Insights on Lulworthiales Inhabiting the Mediterranean Sea and Description of Three Novel Species of the Genus Paralulworthia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Isolates
2.2. Morphological Analysis
2.3. DNA Extraction, PCR Amplification, and Data Assembling
2.4. Sequence Alignment and Phylogenetic Analysis
3. Results
3.1. Phylogenetic Inference
3.2. Taxonomy
3.2.1. Paralulworthia mediterranea sp. nov. A. Poli, E. Bovio, G.C. Varese and V. Prigione
- MYCOBANK: MB841118
- Type: Italy, Tuscany, the Mediterranean Sea, Elba Island (Livorno), Ghiaie, 3–5 m depth, 42°49′04″ N, 10°19′20″ E, from Posidonia oceanica roots, March 2010, R. Mussat-Sartor and N. Nurra, MUT 5417 holotype, living culture permanently preserved in metabolically inactive state by deep-freezing at MUT.
- Additional material examined: Italy, Tuscany, the Mediterranean Sea, Elba Island (Livorno), Ghiaie, 3–5 m depth, 42°49′04″ N, 10°19′20″ E from Posidonia oceanica rhizomes, March 2010, R. Mussat-Sartor and N. Nurra, MUT 654. Italy, Tuscany, the Mediterranean Sea, Elba Island (Livorno), Ghiaie, 3–5 m depth, 42°49′04″ N, 10°19′20″ E from Posidonia oceanica rhizomes, March 2010, R. Mussat-Sartor and N. Nurra, MUT 5080.
- Etymology: In reference to the Mediterranean Sea.
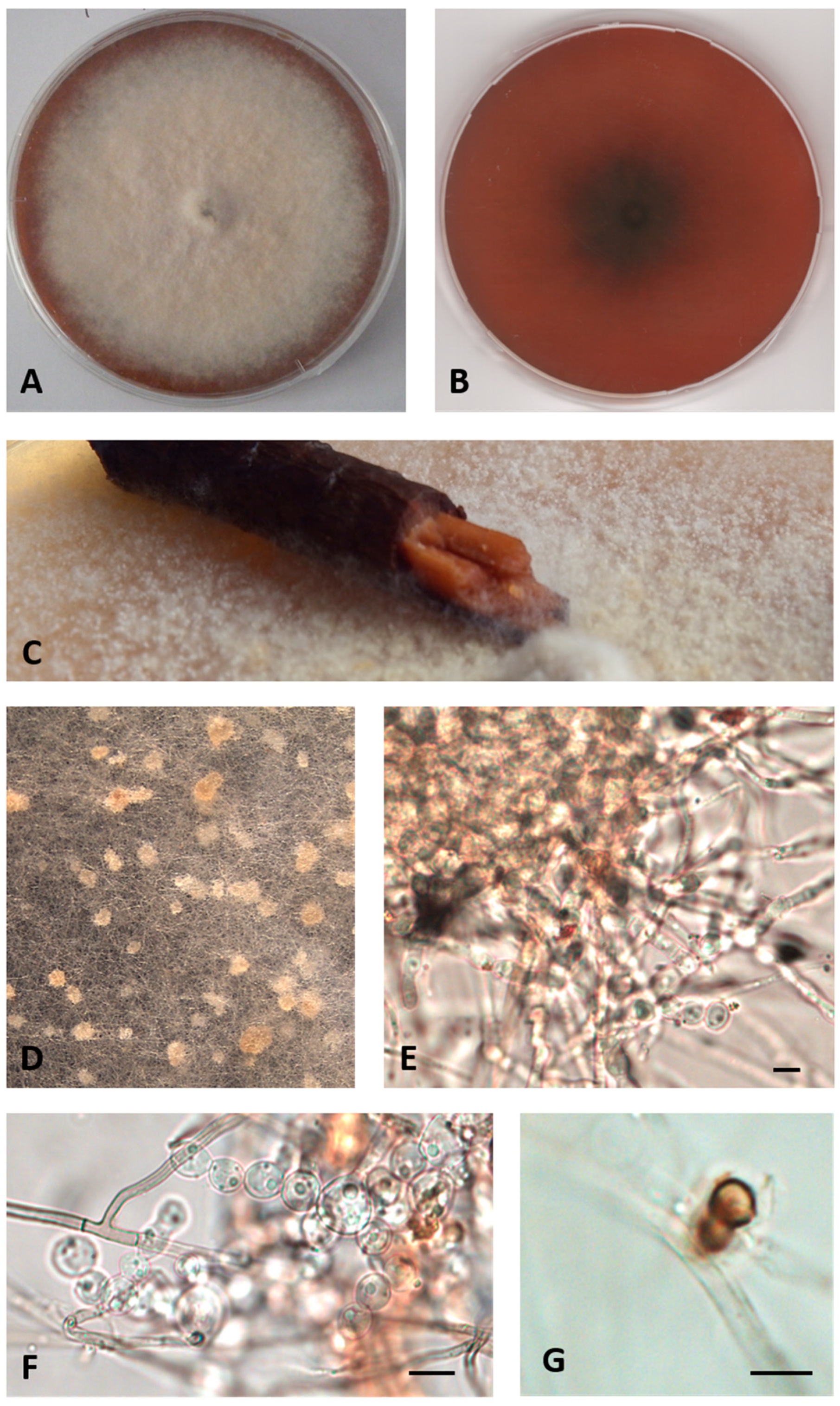
