Characteristics, Risk Factors, and Survival Analysis of Candida auris Cases: Results of One-Year National Surveillance Data from Oman
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical & Demographic Data
2.2. Identification & AST
2.3. Data Analysis
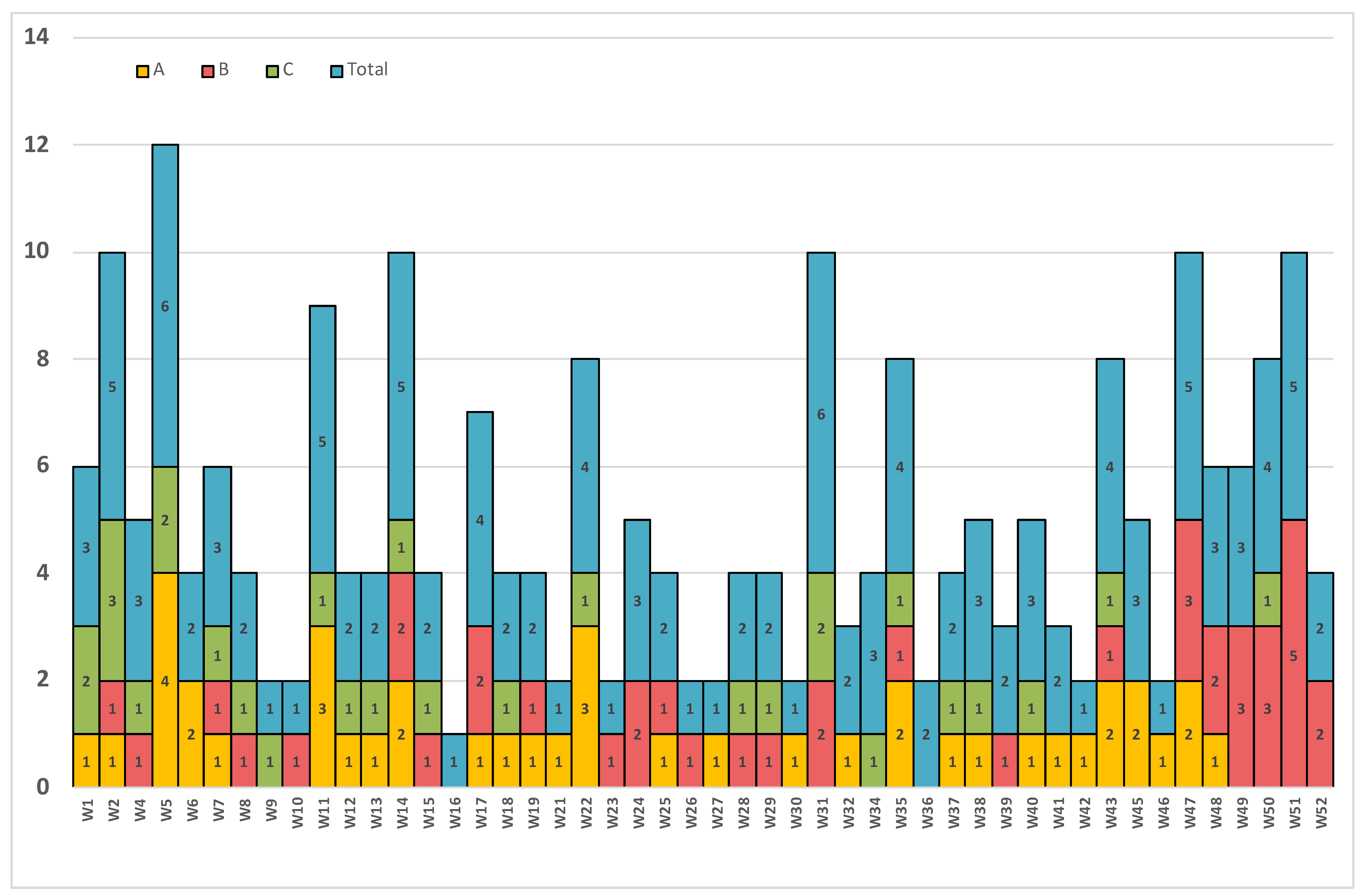
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Satoh, K.; Makimura, K.; Hasumi, Y.; Nishiyama, Y.; Uchida, K.; Yamaguchi, H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol. Immunol. 2009, 53, 41–44. [Google Scholar] [CrossRef]
- Du, H.; Bing, J.; Hu, T.; Ennis, C.L.; Nobile, C.J.; Huang, G. Candida auris: Epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020, 16, e1008921. [Google Scholar] [CrossRef]
- Candida auris: Information for Laboratorians and Health Professionals. Atlanta, Georgia: US Department of Health and Human Services, Centers for Disease Control and Prevention. 2019. Available online: https://www.cdc.gov/fungal/candida-auris/health-professionals.html (accessed on 6 November 2020).
