Abstract
Malassezia species are fastidious and slow-growing yeasts in which isolation from polymicrobial samples is hampered by fast-growing microorganisms. Malassezia selective culture media are needed. Although cycloheximide is often used, some fungi, including the chief human commensal Candida albicans, are resistant to this compound. This study aimed to test whether the macrolide rapamycin could be used in combination with cycloheximide to develop a Malassezia-selective culture medium. Rapamycin susceptibility testing was performed via microdilution assays in modified Dixon against two M. furfur and five Candida spp. The MIC was the lowest concentration that reduced growth by a minimum of 90%. Rapamycin ± cycloheximide 500 mg/L was also added to FastFung solid, and yeast suspensions were inoculated and incubated for 72 h. Rapamycin MICs for Candida spp. ranged from 0.5 to 2 mg/L, except for C. krusei, for which the MIC was >32 mg/L. M. furfur stains were rapamycin-resistant. Rapamycin and cycloheximide supplementation of the FastFung medium effectively inhibited the growth of non-Malassezia yeast, including cycloheximide-resistant C. albicans and C. tropicalis. Based on our findings, this “MalaSelect” medium should be further evaluated on polymicrobial samples for Malassezia isolation and culture.
1. Introduction
The Malassezia genus comprises 18 species that are lipid-dependent yeast commensals on human skin and other warm-blooded vertebrates [1]. Under certain circumstances (e.g., increased humidity and temperature), these yeasts cause common human skin diseases, including pityriasis versicolor, seborrheic dermatitis and folliculitis [2]. They can also cause severe bloodstream infections in neonates or immunocompromised patients hospitalized in intensive care with lipidic parenteral nutrition [3,4,5,6]. Their role in atopic dermatitis and psoriasis was also reported [7,8]. However, data on the interactions between Malassezia species and their hosts remain scarce, especially in humans. This knowledge gap has been widened because the routine use of culture media on which these lipid-dependent yeasts can be cultivated is limited to a few specialized laboratories. In the literature, a variety of lipid-enriched culture media, including Dixon and Leeming–Notman agar and their modified versions, are effective for Malassezia spp. cultivation [9,10,11]. However, these culture media are not selective, in that they also allow the growth of a variety of other fungi, including Candida spp. and molds [12,13,14,15]. Relatively slow-growing Malassezia spp. can remain undetected, overtaken by faster growing and sometimes also more abundant fungal species present in the samples. Therefore, the use of antimycotic compounds without effect on Malassezia spp. seems to be an option for the development of a selective culture medium. Among these types of compounds, cycloheximide is widely used in culture media aimed at isolating and cultivating Malassezia spp. from clinical samples [16,17,18,19]. However, some fungi, especially in the genus Candida (e.g., the most common human opportunistic pathogen C. albicans), are resistant to cycloheximide and might thus interfere with Malassezia isolation. The development of a Malassezia-specific and efficient culture medium is crucial in order to be able to selectively isolate Malassezia spp. from polymicrobial clinical specimens, as these yeast species are detected at relatively higher frequencies by using culture-independent methods in complex polymicrobial niches such as the respiratory and digestive tracts [20,21,22,23,24].
Vézina et al. discovered rapamycin in 1975 [25]. This is a secondary metabolite produced by Streptomyces hygroscopicus and has antifungal activity, especially against Candida albicans. Rapamycin is now commonly used as an immunosuppressive drug [26]. Although its antifungal activity against molds or other yeast species is well known [27,28], the potential activity of rapamycin against Malassezia spp. is poorly studied. However, it was recently reported that M. furfur and M. sympodialis are not sensitive to rapamycin [29]. The aim of this present study was to assess the in vitro antifungal activity of rapamycin against Malassezia spp. and Candida spp. by using a broth microdilution method, and to test whether a solid culture medium supplemented with rapamycin and cycloheximide might allow the growth of Malassezia spp., while inhibiting the growth of Candida spp.
2. Materials and Methods
2.1. Strains
A total of 3 Malassezia species, obtained from the Belgian Co-ordinated Collections of Micro-organisms/Institute of Hygiene and Epidemiology (BCCM/IHEM, Sciensano, Brussels, Belgium), including M. furfur, M. sympodialis and M. pachydermatis, were used to test the antifungal activity of rapamycin. Malassezia species were maintained at 30 °C and subcultured for 5 days before testing on FastFung medium [30,31], composed per liter [pH 6] of 43 g of Schaedler agar, 20 g of peptone, 10 g of glucose, 5 g of ox-bile, 10 g of malt extract, 2 mL oleic acid, 2.5 mL glycerol and 5 mL of Tween 60 (all from Sigma-Aldrich, Saint-Quentin Fallavier, France). Isolates of C. albicans, C. glabrata, C. parapsilosis and C. tropicalis and the C. krusei ATCC 6258 strain were also used. The Candida species were subcultured at 30 °C for 3 days in Sabouraud medium before testing.
