Genomic Analysis of Limosilactobacillus fermentum ATCC 23271, a Potential Probiotic Strain with Anti-Candida Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Whole-Genome Sequencing of Limosilactobacillus fermentum ATCC 23271
2.3. Genome Analysis
2.4. Antagonism Assay
2.5. Interference on Cell Adhesion
2.6. Tolerance to Gastrointestinal Conditions
2.7. Antibiotic Susceptibility Test
2.8. Ethical Aspects
2.9. Statistical Analysis
3. Results
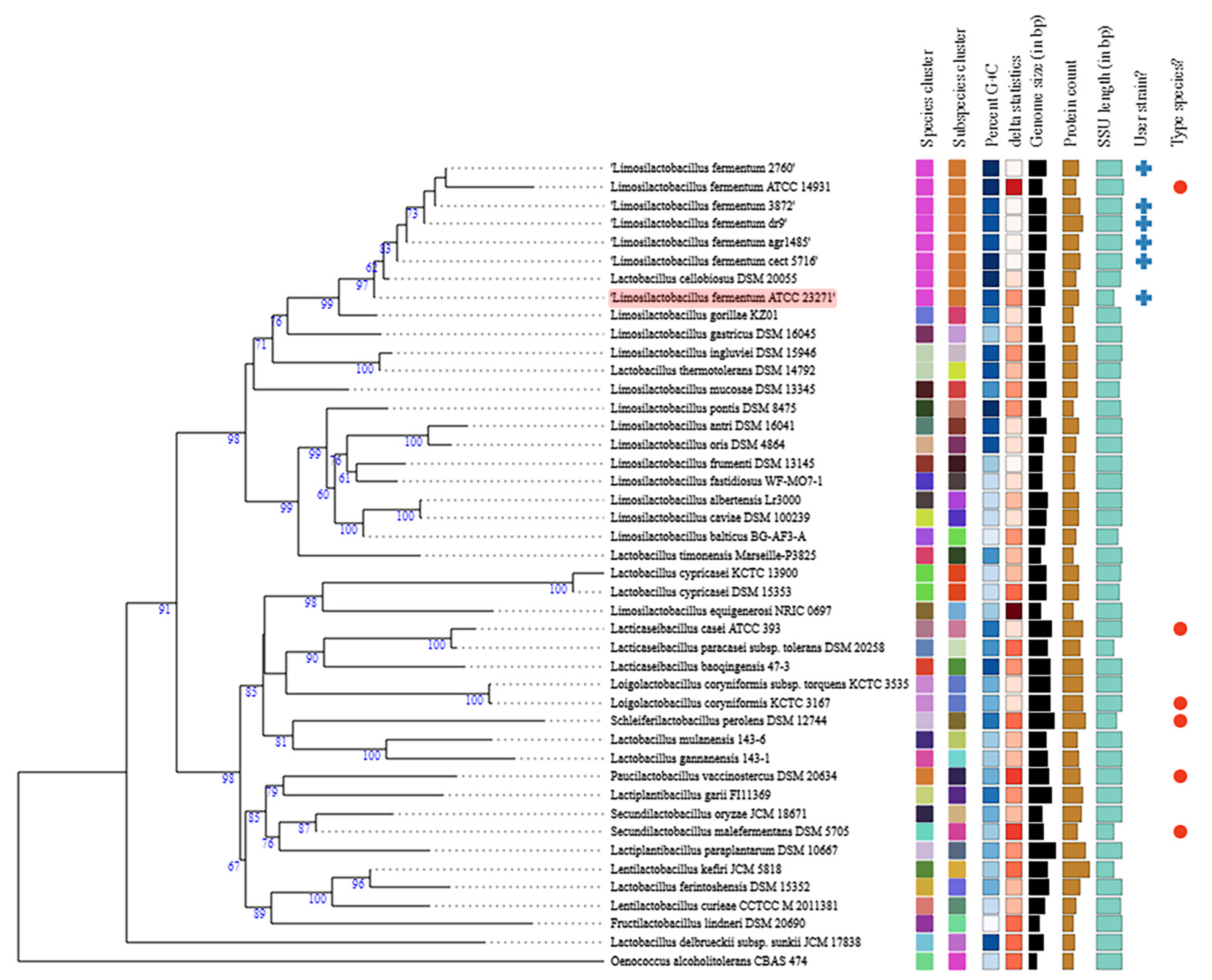
3.1. General Genome Features and Comparative Analysis
3.2. Putative Genes Associated with Probiotic Properties
3.3. Antagonism Activity
3.4. Interference in Pathogen Cell Adhesion
3.5. Tolerance to Bile Salts and Acidic pH
3.6. Antibiotic Susceptibility Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Achkar, J.M.; Fries, B.C. Candida infections of the genitourinary tract. Clin. Microbiol. Rev. 2010, 23, 253–273. [Google Scholar] [CrossRef] [Green Version]
- Millsop, J.W.; Fazel, N. Oral candidiasis. Clin. Dermatol. 2016, 34, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Sobel, J.D. Vaginitis. N. Engl. J. Med. 1997, 337, 1896–1903. [Google Scholar] [CrossRef] [PubMed]
- Pappas, P.G.; Kauffman, C.A.; Andes, D.R.; Clancy, C.J.; Marr, K.A.; Ostrosky-Zeichner, L.; Reboli, A.C.; Schuster, M.G.; Vazquez, J.A.; Walsh, T.J.; et al. Clinical Practice Guideline for the Management of Candidiasis: 2016 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016, 62, e1–e50. [Google Scholar] [CrossRef]
- Owen, M.K.; Clenney, T.L. Management of vaginitis. Am. Fam. Phys. 2004, 70, 2125–2132. [Google Scholar]
- Mroczynska, M.; Brillowska-Dabrowska, A. Review on Current Status of Echinocandins Use. Antibiotics 2020, 9, 227. [Google Scholar] [CrossRef]
- Sobel, J.D.; Sobel, R. Current treatment options for vulvovaginal candidiasis caused by azole-resistant Candida species. Expert Opin. Pharmacother. 2018, 19, 971–977. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Kerry, R.G.; Patra, J.K.; Gouda, S.; Park, Y.; Shin, H.-S.; Das, G. Benefaction of probiotics for human health: A review. J. Food Drug Anal. 2018, 26, 927–939. [Google Scholar] [CrossRef] [Green Version]
- do Carmo, M.S.; Santos, C.I.D.; Araujo, M.C.; Giron, J.A.; Fernandes, E.S.; Monteiro-Neto, V. Probiotics, mechanisms of action, and clinical perspectives for diarrhea management in children. Food Func. 2018, 9, 5074–5095. [Google Scholar] [CrossRef]
- Johnston, B.C.; Ma, S.S.; Goldenberg, J.Z.; Thorlund, K.; Vandvik, P.O.; Loeb, M.; Guyatt, G.H. Probiotics for the prevention of Clostridium difficile-associated diarrhea: A systematic review and meta-analysis. Ann. Intern. Med. 2012, 157, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Feyisetan, O.; Tracey, C.; Hellawell, G.O. Probiotics, dendritic cells and bladder cancer. BJU Int. 2012, 109, 1594–1597. [Google Scholar] [CrossRef] [PubMed]
- Hendler, R.; Zhang, Y. Probiotics in the Treatment of Colorectal Cancer. Medicines 2018, 5, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendoza, L. Potential effect of probiotics in the treatment of breast cancer. Oncol. Rev. 2019, 13, 422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemarajata, P.; Versalovic, J. Effects of probiotics on gut microbiota: Mechanisms of intestinal immunomodulation and neuromodulation. Ther. Adv. Gastroenterol. 2013, 6, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, P.; Curtis, N. The influence of probiotics on vaccine responses—A systematic review. Vaccine 2018, 36, 207–213. [Google Scholar] [CrossRef]
- Goderska, K.; Agudo Pena, S.; Alarcon, T. Helicobacter pylori treatment: Antibiotics or probiotics. Appl. Microbiol. Biotechnol. 2018, 102, 1–7. [Google Scholar] [CrossRef]