- Hori, Y.; Shibuya, K. Role of FKS Gene in the Susceptibility of Pathogenic Fungi to Echinocandins. Med. Mycol. J. 2018, 59, E31–E40. [Google Scholar] [CrossRef] [Green Version]
- Chowdhary, A.; Prakash, A.; Sharma, C.; Kordalewska, M.; Kumar, A.; Sarma, S.; Tarai, B.; Singh, A.; Upadhyaya, G.; Upadhyay, S.; et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: Role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J. Antimicrob. Chemother. 2018, 73, 891–899. [Google Scholar] [CrossRef]
- Fasciana, T.; Cortegiani, A.; Ippolito, M.; Giarratano, A.; Di Quattro, O.; Lipari, D.; Graceffa, D.; Giammanco, A. Candida auris: An Overview of How to Screen, Detect, Test and Control This Emerging Pathogen. Antibiotics 2020, 9, 778. [Google Scholar] [CrossRef]
- Al-Siyabi, T.; Al Busaidi, I.; Balkhair, A.; Al-Muharrmi, Z.; Al-Salti, M.; Al’Adawi, B. First report of Candida auris in Oman: Clinical and microbiological description of five candidemia cases. J. Infect. 2017, 75, 373–376. [Google Scholar] [CrossRef] [PubMed]
- Al Maani, A.; Paul, H.; Al-Rashdi, A.; Al Wahaibi, A.; Al-Jardani, A.; Abri, A.; Albalushi, M.A.H.; Abri; Al Reesi, M.; Al Maqbali, A.; et al. Ongoing Challenges with Healthcare-Associated Candida auris Outbreaks in Oman. J. Fungi 2019, 5, 101. [Google Scholar] [CrossRef] [Green Version]
- Oman Antimicrobial Resistance Surveillance System (OMASS); The First National Annual Antimicrobial Resistance Report (2017); Directorate General for Disease Surveillance and Control, MoH: Muscat, Oman, 2018.
- Oman Antimicrobial Surveillance System (OMASS); The Annual Antimicrobial Resistance Report (2018); Directorate General for Disease Surveillance and Control, MoH: Muscat, Oman, 2019.
- Global Antimicrobial Resistance Surveillance System. April 2018. Available online: https://www.who.int/glass/events/AMR-in-invasive-candida-infections-meeting/en/ (accessed on 28 October 2020).
- Biagi, M.J.; Wiederhold, N.P.; Gibas, C.; Wickes, B.L.; Lozano, V.; Bleasdale, S.C.; Danziger, L. Development of High-Level Echinocandin Resistance in a Patient with Recurrent Candida auris Candidemia Secondary to Chronic Candiduria. Open Forum Infect. Dis. 2019, 6, ofz262. [Google Scholar] [CrossRef] [PubMed]
- Ostrowsky, B.; Greenko, J.; Adams, E.; Quinn, M.; O’Brien, B.; Chaturvedi, V.; Berkow, E.; Vallabhaneni, S.; Forsberg, K.; Chaturvedi, S.; et al. Candida auris Isolates Resistant to Three Classes of Antifungal Medications—New York, 2019. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 6–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antifungal Susceptibility Testing and Interpretation, Candida auris, Centers for Disease Control & Prevention. Available online: https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html (accessed on 28 October 2020).
- Oh, B.J.; Shin, J.H.; Kim, M.-N.; Sung, H.; Lee, K.; Joo, M.Y.; Shin, M.-G.; Suh, S.-P.; Ryang, D.W. Biofilm formation and genotyping of Candida haemulonii, Candida pseudohaemulonii, and a proposed new species (Candida auris) isolates from Korea. Med. Mycol. 2011, 49, 98–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhary, A.; Sharma, C.; Meis, J.F. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017, 13, e1006290. [Google Scholar] [CrossRef] [PubMed]
- Schelenz, S.; Hagen, F.; Rhodes, J.L.; Abdolrasouli, A.; Chowdhary, A.; Hall, A.; Ryan, L.; Shackleton, J.; Trimlett, R.; Meis, J.F.; et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob. Resist. Infect. Control. 2016, 5, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misseri, G.; Ippolito, M.; Cortegiani, A. Global warming “heating up” the ICU through Candida auris infections: The climate changes theory. Crit. Care 2019, 23, 416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casadevall, A.; Kontoyiannis, D.P.; Robert, V. On the Emergence of Candida auris: Climate Change, Azoles, Swamps, and Birds. mBio 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortegiani, A.; Misseri, G.; Fasciana, T.; Giammanco, A.; Giarratano, A.; Chowdhary, A. Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J. Intensiv. Care 2018, 6, 69. [Google Scholar] [CrossRef] [PubMed]
- Spivak, E.S.; Hanson, K.E. Candida auris: An Emerging Fungal Pathogen. J. Clin. Microbiol. 2017, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Division of Foodborne, Waterborne, and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention. Candida auris Clinical Update: September 2017. Available online: https://www.cdc.gov/fungal/candida-auris/c-auris-alert-09-17.html (accessed on 15 November 2020).