2.2. Broth Microdilution
Rapamycin solutions (Sigma-Aldrich, Saint-Quentin-Fallavier, France, Ref. S-015-1mL) at 1 mg/mL in acetonitrile were obtained and stored at −80 °C until use. The rapamycin concentration gradient tested ranged from 0.0625 to 32 mg/L. Broth microdilution method was performed by using modified Dixon broth (3.6% malt extract, 0.6% peptone, 2% ox-bile, 1% Tween 40, 0.2% glycerol, and 0.2% oleic acid, buffered at pH 6) (all from Sigma-Aldrich Saint-Quentin-Fallavier, France). Yeast inoculum suspensions of M. furfur and Candida spp. were prepared in 2 mL sterile saline solution (0.85% NaCl) and standardized spectrophotometrically at 0.5 McFarland (106 colony-forming units [CFU]/mL). These suspensions were diluted 1:10 in sterile distilled water, and a total of 100 µL of the final dilution was transferred into a 96-well microtiter plate containing 100 µL of the medium to achieve a final concentration of 0.5–2.5 × 105 CFU/mL as recommended by EUCAST. Each assay was tested in duplicate. The microtiter plates were incubated at 30 °C and visually read after 24 and 48 h of incubation. The growth of each strain at various rapamycin concentrations, as well as a positive control cultured in rapamycin-free medium, was recorded. The MIC of each strain was defined as the lowest concentration that reduces growth by a minimum of 90% when compared to the control growth.
2.3. Growth Testing on Agar
FastFung medium supplemented with 500 mg/L cycloheximide (CliniSciences, Nanterre, France) was prepared and sterilized by autoclaving at 121 °C for 30 min. After cooling to approximately 56 °C, FastFung medium was partitioned into two equal volumes, and a solution of rapamycin was added to one volume of the FastFung medium. The two media were then distributed in sterile Petri dishes. Malassezia spp. and Candida spp. yeast suspensions at 106 CFU/mL were prepared and further diluted at 105, 104 and 103 in sterile distilled water. For each dilution, 20 µL was plated onto FastFung medium with or without rapamycin. Sterile distilled water was also plated in FastFung medium as a negative control. All agar plates were incubated aerobically at 30 °C and examined daily for 3 days.