- Moayyedi, P.; Ford, A.C.; Talley, N.J.; Cremonini, F.; Foxx-Orenstein, A.E.; Brandt, L.J.; Quigley, E.M. The efficacy of probiotics in the treatment of irritable bowel syndrome: A systematic review. Gut 2010, 59, 325–332. [Google Scholar] [CrossRef]
- Fiocchi, A.; Pawankar, R.; Cuello-Garcia, C.; Ahn, K.; Al-Hammadi, S.; Agarwal, A.; Beyer, K.; Burks, W.; Canonica, G.W.; Ebisawa, M.; et al. World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Probiotics. World Allergy Organ. J. 2015, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Sargin, P.; Ciecko, A.E.; Dew, K.; Rhonda, G.; Jia, S.; Laib, T.; Roethle, M.F.; Hessner, M.J. Probiotic Supplementation with Lactobacillus plantarum 299v Lowers Systemic Inflammation, Reduces Islet ER Stress and Prevents Type 1 Diabetes in BioBreeding Rats. J. Endocrine Soc. 2021, 5 (Suppl. 1), A661–A662. [Google Scholar] [CrossRef]
- Roselli, M.; Pieper, R.; Rogel-Gaillard, C.; de Vries, H.; Bailey, M.; Smidt, H.; Lauridsen, C. Immunomodulating effects of probiotics for microbiota modulation, gut health and disease resistance in pigs. Anim. Feed Sci. Technol. 2017, 233, 104–119. [Google Scholar] [CrossRef] [Green Version]
- Rocha-Ramirez, L.M.; Perez-Solano, R.A.; Castanon-Alonso, S.L.; Moreno Guerrero, S.S.; Ramirez Pacheco, A.; Garcia Garibay, M.; Eslava, C. Probiotic Lactobacillus Strains Stimulate the Inflammatory Response and Activate Human Macrophages. J. Immunol. Res. 2017, 2017, 4607491. [Google Scholar] [CrossRef] [Green Version]
- Vitetta, L.; Saltzman, E.T.; Thomsen, M.; Nikov, T.; Hall, S. Adjuvant Probiotics and the Intestinal Microbiome: Enhancing Vaccines and Immunotherapy Outcomes. Vaccines 2017, 5, 50. [Google Scholar] [CrossRef] [Green Version]
- Hempel, S.; Newberry, S.; Ruelaz, A.; Wang, Z.; Miles, J.; Suttorp, M.J.; Johnsen, B.; Shanman, R.; Slusser, W.; Fu, N. Safety of probiotics used to reduce risk and prevent or treat disease. Evid. Rep./Technol. Assess. 2011, 200, 1–645. [Google Scholar]
- Lee, Y.K. Probiotics microorganisms. In Handbook of Probiotics and Prebiotics, 2nd ed.; Lee, Y.K., Salminen, S., Eds.; John Wiley & Sons: New Jersey, NJ, USA, 2009; pp. 3–24. [Google Scholar]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Archer, A.C.; Halami, P.M. Probiotic attributes of Lactobacillus fermentum isolated from human feces and dairy products. Appl. Microbiol. Biotechnol. 2015, 99, 8113–8123. [Google Scholar] [CrossRef]
- Cox, A.J.; Pyne, D.B.; Saunders, P.U.; Fricker, P.A. Oral administration of the probiotic Lactobacillus fermentum VRI-003 and mucosal immunity in endurance athletes. Br. J. Sports Med. 2010, 44, 222–226. [Google Scholar] [CrossRef] [Green Version]
- Falah, F.; Vasiee, A.; Behbahani, B.A.; Yazdi, F.T.; Moradi, S.; Mortazavi, S.A.; Roshanak, S. Evaluation of adherence and anti-infective properties of probiotic Lactobacillus fermentum strain 4-17 against Escherichia coli causing urinary tract infection in humans. Microb. Pathog. 2019, 131, 246–253. [Google Scholar] [CrossRef]
- Lehri, B.; Seddon, A.; Karlyshev, A. Lactobacillus fermentum 3872 as a potential tool for combatting Campylobacter jejuni infections. Virulence 2017, 8, 1753–1760. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.H.; Yu, B.; Jang, S.H.; Tsen, H.Y. Different probiotic properties for Lactobacillus fermentum strains isolated from swine and poultry. Anaerobe 2007, 13, 107–113. [Google Scholar] [CrossRef]
- Maldonado, J.; Canabate, F.; Sempere, L.; Vela, F.; Sanchez, A.R.; Narbona, E.; Lopez-Huertas, E.; Geerlings, A.; Valero, A.D.; Olivares, M.; et al. Human milk probiotic Lactobacillus fermentum CECT5716 reduces the incidence of gastrointestinal and upper respiratory tract infections in infants. J. Pediatr. Gastroenterol. Nutr. 2012, 54, 55–61. [Google Scholar] [CrossRef]