- Identification of Candida auris, Centers for Disease Control & Prevention. Available online: https://www.cdc.gov/fungal/candida-auris/identification.html (accessed on 6 November 2020).
- Kordalewska, M.; Zhao, Y.; Lockhart, S.R.; Chowdhary, A.; Berrio, I.; Perlin, D.S. Rapid and Accurate Molecular Identification of the Emerging Multidrug-Resistant Pathogen Candida auris. J. Clin. Microbiol. 2017, 55, 2445–2452. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Gaitán, A.C.; Fernández-Pereira, J.; Valentin, E.; Tormo-Mas, M.Á.; Eraso, E.; Pemán, J.; De Groot, P.W.J. Molecular identification of Candida auris by PCR amplification of species-specific GPI protein-encoding genes. Int. J. Med. Microbiol. 2018, 308, 812–818. [Google Scholar] [CrossRef]
- Leach, L.; Zhu, Y.; Chaturvedi, S. Development and Validation of a Real-Time PCR Assay for Rapid Detection of Candida auris from Surveillance Samples. J. Clin. Microbiol. 2018. [Google Scholar] [CrossRef] [Green Version]
- Pierce, J.; Apisarnthanarak, A.; Schellack, N.; Cornistein, W.; Al Maani, A.; Adnan, S.; Stevens, M.P. Global Antimicrobial Stewardship with a Focus on Low- and Middle-Income Countries. Int. J. Infect. Dis. 2020, 96, 621–629. [Google Scholar] [CrossRef]
- Rozwadowski, F.; McAteer, J.; Chow, N.A.; Skrobarcek, K.; Forsberg, K.; Barrett, P.M.; Greeley, R.; Fulton, T.; Wells, J.; Welsh, R.M.; et al. 161. Prevalence and Risk Factors for Candida auris Colonization Among Patients in a Long-term Acute Care Hospital—New Jersey, 2017. Open Forum Infect. Dis. 2018, 5, S14. [Google Scholar] [CrossRef]
- Forsberg, K.; Woodworth, K.; Walters, M.; Berkow, E.L.; Jackson, B.; Chiller, T.; Vallabhaneni, S. Candida auris: The recent emergence of a multidrug-resistant fungal pathogen. Med. Mycol. 2019, 57, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, E.; Quinn, M.; Tsay, S.; Poirot, E.; Chaturvedi, S.; Southwick, K.; Greenko, J.; Fernandez, R.; Al, E.A.E.; Vallabhaneni, S.; et al. Candida auris in Healthcare Facilities, New York, USA, 2013–2017. Emerg. Infect. Dis. 2018, 24, 1816–1824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekyere, J.O. Candida auris: A systematic review and meta-analysis of current updates on an emerging multidrug-resistant pathogen. Microbiologyopen 2018, 7, e00578. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Gaitán, A.; Martínez, H.; Moret, A.M.; Calabuig, E.; Tasias, M.; Alastruey-Izquierdo, A.; Zaragoza, Ó.; Mollar, J.; Frasquet, J.; Salavert-Lletí, M.; et al. Detection and treatment of Candida auris in an outbreak situation: Risk factors for developing colonization and candidemia by this new species in critically ill patients. Expert Rev. Antiinfect. Ther. 2019, 17, 295–305. [Google Scholar] [CrossRef]
- Sayeed, M.A.; Farooqi, J.; Jabeen, K.; Awan, S.; Mahmood, S.F. Clinical spectrum and factors impacting outcome of Candida auris: A single center study from Pakistan. BMC Infect. Dis. 2019, 19, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous Emergence of Multidrug-Resistant Candida auris on 3 Continents Confirmed by Whole-Genome Sequencing and Epidemiological Analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef] [Green Version]