3. Results
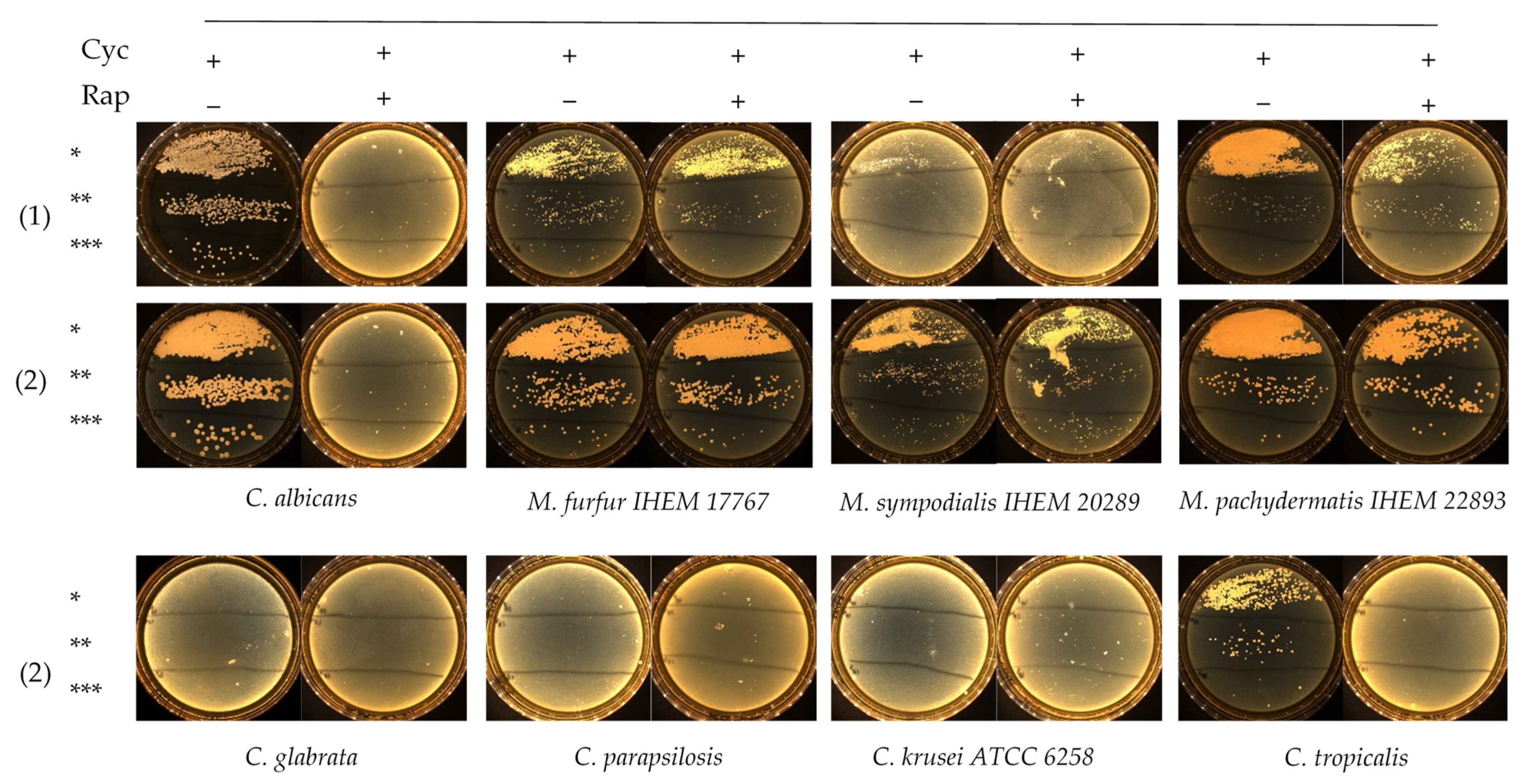
Rapamycin antifungal activity was evaluated in Dixon liquid medium against 2 M. furfur and 5 Candida strains, including C. albicans, C. glabrata, C. tropicalis, C. parapsilosis and C. krusei ATCC 6258. The MIC results are summarized in Table 1. Rapamycin exhibited low MIC values against Candida spp., except for C. krusei ATCC 6258, whose MIC were ≥32 mg/L. MIC after 24 h of incubation were 1- to 2-fold dilutions lower than those recorded after 48 h of incubation (Table 1). Slow-growing M. furfur required 48 h reading time. Both M. furfur strains exhibited high rapamycin MIC ≥ 32 mg/L, suggesting that rapamycin had no significant effect against Malassezia spp. After determination of rapamycin MIC values against M. furfur and Candida spp., we assessed the growth of these isolates on FastFung medium plates supplemented or not with 2 mg/L rapamycin. The plates were examined daily for 3 days. No growth was observed on FastFung medium with cycloheximide, supplemented or not with rapamycin for C. glabrata, C. parapsilosis and C. krusei ATCC 6258 (Figure 1). Cycloheximide (500 mg/L) did not inhibit C. albicans and C. tropicalis in FastFung medium without rapamycin. In FastFung medium supplemented with both cycloheximide and rapamycin, C. albicans and C. tropicalis did not grow after 2 to 3 days of incubation. The growth of M. furfur, M. sympodialis and M. pachydermatis was not altered when these yeasts were cultured in FastFung medium containing cycloheximide, supplemented or not with rapamycin (Figure 1).

Table 1.
Rapamycin in vitro susceptibility testing against M. furfur and Candida spp.
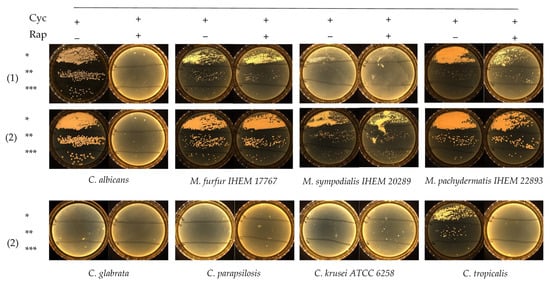
Figure 1.
