W.; Bakker-Woudenberg, I.A. The effects of antifungal agents to conidial and hyphal forms of Aspergillus fumigatus. Med. Mycol. 2010, 48, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Chaieb, K.; Chehab, O.; Zmantar, T.; Rouabhia, M.; Mahdouani, K.; Bakhrouf, A. In vitro effect of pH and ethanol on biofilm formation by clinical ica-positive Staphylococcus epidermidis strains. Ann. Microbiol. 2007, 57, 431–437. [Google Scholar] [CrossRef]
- Guimarães, A.J.; de Cerqueira, M.D.; Zamith-Miranda, D.; Lopez, P.H.; Rodrigues, M.L.; Pontes, B.; Viana, N.B.; DeLeon-Rodriguez, C.M.; Rossi, D.C.P.; Casadevall, A.; et al. Host membrane glycosphingolipids and lipid microdomains facilitate Histoplasma capsulatum internalisation by macrophages. Cell Microbiol. 2019, 21, e12976. [Google Scholar] [CrossRef] [PubMed]
- Cordero, R.J.; Liedke, S.C.; de S Araújo, G.R.; Martinez, L.R.; Nimrichter, L.; Frases, S.; Peralta, J.M.; Casadevall, A.; Rodrigues, M.L.; Nosanchuk, J.D.; et al. Enhanced virulence of Histoplasma capsulatum through transfer and surface incorporation of glycans from Cryptococcus neoformans during co-infection. Sci. Rep. 2016, 6, 21765. [Google Scholar] [CrossRef]
- Ahlmann, M.; Hempel, G. The effect of cyclophosphamide on the immune system: Implications for clinical cancer therapy. Cancer Chemother. Pharmacol. 2016, 78, 661–671. [Google Scholar] [CrossRef]
- Silva, R.L.H.; Rosa-Milani, E.; Brunaldi, M.O.; Maffei, C.M.L. Murine model of invasive pulmonary Aspergillosis: Follow-up of tissue injury, fungal burden and mortality with distinct elastase production strains. J. Mycol. Med. 2019, 29, 112–119. [Google Scholar] [CrossRef]
- Perez-Cantero, A.; Lopez-Fernandez, L.; Guarro, J.; Capilla, J. Azole resistance mechanisms in Aspergillus: Update and recent advances. Int. J. Antimicrob. Agents 2020, 55, 105807. [Google Scholar] [CrossRef]
- Ashu, E.E.; Korfanty, G.A.; Samarasinghe, H.; Pum, N.; You, M.; Yamamura, D.; Xu, J. Widespread amphotericin B-resistant strains of Aspergillus fumigatus in Hamilton, Canada. Infect. Drug Resist. 2018, 11, 1549–1555. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.; El Chazli, Y.; Babu, A.F.; Coste, A.T. Azole Resistance in Aspergillus fumigatus: A Consequence of Antifungal Use in Agriculture? Front. Microbiol. 