- Description: Growing actively on Pinus pinaster wood and Quercus ruber cork, more markedly on the first. Hyphae 2.4–4 μm wide, septate, from hyaline to dematiaceous. Chlamydospores light brown 4–5 × 5–6 μm, unicellular or two-celled often present. Bulbils on the colony surface single or in group, pale yellow or cream colored, becoming ochre with age, nearly spherical, 150–400 μm diameter, formed by swollen cells (10–15 μm diameter) (Figure 2).
- Sexual morph not observed. Asexual morph with differentiated conidiogenesis not observed.
- Colony description: Colony growing on MEASW, reaching 57–70 mm diameter after 14 days at 21 °C, mycelium feltrose, becoming granular with age due to the presence of bulbils, with irregular edges, beige, sometimes with greyish shades at the edges; reverse from amber to dark orange. A yellowish brown colored diffusible pigment was often present (Figure 2).
3.2.2. Paralulworthia candida sp. nov. A. Poli, E. Bovio, V. Prigione and G.C. Varese
- MYCOBANK: 841116
- Type: Italy, Tuscany, the Mediterranean Sea, Elba Island (Livorno), Ghiaie, 3–5 m depth, 42°49′04″ N, 10°19′20″ E, from Posidonia oceanica roots, March 2010, R. Mussat-Sartor and N. Nurra, MUT 5430 holotype, living culture permanently preserved in metabolically inactive state by deep-freezing at MUT.
- Etymology: In reference to the colony color.