Growth testing via serial inocula dilution of Candida spp. and Malassezia spp. on the FastFung medium without (−) or with (+) 2 mg/L rapamycin (Rap) and 500 mg/L cycloheximide (Cyc). Culture at 48 h (1) and 72 h (2) of incubation. * 105 CFU/mL, ** 104 CFU/mL and *** 103 CFU/mL.
4. Discussion
The development of selective culture media is an important challenge in medical mycology for isolation and culture of pathogens from clinical specimens. The use of antimicrobial agents remains the main strategy for the inhibition of undesirable microorganisms in culture. Here, we evaluated the in vitro antifungal activity of rapamycin against 2 M. furfur and 5 Candida spp. and found that M. furfur is resistant, with MIC values ≥ 32 mg/L (Table 1). However, rapamycin showed higher activity against Candida spp., with MIC values ranging from 0.5 to 2 mg/L, except C. krusei ATCC 6258 (Table 1). This higher activity of rapamycin against Candida spp., especially C. albicans, is consistent with results reported in the literature [25,27,28]. MIC values ≥ 32 mg/L recorded for C. krusei ATCC 6258 deserve further investigation to determine if rapamycin does not affect C. krusei species or whether our strain is resistant.
Growth testing on agar showed that cycloheximide and rapamycin have dual negative effects against Candida spp. in culture. Cycloheximide inhibited the growth of C. glabrata, C. parapsilosis and C. krusei ATCC 6258, whereas C. albicans and C. tropicalis were able to grow in FastFung medium supplemented with cycloheximide (Figure 1). The effects of cycloheximide against certain Candida spp. are already known [12,14,15]. By adding rapamycin, we were able to inhibit all Candida spp. tested, as no growth was observed in culture (Figure 1). No effect of cycloheximide against Malassezia species was observed, which is in agreement with the literature [17,18].
One of the most striking findings was the resistance of Malassezia spp. to rapamycin. Both strains of M. furfur showed resistance, with MIC values ≥ 32 mg/L (Table 1). These results were confirmed by testing M. furfur, M. sympodialis and M. pachydermatis in FastFung medium supplemented with rapamycin (Figure 1). Our findings are particularly interesting, as rapamycin inhibits a broad spectrum of fungi, including filamentous fungi and yeasts [28]. These results show that rapamycin can be a good candidate in selective agents. For example, it is well known that Candida spp. grow quickly, whereas Malassezia spp. grow slowly. Rapamycin allows the growth of Malassezia spp. by limiting the growth of Candida spp. when samples with multiple fungal species are analyzed. On the other hand, rapamycin can prevent contamination during subculturing. Contamination problems are frequent in slow-growing fungi [32].
5. Conclusions
Based on our findings, we propose the use of culture media such as FastFung, supplemented with cycloheximide and antibiotics, including rapamycin, for isolation and culture of Malassezia species from polymicrobial samples. There is no standardization for susceptibility testing of Malassezia, and only a very limited number of strains have been tested, so this result warrants further investigation. In the future, it will be interesting to test this selective medium with clinical specimens of varied origins, including stool.
Author Contributions
A.A., conceptualization; data curation; investigation; formal analysis; methodology; visualization; writing the original draft. S.R., conceptualization; methodology; investigation; formal analysis; resources; supervision; writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Institut Hospitalo-Universitaire (IHU) Méditerranée Infection, the French National Research Agency under the program “Investissements d’avenir”, reference ANR-10-IAHU-03, the Région Provence Alpes Côte d’Azur and European funding FEDER PRIMI.
Acknowledgments
We thank Dirk Stubbe for graciously providing us with the Malassezia strains from the fungal collection BCCM/IHEM, Sciensano, Brussels, Belgium.
Conflicts of Interest
The authors declare no conflict of interests.