- Mikelsaar, M.; Zilmer, M. Lactobacillus fermentum ME-3–An antimicrobial and antioxidative probiotic. Microb. Ecol. Health Dis. 2009, 21, 1–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, C.L.; Thorsen, L.; Schwan, R.F.; Jespersen, L. Strain-specific probiotics properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus brevis isolates from Brazilian food products. Food Microbiol. 2013, 36, 22–29. [Google Scholar] [CrossRef] [Green Version]
- Pereira, D.I.; McCartney, A.L.; Gibson, G.R. An in vitro study of the probiotic potential of a bile-salt-hydrolyzing Lactobacillus fermentum strain, and determination of its cholesterol-lowering properties. Appl. Environ. Microbiol. 2003, 69, 4743–4752. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Hong, K.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Lactobacillus fermentum and its potential immunomodulatory properties. J. Funct. Foods 2019, 56, 21–32. [Google Scholar] [CrossRef]
- do Carmo, M.S.; Noronha, F.M.; Arruda, M.O.; Costa, E.P.; Bomfim, M.R.; Monteiro, A.S.; Ferro, T.A.; Fernandes, E.S.; Giron, J.A.; Monteiro-Neto, V. Lactobacillus fermentum ATCC 23271 Displays In vitro Inhibitory Activities against Candida spp. Front. Microbiol. 2016, 7, 1722. [Google Scholar] [CrossRef] [Green Version]
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2015, 31, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Wang, Y.; Coleman-Derr, D.; Chen, G.; Gu, Y.Q. OrthoVenn: A web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2015, 43, W78–W84. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosentino, S.; Voldby Larsen, M.; Møller Aarestrup, F.; Lund, O. PathogenFinder-distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef] [Green Version]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [Green Version]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [Green Version]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef] [Green Version]
- Kaewnopparat, S.; Dangmanee, N.; Kaewnopparat, N.; Srichana, T.; Chulasiri, M.; Settharaksa, S. In vitro probiotic properties of Lactobacillus fermentum SK5 isolated from vagina of a healthy woman. Anaerobe 2013, 22, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, C.; do Carmo, M.S.; Melo, B.O.; Alves, M.S.; Dos Santos, C.I.; Monteiro, S.G.; Bomfim, M.R.Q.; Fernandes, E.S.; Monteiro-Neto, V. In Vitro Antimicrobial Activity and Probiotic Potential of Bifidobacterium and Lactobacillus against Species of Clostridium. Nutrients 2019, 11, 448. [Google Scholar] [CrossRef] [Green Version]
- Charteris, W.P.; Kelly, P.M.; Morelli, L.; Collins, J.K. Antibiotic susceptibility of potentially probiotic Lactobacillus species. J. Food Prot. 1998, 61, 1636–1643. [Google Scholar] [CrossRef]
- Al-Madboly, L.A.; Khedr, E.G.; Ali, S.M. Optimization of Reduced Glutathione Production by a Lactobacillus plantarum Isolate Using Plackett-Burman and Box-Behnken Designs. Front. Microbiol. 2017, 8, 772. [Google Scholar] [CrossRef]
- Ianniello, R.G.; Zheng, J.; Zotta, T.; Ricciardi, A.; Ganzle, M.G. Biochemical analysis of respiratory metabolism in the heterofermentative Lactobacillus spicheri and Lactobacillus reuteri. J. Appl. Microbiol. 2015, 119, 763–775. [Google Scholar] [CrossRef]
- Walter, J.; Heng, N.C.; Hammes, W.P.; Loach, D.M.; Tannock, G.W.; Hertel, C. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 2003, 69, 2044–2051. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Ji, H.; Zhang, D.; Liu, H.; Wang, S.; Wang, J.; Wang, Y. Complete Genome Sequencing of Lactobacillus plantarum ZLP001, a Potential Probiotic That Enhances Intestinal Epithelial Barrier Function and Defense Against Pathogens in Pigs. Front. Physiol. 2018, 9, 1689. [Google Scholar] [CrossRef]