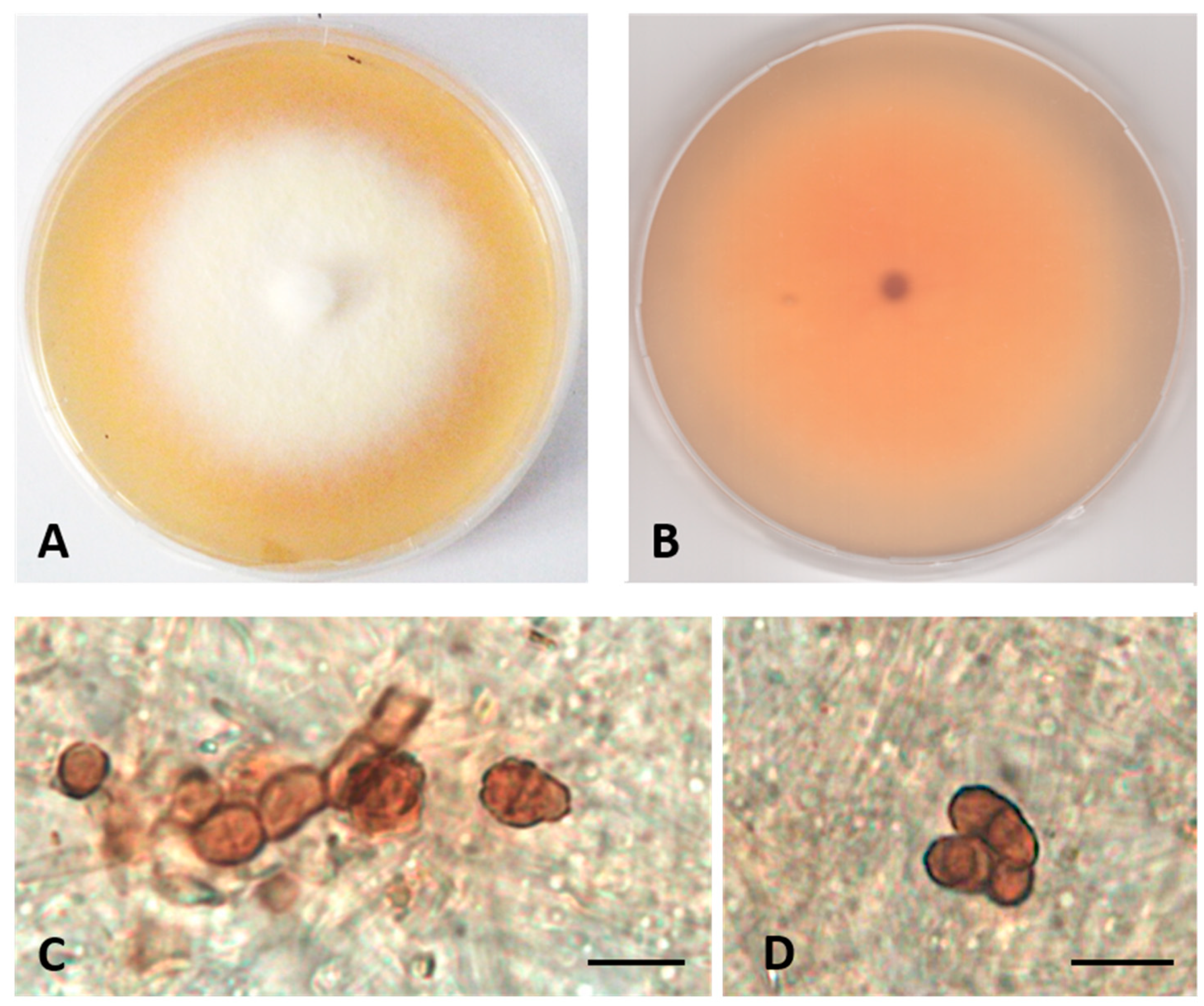
- Description: Poor colonization of Pinus pinaster wood and Quercus ruber cork. Hyphae 2.2–4.2 μm wide, septate, hyaline. Chlamydospores abundant, brown, globose, or subglobose, from unicellular (5–7 × 5–8 μm) to eight-cellular (8–13 μm diameter), in the shape of a sarcina (Figure 3).
- Sexual morph not observed. Asexual morph with differentiated conidiogenesis not observed.
- Colony description. Growing on MEASW, reaching 27–32 mm diameter after 14 days at 21 °C, mycelium floccose, white with yellowish shades in the center, submerged edges giving a beige halo to the colony; reverse light orange. A pinkish colored diffusible pigment present (Figure 3).