References
- Lorch, J.M.; Palmer, J.M.; Vanderwolf, K.J.; Schmidt, K.Z.; Verant, M.L.; Weller, T.J.; Blehert, D.S. Malassezia vespertilionis sp. nov.: A new cold-tolerant species of yeast isolated from bats. Pers. Mol. Phylogeny Evol. Fungi 2018, 41, 56–70. [Google Scholar] [CrossRef]
- Gaitanis, G.; Magiatis, P.; Hantschke, M.; Bassukas, I.D.; Velegraki, A. The Malassezia Genus in Skin and Systemic Diseases. Clin. Microbiol. Rev. 2012, 25, 106–141. [Google Scholar] [CrossRef]
- Chryssanthou, E.; Broberger, U.; Petrini, B. Malassezia pachydermatis fungaemia in a neonatal intensive care unit. Acta Paediatr. 2007, 90, 323–327. [Google Scholar] [CrossRef]
- Al-Sweih, N.; Ahmad, S.; Joseph, L.; Khan, S.; Khan, Z. Malassezia pachydermatis fungemia in a preterm neonate resistant to fluconazole and flucytosine. Med. Mycol. Case Rep. 2014, 5, 9–11. [Google Scholar] [CrossRef]
- Iatta, R.; Cafarchia, C.; Cuna, T.; Montagna, O.; Laforgia, N.; Gentile, O.; Rizzo, A.; Boekhout, T.; Otranto, D.; Montagna, M.T. Bloodstream infections by Malassezia and Candida species in critical care patients. Med. Mycol. 2014, 52, 264–269. [Google Scholar] [CrossRef]
- Rhimi, W.; Theelen, B.; Boekhout, T.; Otranto, D.; Cafarchia, C. Malassezia spp. Yeasts of Emerging Concern in Fungemia. Front. Cell. Infect. Microbiol. 2020, 10, 370. [Google Scholar] [CrossRef]
- Celakovska, J.; Vankova, R.; Bukac, J.; Cermakova, E.; Andrys, C.; Krejsek, J. Atopic Dermatitis and Sensitisation to Molecular Components of Alternaria, Cladosporium, Penicillium, Aspergillus, and Malassezia—Results of Allergy Explorer ALEX 2. J. Fungi 2021, 7, 183. [Google Scholar] [CrossRef]
- Gomez-Moyano, E.; Crespo-Erchiga, V.; Martínez-Pilar, L.; Diaz, D.G.; Martínez-García, S.; Navarro, M.L.; Casaño, A.V. Do Malassezia species play a role in exacerbation of scalp psoriasis? J. Mycol. Méd. 2014, 24, 87–92. [Google Scholar] [CrossRef]
- Chua, K.B.; Chua, I.L.; Chua, I.E.; Chong, K.H.; Chua, K.H. A modified mycological medium for isolation and culture of Malassezia furfur. Malays. J. Pathol. 2005, 27, 99–105. [Google Scholar]
- Kaneko, T.; Makimura, K.; Onozaki, M.; Ueda, K.; Yamada, Y.; Nishiyama, Y.; Yamaguchi, H. Vital growth factors of Malassezia species on modified CHROMagar Candida. Med. Mycol. 2005, 43, 699–704. [Google Scholar] [CrossRef]
- Kaneko, T.; Makimura, K.; Sugita, T.; Yamaguchi, H. Tween 40-based precipitate production observed on modified chromogenic agar and development of biological identification kit for Malassezia species. Med. Mycol. 2006, 44, 227–231. [Google Scholar] [CrossRef]
- Aridogan, I.A.; Ilkit, M.; Izol, V.; Ates, A.; Demirhindi, H. Glans penis and prepuce colonisation of yeast fungi in a paediatric population: Pre- and postcircumcision results. Mycoses 2009, 52, 49–52. [Google Scholar] [CrossRef]
- Hamad, I.; Ranque, S.; Azhar, E.; Yasir, M.; Jiman-Fatani, A.A.; Tissot-Dupont, H.; Raoult, D.; Bittar, F. Culturomics and Amplicon-based Metagenomic Approaches for the Study of Fungal Population in Human Gut Microbiota. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Iskit, S.; Ilkit, M.; Turaç-Biçer, A.; Demirhindi, H.; Türker, M. Effect of circumcision on genital colonization of Malassezia spp. in a pediatric population. Med. Mycol. 2006, 44, 113–117. [Google Scholar] [CrossRef]
- Mayser, P.; Schütz, M.; Schuppe, H.-C.; Jung, A.; Schill, W.-B. Frequency and spectrum of Malassezia yeasts in the area of the prepuce and glans penis. BJU Int. 2001, 88, 554–558. [Google Scholar] [CrossRef]
- Singh, S.; Banerjee, T.; Tilak, R.; Chaudhary, R. Prevalence of different Malassezia species in pityriasis versicolor in central India. Indian J. Dermatol. Venereol. Leprol. 2010, 76, 159–164. [Google Scholar] [CrossRef]