- Taori, S.K.; Khonyongwa, K.; Hayden, I.; Athukorala, G.D.A.; Letters, A.; Fife, A.; Desai, N.; Borman, A.M. Candida auris outbreak: Mortality, interventions and cost of sustaining control. J. Infect. 2019, 79, 601–611. [Google Scholar] [CrossRef]
- Arensman, K.; Miller, J.L.; Chiang, A.; Mai, N.; Levato, J.; Lachance, E.; Anderson, M.; Beganovic, M.; Pena, J.D. Clinical Outcomes of Patients Treated for Candida auris Infections in a Multisite Health System, Illinois, USA. Emerg. Infect. Dis. 2020, 26, 876–880. [Google Scholar] [CrossRef]
- Alfouzan, W.; Dhar, R.; Albarrag, A.; Al-Abdely, H. The emerging pathogen Candida auris: A focus on the Middle-Eastern countries. J. Infect. Public Health 2019, 12, 451–459. [Google Scholar] [CrossRef]
- Chowdhary, A.; Kumar, V.A.; Sharma, C.; Prakash, A.; Agarwal, K.; Babu, R.; Dinesh, K.R.; Karim, S.; Singh, S.K.; Hagen, F.; et al. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 919–926. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, J.F.; Gade, L.; Chow, N.A.; Loparev, V.N.; Juieng, P.; Berkow, E.L.; Farrer, R.A.; Litvintseva, A.P.; Cuomo, C.A. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Categories | Overall (N = 108) |
|---|---|
| Age group: ≥65 years | 40 (37%) |
| Males | 72 (66.7%) |
| Omani | 85 (78.7%) |
| Candidemia | 42 (38.9%) |
| Infection (not colonized) | 61 (56.5%) |
| Length of stay (months): <3 m | 66 (61.1%) |
| Comorbidities | 74 (68.5%) |
| Length of stay (months) after diagnosis: Mean (SD) | 1.7 (2.8) |
| ICU admission | 85 (78.7%) |
| Died (out of total patients) | 42 (38.9%) |
| Antifungal | Fluconazole | Voriconazole | Caspofungin | Micafungin | Amphotericin B | Flucytosine |
|---|---|---|---|---|---|---|
| MIC | ≤8 µg/mL 4 (5.2%) | ≤1 µg/mL 68 (88.3%) | <1 µg/mL 76 (98.7%) | ≤1 µg/mL 75 (97.4%) | <1 µg/mL 3 (3.9%) | ≤8 µg/mL 70 (90.9%) |
| MIC | 16 µg/mL 34 (44.2%) | 2 µg/mL 7 (9.1%) | 1 µg/mL 0 (0.0%) | 2 µg/mL 0 (0.0%) | 1 µg/mL 9 (11.7%) | 16 µg/mL 1 (1.3%) |
| MIC | ≥32 µg/mL 39 (50.6%) | ≥4 µg/mL 2 (2.6%) | ≥2 µg/mL 1 (1.3%) | ≥4 µg/mL 2 (2.6%) | ≥2 µg/mL 65 (84.4%) | ≥32 µg/mL 6 (7.8%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Rashdi, A.; Al-Maani, A.; Al-Wahaibi, A.; Alqayoudhi, A.; Al-Jardani, A.; Al-Abri, S. Characteristics, Risk Factors, and Survival Analysis of Candida auris Cases: Results of One-Year National Surveillance Data from Oman. J. Fungi 2021, 7, 31. https://doi.org/10.3390/jof7010031
Al-Rashdi A, Al-Maani A, Al-Wahaibi A, Alqayoudhi A, Al-Jardani A, Al-Abri S. Characteristics, Risk Factors, and Survival Analysis of Candida auris Cases: Results of One-Year National Surveillance Data from Oman. Journal of Fungi. 2021; 7(1):31. https://doi.org/10.3390/jof7010031
Chicago/Turabian StyleAl-Rashdi, Azza, Amal Al-Maani, Adil Al-Wahaibi, Abdullah Alqayoudhi, Amina Al-Jardani, and Seif Al-Abri. 2021. "Characteristics, Risk Factors, and Survival Analysis of Candida auris Cases: Results of One-Year National Surveillance Data from Oman" Journal of Fungi 7, no. 1: 31. https://doi.org/10.3390/jof7010031
APA StyleAl-Rashdi, A., Al-Maani, A., Al-Wahaibi, A., Alqayoudhi, A., Al-Jardani, A., & Al-Abri, S. (2021). Characteristics, Risk Factors, and Survival Analysis of Candida auris Cases: Results of One-Year National Surveillance Data from Oman. Journal of Fungi, 7(1), 31. https://doi.org/10.3390/jof7010031