3.2.3. Paralulworthia elbensis sp. nov. A. Poli, E. Bovio, V. Prigione and G.C. Varese
- MYCOBANK: MB841117
- Type: Italy, Tuscany, the Mediterranean Sea, Elba Island (Livorno), Margidore,14–15 m depth, 42°45′29″ N, 10°18′24″ E, from Posidonia oceanica roots, March 2010, R. Mussat-Sartor and N. Nurra, MUT 5422 holotype, living culture permanently preserved in metabolically inactive state by deep-freezing at MUT.
- Additional material examined: Italy, Tuscany, the Mediterranean Sea, Elba Island (Livorno), Ghiaie, 3–5 m depth, 42°49′04″ N, 10°19′20″ E, from Posidonia oceanica roots, March 2010, R. Mussat-Sartor and N. Nurra, MUT 377. Italy, Tuscany, the Mediterranean Sea, Elba Island (Livorno), Margidore,14–15 m depth, 42°45′29″ N, 10°18′24″ E, from Posidonia oceanica roots, March 2010, R. Mussat-Sartor and N. Nurra, MUT 5438. Italy, Tuscany, the Mediterranean Sea, Elba Island (Livorno), Margidore, 14–15 m depth, 42°45′29″ N, 10°18′24″ E, from Posidonia oceanica roots, March 2010, R. Mussat-Sartor and N. Nurra, MUT 5461.
- Etymology: In reference to the location of isolation.
- Description: Poor colonization of Pinus pinaster wood and Quercus ruber cork. Hyphae 2.6–4.5 μm wide, septate, hyaline. Chlamydospores abundant, brown, single or in chains, from globose to ellipsoidal, unicellular (5–7 × 6–7 μm) or multicellular (8–11 × 9–12 μm diameter) (Figure 4).
- Sexual morph not observed. Asexual morph with differentiated conidiogenesis not observed.
- Colony description. Growing on MEASW, reaching 35–37 mm diameter after 14 days at 21 °C, mycelium feltrose, white with yellowish shades, submerged edges; reverse light orange (Figure 4).
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding

Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, E.B.G.; Pang, K.-L.; Abdel-Wahab, M.A.; Scholz, B.; Hyde, K.D.; Boekhout, T.; Ebel, R.; Rateb, M.E.; Henderson, L.; Sakayaroj, J.; et al. An online resource for marine fungi. Fungal Divers. 2019, 96, 347–433. [Google Scholar] [CrossRef]
- Jones, E.B.G.; Suetrong, S.; Sakayaroj, J.; Bahkali, A.H.; Abdel-Wahab, M.A.; Boekhout, T.; Pang, K.L. Classification of marine Ascomycota, Basidiomycota, Blastocladiomycota and Chytridiomycota. Fungal Divers. 2015, 73, 1–72. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Bhat, J.D.; Dayarathne, M.C.; Huang, S.K.; Norphanphoun, C.; Senanayake, I.C.; Perera, R.H.; et al. Families of Sordariomycetes. Fungal Divers. 2016, 79, 1–317. [Google Scholar] [CrossRef]
- Jones, E.G.; Pang, K.-L. (Eds.) Marine Fungi and Fungal-like Organisms; Walter de Gruyter: Berlin, Germany, 2012. [Google Scholar]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Huang, S.K.; Abdel-Wahab, M.A.; Daranagama, D.A.; Dayarathne, M.; D’Souza, M.J.; Goonasekara, I.D.; et al. Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 2015, 72, 199–301. [Google Scholar] [CrossRef]
- Kohlmeyer, J.; Spatafora, J.W.; Volkmann-Kohlmeyer, B. Lulworthiales, a new order of marine Ascomycota. Mycologia 2000, 92, 453–458. [Google Scholar] [CrossRef]
- Campbell, J.; Volkmann-Kohlmeyer, B.; Grafenhan, T.; Spatafora, J.W.; Kohlmeyer, J. A re-evaluation of Lulworthiales: Relationships based on 18S and 28S rDNA. Mycol. Res. 2005, 109, 556–568. [Google Scholar] [CrossRef] [Green Version]