- Martínez-Abad, B.; Garrote, J.A.; Bernardo, D.; Montalvillo, E.; Escudero-Hernández, C.; Vázquez, E.; Rueda, R.; Arranz, E. Differential immunomodulatory effects of Lactobacillus rhamnosus DR20, Lactobacillus fermentum CECT 5716 and Bifidobacterium animalis subsp. lactis on monocyte-derived dendritic cells. J. Funct. Foods 2016, 22, 300–312. [Google Scholar] [CrossRef]
- Olivares, M.; Diaz-Ropero, M.P.; Sierra, S.; Lara-Villoslada, F.; Fonolla, J.; Navas, M.; Rodriguez, J.M.; Xaus, J. Oral intake of Lactobacillus fermentum CECT5716 enhances the effects of influenza vaccination. Nutrition 2007, 23, 254–260. [Google Scholar] [CrossRef]
- Maldonado-Lobon, J.A.; Diaz-Lopez, M.A.; Carputo, R.; Duarte, P.; Diaz-Ropero, M.P.; Valero, A.D.; Sanudo, A.; Sempere, L.; Ruiz-Lopez, M.D.; Banuelos, O.; et al. Lactobacillus fermentum CECT 5716 Reduces Staphylococcus Load in the Breastmilk of Lactating Mothers Suffering Breast Pain: A Randomized Controlled Trial. Breastfeed. Med. 2015, 10, 425–432. [Google Scholar] [CrossRef]
- Hor, Y.Y.; Lew, L.C.; Jaafar, M.H.; Lau, A.S.; Ong, J.S.; Kato, T.; Nakanishi, Y.; Azzam, G.; Azlan, A.; Ohno, H.; et al. Lactobacillus sp. improved microbiota and metabolite profiles of aging rats. Pharmacol. Res. 2019, 146, 104312. [Google Scholar] [CrossRef]
- Hor, Y.Y.; Ooi, C.H.; Lew, L.C.; Jaafar, M.H.; Lau, A.S.; Lee, B.K.; Azlan, A.; Choi, S.B.; Azzam, G.; Liong, M.T. The molecular mechanisms of probiotic strains in improving ageing bone and muscle of d-galactose-induced ageing rats. J. Appl. Microbiol. 2021, 130, 1307–1322. [Google Scholar] [CrossRef] [PubMed]
- Lew, L.C.; Hor, Y.Y.; Jaafar, M.H.; Lau, A.S.Y.; Ong, J.S.; Chuah, L.O.; Yap, K.P.; Azzam, G.; Azlan, A.; Liong, M.T. Lactobacilli modulated AMPK activity and prevented telomere shortening in ageing rats. Benef. Microbes 2019, 10, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Lehri, B.; Seddon, A.; Karlyshev, A. Discovery and characterisation of a novel plasmid of a probiotic strain Lactobacillus fermentum 3872. In Proceedings of the International Conference on Antimicrobial Research, Madrid, Spain, 1–3 October 2014. [Google Scholar]
- Anderson, R.C.; Cookson, A.L.; McNabb, W.C.; Kelly, W.J.; Roy, N.C. Lactobacillus plantarum DSM 2648 is a potential probiotic that enhances intestinal barrier function. FEMS Microbiol. Lett. 2010, 309, 184–192. [Google Scholar] [CrossRef]
- Archer, A.C.; Kurrey, N.K.; Halami, P.M. In vitro adhesion and anti-inflammatory properties of native Lactobacillus fermentum and Lactobacillus delbrueckii spp. J. Appl. Microbiol. 2018, 125, 243–256. [Google Scholar] [CrossRef]
- Rogosa, M.; Wiseman, R.; Mitchell, J.A.; Disraely, M.; Beaman, A. Species differentiation of oral lactobacilli from man including descriptions of Lactobacillus salivarius nov spec and Lactobacillus cellobiosus nov spec. J. Bacteriol. 1953, 65, 681–699. [Google Scholar] [CrossRef] [Green Version]
- Dellaglio, F.; Torriani, S.; Felis, G.E. Reclassification of Lactobacillus cellobiosus Rogosa et al. 1953 as a later synonym of Lactobacillus fermentum Beijerinck 1901. Int. J. Syst. Evol. Microbiol. 2004, 54, 809–812. [Google Scholar] [CrossRef] [Green Version]
- Tsuchida, S.; Nezuo, M.; Tsukahara, M.; Ogura, Y.; Hayashi, T.; Ushida, K. Draft Genome Sequence of Lactobacillus gorillae Strain KZ01T, Isolated from a Western Lowland Gorilla. Genome Announc. 2015, 3, e01196-15. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, L.C.; Saraiva, T.D.; Silva, W.M.; Pereira, U.P.; Campos, B.C.; Benevides, L.J.; Rocha, F.S.; Figueiredo, H.C.; Azevedo, V.; Soares, S.C. Analyses of the probiotic property and stress resistance-related genes of Lactococcus lactis subsp. lactis NCDO 2118 through comparative genomics and in vitro assays. PLoS ONE 2017, 12, e0175116. [Google Scholar] [CrossRef]