2017, 8, 1024. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.; Messina, F.; Marin, E.; Arechavala, A.; Depardo, R.; Walker, L.; Negroni, R.; Santiso, G. Antifungal Resistance in Clinical Isolates of Aspergillus spp.: When Local Epidemiology Breaks the Norm. J. Fungi 2019, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Latge, J.P.; Mouyna, I.; Tekaia, F.; Beauvais, A.; Debeaupuis, J.P.; Nierman, W. Specific molecular features in the organization and biosynthesis of the cell wall of Aspergillus fumigatus. Med. Mycol. 2005, 43 (Suppl. 1), S15–S22. [Google Scholar] [CrossRef]
- Hohl, T.M.; Van Epps, H.L.; Rivera, A.; Morgan, L.A.; Chen, P.L.; Feldmesser, M.; Pamer, E.G. Aspergillus fumigatus triggers inflammatory responses by stage-specific beta-glucan display. PLoS Pathog. 2005, 1, e30. [Google Scholar] [CrossRef] [PubMed]
- Beauvais, A.; Latge, J.P. Aspergillus Biofilm In Vitro and In Vivo. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Chaturvedi, A.K.; Kavishwar, A.; Shiva Keshava, G.B.; Shukla, P.K. Monoclonal immunoglobulin G1 directed against Aspergillus fumigatus cell wall glycoprotein protects against experimental murine aspergillosis. Clin. Diagn. Lab. Immunol. 2005, 12, 1063–1068. [Google Scholar] [CrossRef]
- Reichhardt, C.; Joubert, L.M.; Clemons, K.V.; Stevens, D.A.; Cegelski, L. Integration of electron microscopy and solid-state NMR analysis for new views and compositional parameters of Aspergillus fumigatus biofilms. Med. Mycol. 2019, 57, S239–S244. [Google Scholar] [CrossRef]
- Dagenais, T.R.; Keller, N.P. Pathogenesis of Aspergillus fumigatus in Invasive Aspergillosis. Clin. Microbiol. Rev. 2009, 22, 447–465. [Google Scholar] [CrossRef]
- Noris, M.; Remuzzi, G. Overview of complement activation and regulation. Semin. Nephrol. 2013, 33, 479–492. [Google Scholar] [CrossRef]
- Nicola, A.M.; Casadevall, A.; Goldman, D.L. Fungal killing by mammalian phagocytic cells. Curr. Opin. Microbiol. 2008, 11, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Gersuk, G.M.; Underhill, D.M.; Zhu, L.; Marr, K.A. Dectin-1 and TLRs permit macrophages to distinguish between different Aspergillus fumigatus cellular states. J. Immunol. 2006, 176, 3717–3724. [Google Scholar] [CrossRef] [PubMed]