- Talaee, R.; Katiraee, F.; Ghaderi, M.; Erami, M.; Alavi, A.K.; Nazeri, M. Molecular Identification and Prevalence of Malassezia Species in Pityriasis Versicolor Patients from Kashan, Iran. Jundishapur J. Microbiol. 2014, 7, e11561. [Google Scholar] [CrossRef]
- Zeinali, E.; Sadeghi, G.; Yazdinia, F.; Shams-Ghahfarokhi, M.; Razzaghi-Abyaneh, M. Clinical and epidemiological features of the genus Malassezia in Iran. Iran. J. Microbiol. 2014, 6, 354–360. [Google Scholar]
- Zhao, Y.; Li, L.; Wang, J.-J.; Kang, K.F.; Zhang, Q.Q. Cutaneous malasseziasis: Four case reports of atypical dermatitis and onychomycosis caused by Malassezia. Int. J. Dermatol. 2010, 49, 141–145. [Google Scholar] [CrossRef]
- Cleland, E.J.; Bassiouni, A.; Boase, S.; Dowd, S.; Vreugde, S.; Wormald, P.-J. The fungal microbiome in chronic rhinosinusitis: Richness, diversity, postoperative changes and patient outcomes. Int. Forum Allergy Rhinol. 2014, 4, 259–265. [Google Scholar] [CrossRef]
- Hamad, I.; Abdallah, R.A.; Ravaux, I.; Mokhtari, S.; Tissot-Dupont, H.; Michelle, C.; Stein, A.; Lagier, J.-C.; Raoult, D.; Bittar, F. Metabarcoding analysis of eukaryotic microbiota in the gut of HIV-infected patients. PLoS ONE 2018, 13, e0191913. [Google Scholar] [CrossRef]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef]
- Willger, S.D.; Grim, S.L.; Dolben, E.L.; Shipunova, A.; Hampton, T.H.; Morrison, H.G.; Filkins, L.M.; O’Toole, G.A.; Moulton, L.A.; Ashare, A.; et al. Characterization and quantification of the fungal microbiome in serial samples from individuals with cystic fibrosis. Microbiome 2014, 2, 40. [Google Scholar] [CrossRef]
- Van Woerden, H.C.; Gregory, C.; Brown, R.; Marchesi, J.R.; Hoogendoorn, B.; Matthews, I.P. Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: A community based case control study. BMC Infect. Dis. 2013, 13, 69. [Google Scholar] [CrossRef]
- Vézina, C.; Kudelski, A.; Sehgal, S.N. Rapamycin (AY-22,989), A new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. 1975, 28, 721–726. [Google Scholar] [CrossRef]
- Thomson, A.W.; Starzl, T.E. New Immunosuppressive Drugs: Mechanistic Insights and Potential Therapeutic Advances. Immunol. Rev. 1993, 136, 71–97. [Google Scholar] [CrossRef]
- Cruz, M.C.; Goldstein, A.L.; Blankenship, J.; Del Poeta, M.; Perfect, J.R.; McCusker, J.H.; Bennani, Y.L.; Cardenas, M.E.; Heitman, J. Rapamycin and Less Immunosuppressive Analogs Are Toxic to Candida albicans and Cryptococcus neoformans via FKBP12-Dependent Inhibition of TOR. Antimicrob. Agents Chemother. 2001, 45, 3162–3170. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.K.; Griffith, S.; Kojima, I.; Demain, A.L. Antifungal activities of rapamycin and its derivatives, prolylrapamycin, 32-desmethylrapamycin, and 32-desmethoxyrapamycin. J. Antibiot. 1998, 51, 487–491. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ianiri, G.; Clancey, S.A.; Lee, S.C.; Heitman, J. FKBP12-Dependent Inhibition of Calcineurin Mediates Immunosuppressive Antifungal Drug Action in Malassezia. mBio 2017, 8, e01752-17. [Google Scholar] [CrossRef] [PubMed]
- Bittar, F.; Gouriet, F.; Khelaifia, S.; Raoult, D.; Ranque, S. FastFung: A novel medium for the culture and isolation of fastidious fungal species from clinical samples. J. Microbiol. Methods 2020, 180, 106108. [Google Scholar] [CrossRef] [PubMed]
- Abdillah, A.; Khelaifia, S.; Raoult, D.; Bittar, F.; Ranque, S. Comparison of Three Skin Sampling Methods and Two Media for Culturing Malassezia Yeast. J. Fungi 2020, 6, 350. [Google Scholar] [CrossRef] [PubMed]
- Gümral, R.; Döğen, A.; Ilkit, M.M. Comparison of the contamination rates of culture media used for isolation and identification of dermatophytes. Turk. J. Med. Sci. 2015, 45, 587–592. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).