- Jayashree, S.; Pooja, S.; Pushpanathan, M.; Rajendhran, J.; Gunasekaran, P. Identification and characterization of bile salt hydrolase genes from the genome of Lactobacillus fermentum MTCC 8711. Appl. Biochem. Biotechnol. 2014, 174, 855–866. [Google Scholar] [CrossRef]
- John, G.S.; Brot, N.; Ruan, J.; Erdjument-Bromage, H.; Tempst, P.; Weissbach, H.; Nathan, C. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc. Natl. Acad. Sci. USA 2001, 98, 9901–9906. [Google Scholar] [CrossRef] [Green Version]
- Skaar, E.P.; Tobiason, D.M.; Quick, J.; Judd, R.C.; Weissbach, H.; Etienne, F.; Brot, N.; Seifert, H.S. The outer membrane localization of the Neisseria gonorrhoeae MsrA/B is involved in survival against reactive oxygen species. Proc. Natl. Acad. Sci. USA 2002, 99, 10108–10113. [Google Scholar] [CrossRef] [Green Version]
- Brot, N.; Weissbach, H. Peptide methionine sulfoxide reductase: Biochemistry and physiological role. Biopolymers 2000, 55, 288–296. [Google Scholar] [CrossRef]
- Ruas-Madiedo, P.; Gueimonde, M.; Margolles, A.; de los Reyes-Gavilan, C.G.; Salminen, S. Exopolysaccharides produced by probiotic strains modify the adhesion of probiotics and enteropathogens to human intestinal mucus. J. Food Prot. 2006, 69, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Kapse, N.G.; Engineer, A.S.; Gowdaman, V.; Wagh, S.; Dhakephalkar, P.K. Functional annotation of the genome unravels probiotic potential of Bacillus coagulans HS243. Genomics 2019, 111, 921–929. [Google Scholar] [CrossRef]
- Heinemann, C.; van Hylckama Vlieg, J.E.; Janssen, D.B.; Busscher, H.J.; van der Mei, H.C.; Reid, G. Purification and characterization of a surface-binding protein from Lactobacillus fermentum RC-14 that inhibits adhesion of Enterococcus faecalis 1131. FEMS Microbiol. Lett. 2000, 190, 177–180. [Google Scholar] [CrossRef]
- Varma, P.; Dinesh, K.R.; Menon, K.K.; Biswas, R. Lactobacillus fermentum isolated from human colonic mucosal biopsy inhibits the growth and adhesion of enteric and foodborne pathogens. J. Food Sci. 2010, 75, M546–M551. [Google Scholar] [CrossRef]
- Marcotte, H.; Ferrari, S.; Cesena, C.; Hammarstrom, L.; Morelli, L.; Pozzi, G.; Oggioni, M.R. The aggregation-promoting factor of Lactobacillus crispatus M247 and its genetic locus. J. Appl. Microbiol. 2004, 97, 749–756. [Google Scholar] [CrossRef]
- Martín, R.; Sánchez, B.; Suárez, J.E.; Urdaci, M.C. Characterization of the adherence properties of human Lactobacilli strains to be used as vaginal probiotics. FEMS Microbiol. Lett. 2012, 328, 166–173. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Wannun, P.; Piwat, S.; Teanpaisan, R. Purification, Characterization, and Optimum Conditions of Fermencin SD11, a Bacteriocin Produced by Human Orally Lactobacillus fermentum SD11. Appl. Biochem. Biotechnol. 2016, 179, 572–582. [Google Scholar] [CrossRef]
- Nilsen, T.; Nes, I.F.; Holo, H. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 2003, 69, 2975–2984. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority (EFSA). Introduction of a Qualified Presumption of Safety (QPS) approach for assessment of selected microorganisms referred to EFSA-Opinion of the Scientific Committee. EFSA J. 2007, 5, 587. [Google Scholar] [CrossRef]
- Nawaz, M.; Wang, J.; Zhou, A.; Ma, C.; Wu, X.; Moore, J.E.; Millar, B.C.; Xu, J. Characterization and transfer of antibiotic resistance in lactic acid bacteria from fermented food products. Curr. Microbiol. 2011, 62, 1081–1089. [Google Scholar] [CrossRef]