- Rapaka, R.R.; Goetzman, E.S.; Zheng, M.; Vockley, J.; McKinley, L.; Kolls, J.K.; Steele, C. Enhanced defense against Pneumocystis carinii mediated by a novel Dectin-1 receptor Fc fusion protein. J. Immunol. 2007, 178, 3702–3712. [Google Scholar] [CrossRef] [PubMed]
- Zimmerli, S.; Knecht, U.; Leib, S.L. A model of cerebral aspergillosis in non-immunosuppressed nursing rats. Acta Neuropathol. 2007, 114, 411–418. [Google Scholar] [CrossRef][Green Version]
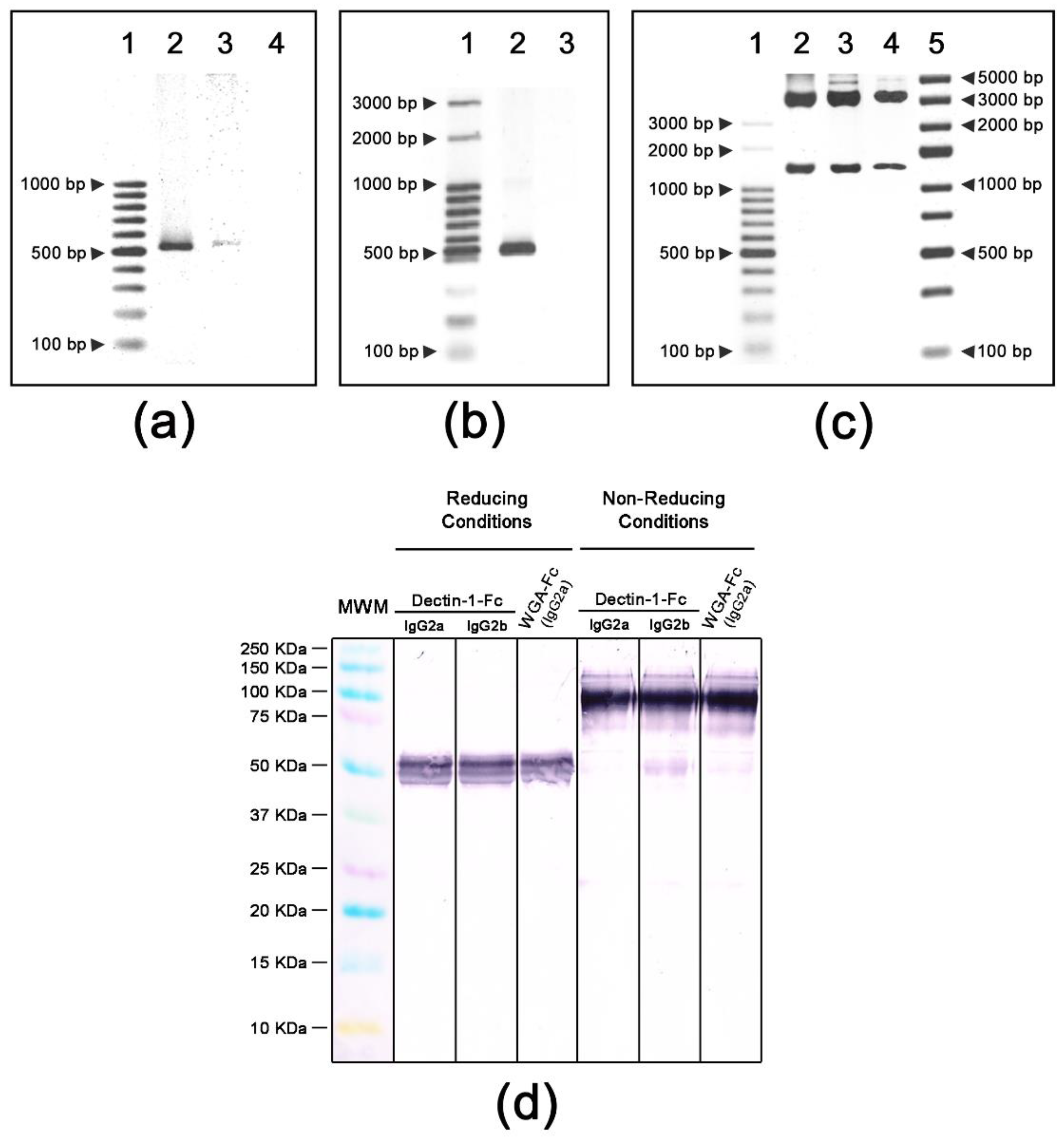

| PCR Oligonucleotides | |
|---|---|
| Dectin-1 primers | |
| Dectin-1 foward | 5′-TTT GAA TTC GCA CAA TTC AGG GAG AAA TCC-3 |
| Dectin-1 reverse | 5′-TTT AGA TCT CAG TTC CTT CTC ACA GAT AC-3 |
| WGA primers | |
| WGA forward | 5′-TTT GAA TTC GCA GAG GTG CGG CGA GC-3 |
| WGA reverse | 5′-TTT CCA TGG CAG CGT CAC AGC CGC C-3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodriguez-de la Noval, C.; Ruiz Mendoza, S.; de Souza Gonçalves, D.; da Silva Ferreira, M.; Honorato, L.; Peralta, J.M.; Nimrichter, L.; Guimarães, A.J. Protective Efficacy of Lectin-Fc(IgG) Fusion Proteins In Vitro and in a Pulmonary Aspergillosis In Vivo Model. J. Fungi 2020, 6, 250. https://doi.org/10.3390/jof6040250
Rodriguez-de la Noval C, Ruiz Mendoza S, de Souza Gonçalves D, da Silva Ferreira M, Honorato L, Peralta JM, Nimrichter L, Guimarães AJ. Protective Efficacy of Lectin-Fc(IgG) Fusion Proteins In Vitro and in a Pulmonary Aspergillosis In Vivo Model. Journal of Fungi. 2020; 6(4):250. https://doi.org/10.3390/jof6040250
Chicago/Turabian StyleRodriguez-de la Noval, Claudia, Susana Ruiz Mendoza, Diego de Souza Gonçalves, Marina da Silva Ferreira, Leandro Honorato, José Mauro Peralta, Leonardo Nimrichter, and Allan J. Guimarães. 2020. "Protective Efficacy of Lectin-Fc(IgG) Fusion Proteins In Vitro and in a Pulmonary Aspergillosis In Vivo Model" Journal of Fungi 6, no. 4: 250. https://doi.org/10.3390/jof6040250
APA StyleRodriguez-de la Noval, C., Ruiz Mendoza, S., de Souza Gonçalves, D., da Silva Ferreira, M., Honorato, L., Peralta, J. M., Nimrichter, L., & Guimarães, A. J. (2020). Protective Efficacy of Lectin-Fc(IgG) Fusion Proteins In Vitro and in a Pulmonary Aspergillosis In Vivo Model. Journal of Fungi, 6(4), 250. https://doi.org/10.3390/jof6040250