- Palmer, K.L.; Gilmore, M.S. Multidrug-resistant enterococci lack CRISPR-cas. mBio 2010, 1, e00227–e00310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClelland, R.S.; Richardson, B.A.; Hassan, W.M.; Graham, S.M.; Kiarie, J.; Baeten, J.M.; Mandaliya, K.; Jaoko, W.; Ndinya-Achola, J.O.; Holmes, K.K. Prospective study of vaginal bacterial flora and other risk factors for vulvovaginal candidiasis. J. Infect. Dis. 2009, 199, 1883–1890. [Google Scholar] [CrossRef] [Green Version]
- Pramanick, R.; Mayadeo, N.; Warke, H.; Begum, S.; Aich, P.; Aranha, C. Vaginal microbiota of asymptomatic bacterial vaginosis and vulvovaginal candidiasis: Are they different from normal microbiota? Microb. Pathog. 2019, 134, 103599. [Google Scholar] [CrossRef] [PubMed]
- Willems, H.M.E.; Ahmed, S.S.; Liu, J.; Xu, Z.; Peters, B.M. Vulvovaginal Candidiasis: A Current Understanding and Burning Questions. J. Fungi 2020, 6, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuochi, V.; Cardile, V.; Petronio Petronio, G.; Furneri, P.M. Biological properties and production of bacteriocins-like-inhibitory substances by Lactobacillus sp. strains from human vagina. J. Appl. Microbiol. 2019, 126, 1541–1550. [Google Scholar] [CrossRef]
- Itapary Dos Santos, C.; Ramos Franca, Y.; Duarte Lima Campos, C.; Quaresma Bomfim, M.R.; Oliveira Melo, B.; Assuncao Holanda, R.; Santos, V.L.; Gomes Monteiro, S.; Buozzi Moffa, E.; Souza Monteiro, A.; et al. Antifungal and Antivirulence Activity of Vaginal Lactobacillus Spp. Products against Candida Vaginal Isolates. Pathogens 2019, 8, 150. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Wang, Q.; Yang, E.; Yan, L.; Li, T.; Zhuang, H. Antimicrobial Compounds Produced by Vaginal Lactobacillus crispatus Are Able to Strongly Inhibit Candida albicans Growth, Hyphal Formation and Regulate Virulence-related Gene Expressions. Front. Microbiol. 2017, 8, 564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
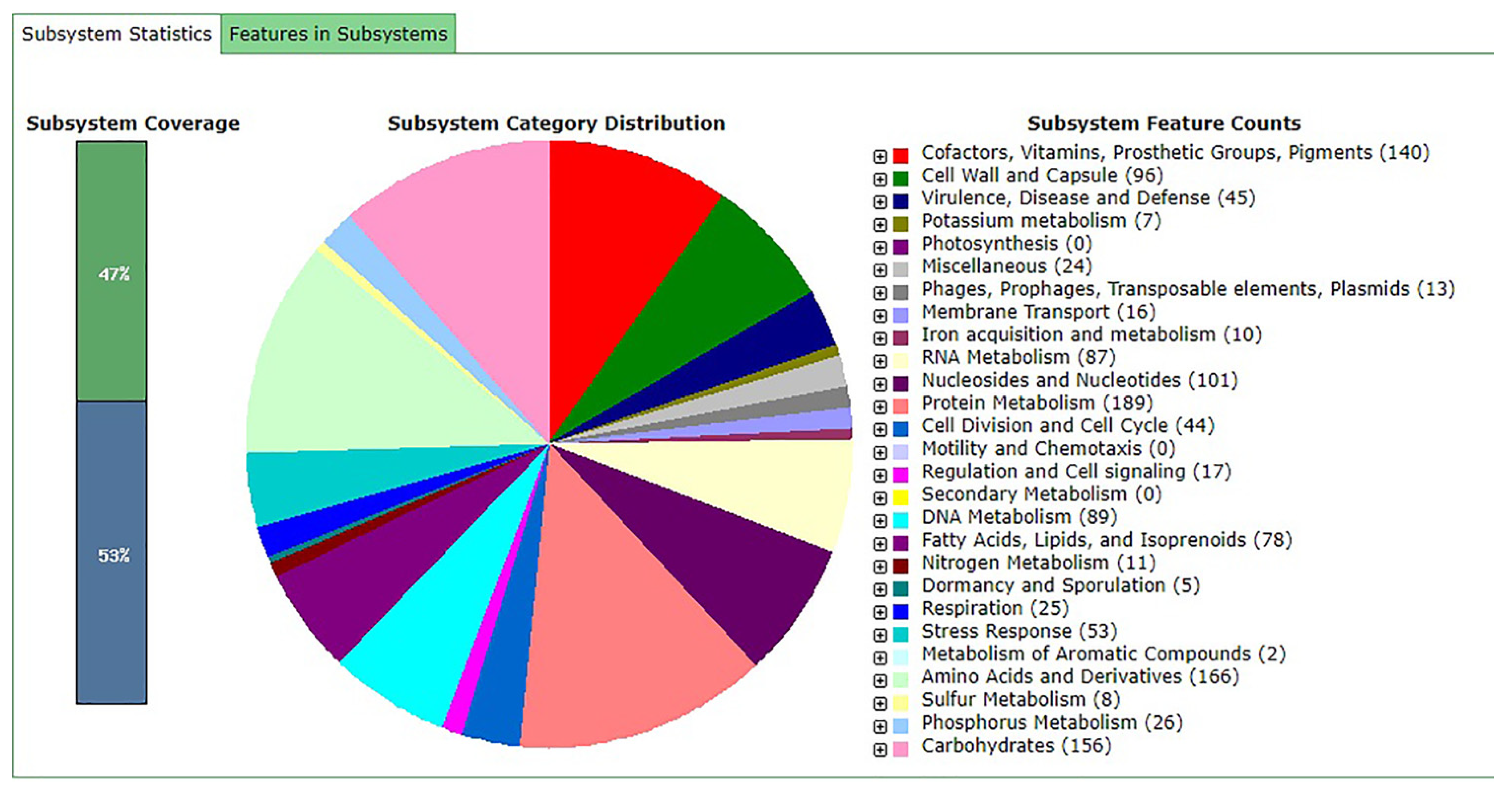
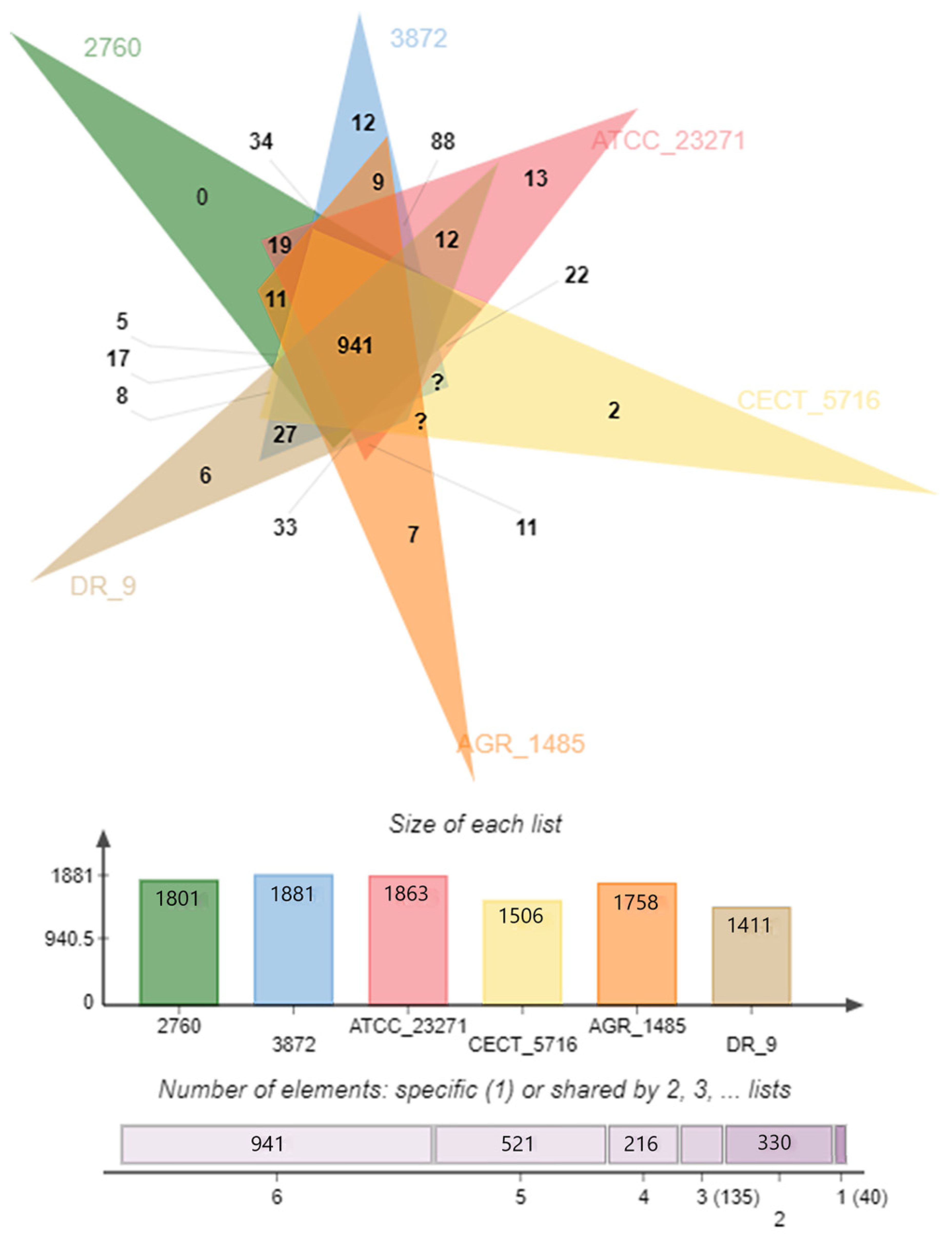
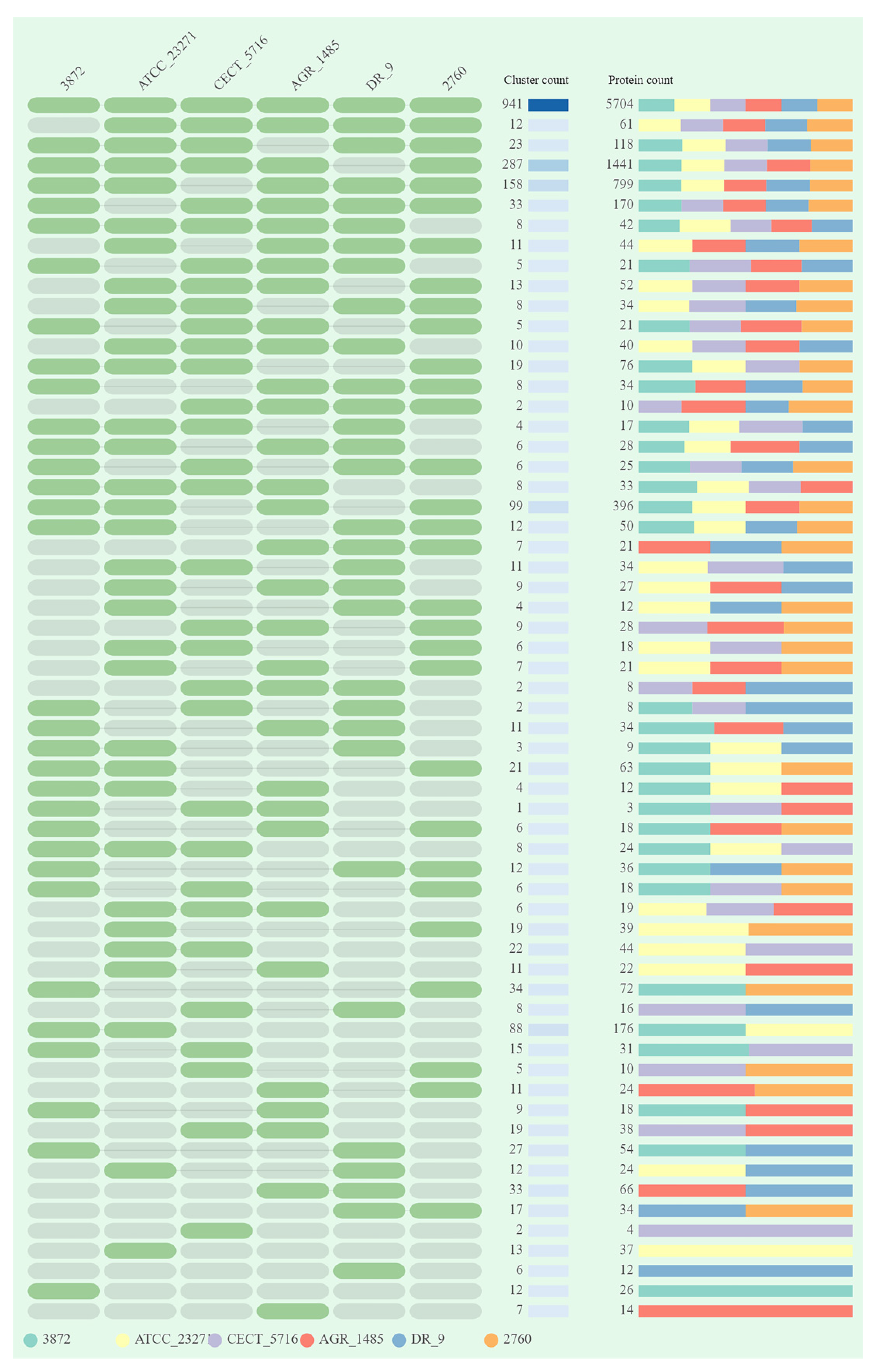
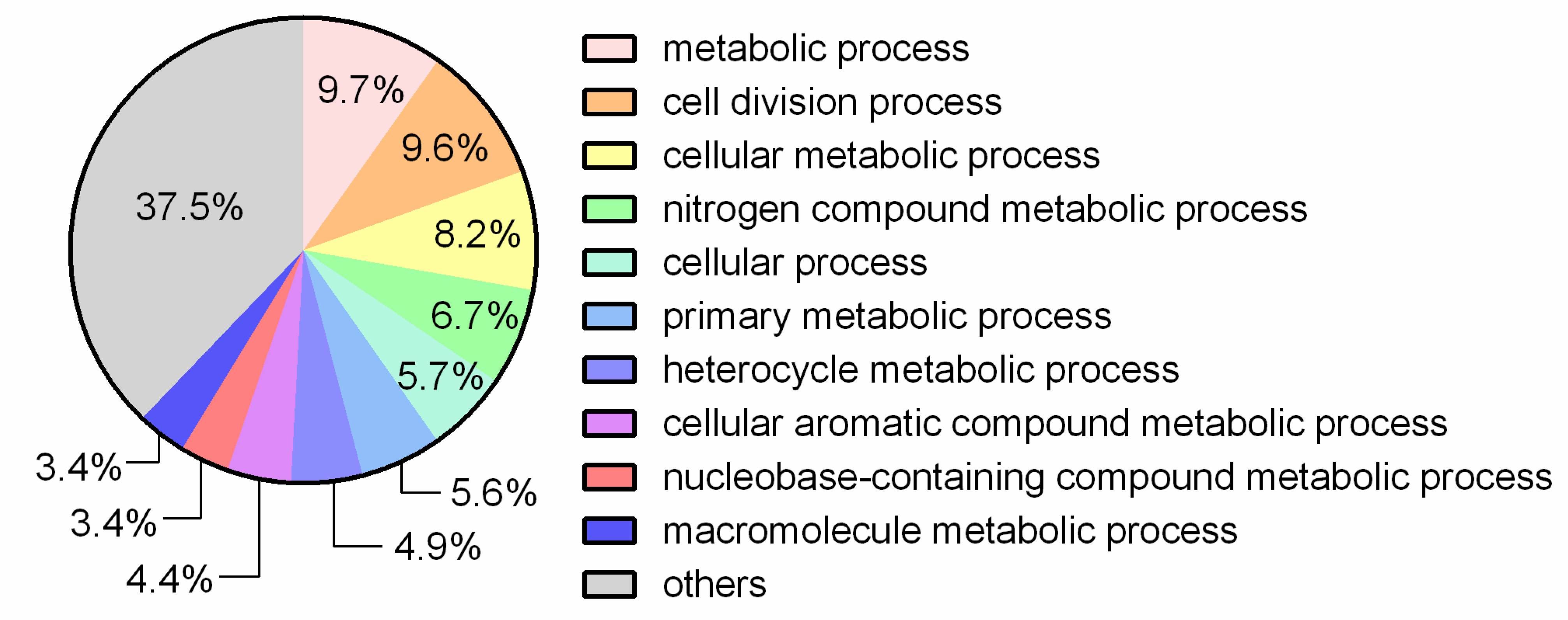
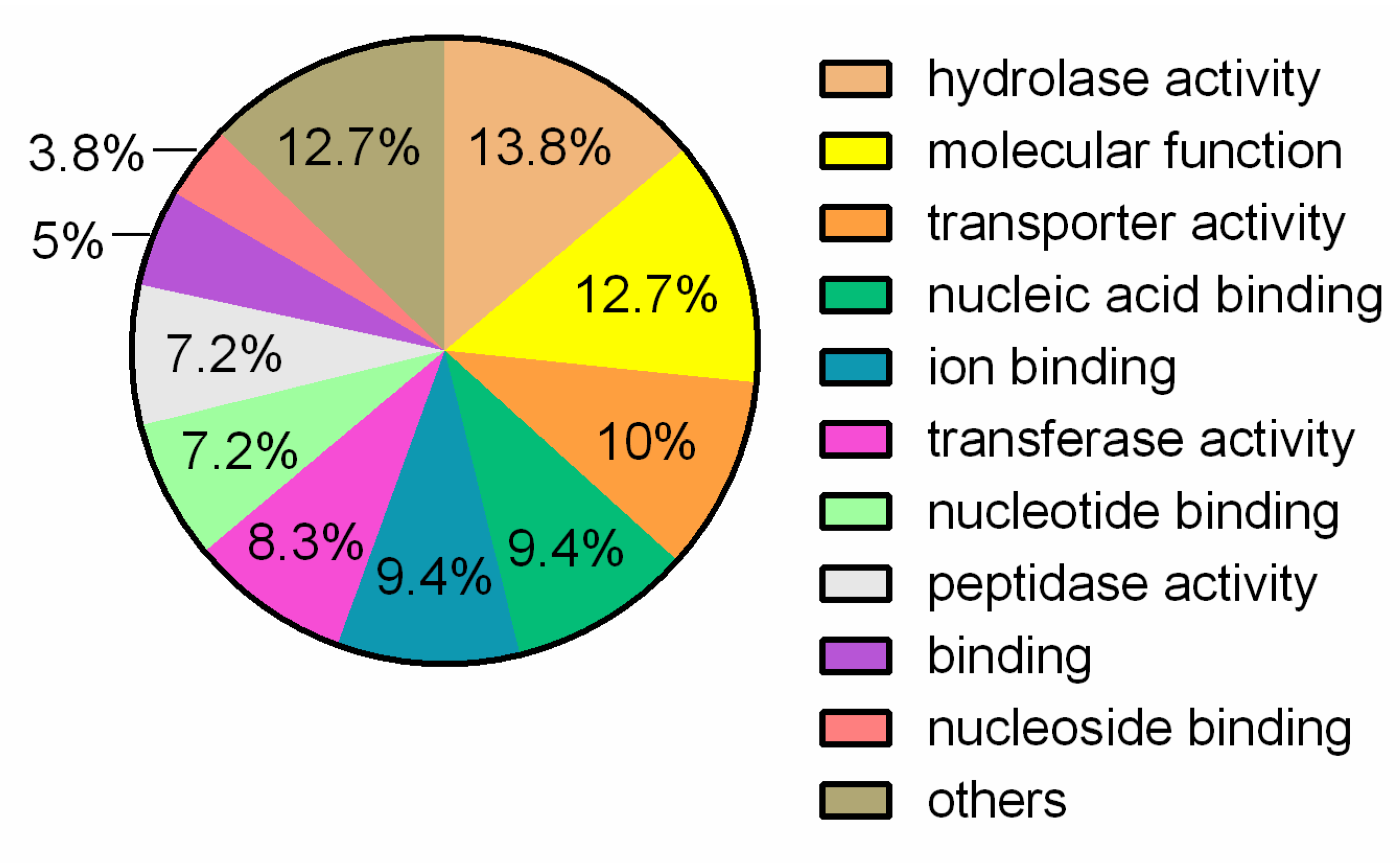



| Attribute * | Indicators |
|---|---|
| Genome size (bp) | 2,193,335 |
| G + C content (%) | 50.9 |
| N50 | 33,843 |
| L50 | 22 |
| Number of contigs (with PEGs) | 223 |
| Number of subsystems | 312 |
| Prophage-like clusters | 2 |
| CDS | 2387 |
| CDS (with proteins) | 2123 |
| N° of RNAs | 79 |
| N° of tmRNA operons | 1 |
| N° of tRNA | 55 |
| N° of rRNA | 21 |
| N° of CRISPR loci | 2 |
| RAST/BLAST Description | Query Length | Accession Length | Query Cover | E Value | Per Ident | Accession |
|---|---|---|---|---|---|---|
| ATP synthase F0 sector subunit a/F0F1 ATP synthase subunit A | 711 pb | 236 aa | 100% | 7 × 10−167 | 100% | WP_003682740.1 |
| ATP synthase F0 sector subunit b/F0F1 ATP synthase subunit B | 507 pb | 168 aa | 100% | 6 × 10−116 | 100% | WP_054173734.1 |
| ATP synthase F0 sector subunit c/MULTISPECIES: F0F1 ATP synthase subunit C | 213 pb | 70 aa | 100% | 1 × 10–38 | 100% | WP_003682741.1 |
| ATP synthase alpha chain/F0F1 ATP synthase subunit alpha | 1539 pb | 512 aa | 100% | 0.0 | 99.80% | WP_086439482.1 |
| ATP synthase beta chain/F0F1 ATP synthase subunit beta | 1422 pb | 473 aa | 100% | 0.0 | 99.79% | WP_057194567.1 |
| ATP synthase delta chain/F0F1 ATP synthase subunit delta | 546 pb | 181 aa | 100% | 3 × 10–126 | 99.45% | WP_057194565.1 |
| ATP synthase gamma chain/F0F1 ATP synthase subunit gamma | 936 pb | 311 aa | 100% | 0.0 | 99.68% | WP_088460387.1 |
| ATP synthase epsilon chain/ | 423 pb | 140 aa | 100% | 3 × 10–95 | 99.29% | WP_003685876.1 |
| L-lactate dehydrogenase (EC 1.1.1.27)/L-lactate dehydrogenase | 954 bp | 317 aa | 100% | 0.0 | 99.68% | WP_012391154.1 |
| L-lactate dehydrogenase (EC 1.1.1.27)/L-lactate dehydrogenase | 942 bp | 313 aa | 100% | 0.0 | 100% | WP_138464682.1 |
| L-lactate dehydrogenase (EC 1.1.1.27)/L-lactate dehydrogenase | 933 bp | 310 aa | 100% | 0.0 | 100% | WP_046948611.1 |
| PTS system, cellobiose-specific IIC component/PTS system oligo-beta-mannoside-specific EIIC component | 1311 bp | 436 aa | 100% | 0.0 | 100% | QIX58800.1 |
| ATP-dependent Clp protease ATP-binding subunit ClpX/ATP-dependent Clp protease ATP-binding subunit ClpX | 1251 bp | 416 aa | 100% | 0.0 | 99.52% | WP_096493701.1 |
| Glucose-6-phosphate isomerase (EC 5.3.1.9)/glucose-6-phosphate isomerase | 525 bp | 174 aa | 100% | 1 × 10–124 | 99.43% | RGW51862.1 |
| Glucose-6-phosphate isomerase (EC 5.3.1.9)/glucose-6-phosphate isomerase | 1353 bp | 450 aa | 100% | 0.0 | 99.56% | WP_021815746.1 |
| GTP pyrophosphokinase (EC 2.7.6.5)/GTP pyrophosphokinase | 612 bp | 203 aa | 100% | 8 × 10–150 | 99.51% | WP_100184301.1 |
| Pyruvate kinase/pyruvate kinase | 1422 bp | 473 aa | 100% | 0.0 | 100% | WP_003684953.1 |
| Arginine/ornithine antiporter ArcD/Amino acid transporter | 1419 bp | 472 aa | 100% | 0.0 | 100% | AOR74635.1 |
| Phosphoglycerate mutase/2,3-bisphosphoglycerate-dependent phosphoglycerate mutase | 678 bp | 225 aa | 100% | 4 × 10–166 | 100% | WP_004562727.1 |
| Choloylglycine hydrolase/choloylglycine hydrolase family protein | 978 bp | 325 aa | 100% | 0.0 | 100% | WP_035436617.1 |
| CTP synthase/CTP synthase | 1602 bp | 533 aa | 100% | 0.0 | 99.81% | WP_003684004.1 |
| RAST/BLAST Description | Function | Query Length | Accession Length | Query Cover | E Value | Per Ident | Accession |
|---|---|---|---|---|---|---|---|
| Aggregation substance precursor | Aggregation | 1842 pb | 613 aa | 99% | 0.0 | 99.84% | AKM50933.1 |
| LysM peptidoglycan-binding domain-containing protein | 591 pb | 196 aa | 99% | 9 × 10–85 | 100.00% | WP_021815732.1 | |
| LysM peptidoglycan-binding domain-containing protein | 816 pb | 315 aa | 48% | 1 × 10–40 | 100.00% | WP_168183590.1 | |
| Exopolysaccharide biosynthesis polyprenyl glycosylphosphotransferase | Exopolysaccharide production | 666 pb | 229 aa | 99% | 3 × 10–159 | 98.64% | WP_104877738.1 |
| Exopolysaccharide biosynthesis protein | 771 pb | 256 aa | 99% | 0.0 | 99.22% | WP_163601282.1 | |
| CpsD/CapB family tyrosine-protein kinase | 741 pb | 247 aa | 91% | 3 × 10–160 | 99.12% | WP_062813522.1 | |
| Exopolysaccharide biosynthesis protein | 771 pb | 256 aa | 99% | 5 × 10–174 | 99.61% | KPH03198.1 | |
| Fibronectin-binding domain-containing protein | Adhesion | 1692 pb | 563 aa | 99% | 0.0 | 99.82% | WP_103205388.1 |
| Algorithms * | Results |
|---|---|
| SVM | 1.000 |
| DA | 1.000 |
| RF | 0.957 |
| ANN | NAMP |
| RAST/BLAST Description | Query Length | Accession Length | Query Cover | E Value | Per Ident | Accession |
|---|---|---|---|---|---|---|
| Penicillin-binding protein | 1017 | 338 | 99% | 0.0 | 99.41% | EQC60084.1 |
| Class A beta-lactamase-related serine hydrolase | 1020 | 339 | 99% | 0.0 | 99.12% | MBD9348952.1 |
| Cation diffusion facilitator family transporter | 906 | 301 | 99% | 0.0 | 100.00% | WP_015639412.1 |
| Elongation factor G | 2085 | 694 | 99% | 0.0 | 99.86% | KPH03387.1 |
| DNA topoisomerase IV subunit B | 1998 | 665 | 99% | 0.0 | 99.85% | WP_003683141.1 |
| Topoisomerase IV subunit A | 2478 | 825 | 99% | 0.0 | 100.00% | BAG27240.1 |
| MerR family transcriptional regulator | 423 | 140 | 99% | 1 × 10–97 | 100.00% | WP_003682036.1 |
| Multidrug resistance protein MdtG | 342 | 113 | 99% | 2 × 10–61 | 99.12% | QIX57855.1 |
| MFS transporter | 123 | 160 | 97% | 2 × 10–17 | 97.50% | WP_155762340.1 |
| GTP-binding protein | 1935 | 644 | 99% | 0.0 | 100.00% | WP_112296957.1 |
| Multidrug transporter | 1227 | 413 | 99% | 0.0 | 99.75% | AKM51464.1 |
| DNA topoisomerase (ATP-hydrolyzing) subunit B | 1950 | 649 | 99% | 0.0 | 99.85% | WP_023465959.1 |
| DNA gyrase subunit A | 2511 | 836 | 99% | 0.0 | 99.76% | WP_160229810.1 |
| MerR family transcriptional regulator | 423 | 151 | 99% | 3 × 10–98 | 100.00% | CDI69999.1 |
| Multidrug transporter MatE | 1317 | 438 | 99% | 0.0 | 99.77% | WP_042513988.1 |
| Heavy metal translocating P-type ATPase | 1929 | 642 | 99% | 0.0 | 99.84% | WP_112297009.1 |
| MATE family efflux transporter | 1320 | 439 | 99% | 0.0 | 97.95% | WP_135252410.1 |
| FAD-dependent oxidoreductase | 1350 | 449 | 99% | 0.0 | 100.00% | WP_100184414.1 |
| Cation transporter | 552 | 207 | 99% | 4 × 10–98 | 100.00% | WP_114684362.1 |
| Pathogen | Inhibition Zone in mm ± SD * |
|---|---|
| E. faecalis (ATCC 29212) | In situ |
| E. coli enteroaggregative 17.2 | In situ |
| Salmonella enterica (ATCC 13076) | In situ |
| C. albicans (ATCC 90028) | 17 ± 1.41 |
| C. albicans (SC 5314) | 16.5 ± 1.12 |
| C. albicans 44 | 14 ± 1.4 |
| C. albicans CAS | 20 ± 0 |
| C. krusei (ATCC 6258) | − |
| C. krusei GJFD | − |
| Candida parapsilosis (ATCC 22019) | 26.5 ± 2.1 |
| C. parapsilosis FSG | 13.5 ± 2.1 |
| C. parapsilosis RCL | 14.5 ± 0.7 |
| Conditions | % of L. fermentum Survival (±SD) 1 | p-Value 2 |
|---|---|---|
| pH 2.0 | 60.88 ± 0.9569 | 0.0043 |
| pH 4.0 | 105.0 ± 5909 | |
| Bile salt 0.5% | 109.7 ± 4434 | 0.0348 |
| Bile salt 1% | 94.13 ± 1244 |
| Antibiotics | L. fermentum ATCC 23271 | |
|---|---|---|
| Zone Inhibition in mm | Interpretation * | |
| Cefazolin | 22 | Susceptible |
| Chloramphenicol | 34 | |
| Ciprofloxacin | 21 | |
| Clindamycin | 31 | |
| Erythromycin | 39 | |
| Linezolid | 40 | |
| Nitrofurantoin | 30 | |
| Rifampicin | 34 | |
| Penicillin G | 41 | |
| Tetracycline | 38 | |
| Tigecillin | 40 | |
| Cefoxitin | 15 | Moderately susceptible |
| Norfloxacin | 14 | |
| Gentamycin | 12 | Resistant |
| Sulfazotrim | 0 | |
| Vancomycin | 0 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
dos Santos, C.I.; Campos, C.D.L.; Nunes-Neto, W.R.; do Carmo, M.S.; Nogueira, F.A.B.; Ferreira, R.M.; Costa, E.P.S.; Gonzaga, L.F.; Araújo, J.M.M.; Monteiro, J.M.; et al. Genomic Analysis of Limosilactobacillus fermentum ATCC 23271, a Potential Probiotic Strain with Anti-Candida Activity. J. Fungi 2021, 7, 794. https://doi.org/10.3390/jof7100794
dos Santos CI, Campos CDL, Nunes-Neto WR, do Carmo MS, Nogueira FAB, Ferreira RM, Costa EPS, Gonzaga LF, Araújo JMM, Monteiro JM, et al. Genomic Analysis of Limosilactobacillus fermentum ATCC 23271, a Potential Probiotic Strain with Anti-Candida Activity. Journal of Fungi. 2021; 7(10):794. https://doi.org/10.3390/jof7100794
Chicago/Turabian Styledos Santos, Camilla I., Carmem D. L. Campos, Wallace R. Nunes-Neto, Monique S. do Carmo, Flávio A. B. Nogueira, Rômulo M. Ferreira, Ennio P. S. Costa, Laoane F. Gonzaga, Jéssica M. M. Araújo, Joveliane M. Monteiro, and et al. 2021. "Genomic Analysis of Limosilactobacillus fermentum ATCC 23271, a Potential Probiotic Strain with Anti-Candida Activity" Journal of Fungi 7, no. 10: 794. https://doi.org/10.3390/jof7100794
APA Styledos Santos, C. I., Campos, C. D. L., Nunes-Neto, W. R., do Carmo, M. S., Nogueira, F. A. B., Ferreira, R. M., Costa, E. P. S., Gonzaga, L. F., Araújo, J. M. M., Monteiro, J. M., Monteiro, C. R. A. V., Platner, F. S., Figueiredo, I. F. S., Holanda, R. A., Monteiro, S. G., Fernandes, E. S., Monteiro, A. S., & Monteiro-Neto, V. (2021). Genomic Analysis of Limosilactobacillus fermentum ATCC 23271, a Potential Probiotic Strain with Anti-Candida Activity. Journal of Fungi, 7(10), 794. https://doi.org/10.3390/jof7100794