- Poli, A.; Bovio, E.; Ranieri, L.; Varese, G.C.; Prigione, V. Fungal Diversity in the Neptune Forest: Comparison of the Mycobiota of Posidonia oceanica, Flabellia petiolata, and Padina pavonica. Front. Microbiol. 2020, 11, 933. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, M.F.M.; Abreu, A.C.; Hilario, S.; Alves, A. Diversity of marine fungi associated with wood baits in the estuary Ria de Aveiro, with descriptions of Paralulworthia halima, comb. nov., Remispora submersa, sp. nov., and Zalerion pseudomaritima, sp. nov. Mycologia 2021, 113, 664–683. [Google Scholar] [CrossRef]
- Raghukumar, S. Fungi in Coastal and Oceanic Marine Ecosystems: Marine Fungi; Springer: Cham, Switzerland, 2017. [Google Scholar]
- Campbell, J.; Inderbitzin, P.; Kohlmeyer, J.; Volkmann-Kohlmeyer, B. Koralionastetales, a new order of marine Ascomycota in the Sordariomycetes. Mycol. Res. 2009, 113, 373–380. [Google Scholar] [CrossRef]
- Panno, L.; Bruno, M.; Voyron, S.; Anastasi, A.; Gnavi, G.; Miserere, L.; Varese, G.C. Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia oceanica. New Biotechnol. 2013, 30, 685–694. [Google Scholar] [CrossRef]
- Garzoli, L.; Gnavi, G.; Tamma, F.; Tosi, S.; Varese, G.C.; Picco, A.M. Sink or swim: Updated knowledge on marine fungi associated with wood substrates in the Mediterranean Sea and hints about their potential to remediate hydrocarbons. Prog. Oceanogr. 2015, 137, 140–148. [Google Scholar] [CrossRef]
- Abdel-Wahab, M.A.; Hodhod, M.S.; Bahkali, A.H.A.; Jones, E.B.G. Marine fungi of Saudi Arabia. Bot. Mar. 2014, 57, 323–335. [Google Scholar] [CrossRef]
- Dayarathne, M.C.; Wanasinghe, D.N.; Devadatha, B.; Abeywickrama, P.; Jones, E.B.G.; Chomnunti, P.; Sarma, V.V.; Hyde, K.D.; Lumyong, S.; McKenzie, E.H.C. Modern taxonomic approaches to identifying diatrypaceous fungi from marine habitats, with a novel genus Halocryptovalsa Dayarathne & KD Hyde, gen. nov. Cryptogam. Mycol. 2020, 41, 21–67. [Google Scholar] [CrossRef]
- Poli, A.; Bovio, E.; Ranieri, L.; Varese, G.C.; Prigione, V. News from the Sea: A New Genus and Seven New Species in the Pleosporalean Families Roussoellaceae and Thyridariaceae. Diversity 2020, 12, 144. [Google Scholar] [CrossRef] [Green Version]
- Bovio, E.; Gnavi, G.; Prigione, V.; Spina, F.; Denaro, R.; Yakimov, M.; Calogero, R.; Crisafi, F.; Varese, G.C. The culturable mycobiota of a Mediterranean marine site after an oil spill: Isolation, identification and potential application in bioremediation. Sci. Total Environ. 2017, 576, 310–318. [Google Scholar] [CrossRef]
- Panebianco, C.; Tam, W.Y.; Jones, E.B.G. The effect of pre-inoculation of balsa wood by selected marine fungi and their effect on subsequent colonisation in the sea. Fungal Divers. 2002, 10, 77–88. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: London, UK, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matheny, P.B.; Wang, Z.; Binder, M.; Curtis, J.M.; Lim, Y.W.; Nilsson, R.H.; Hughes, K.W.; Hofstetter, V.; Ammirati, J.F.; Schoch, C.L.; et al. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol. Phylogenet. Evol. 2007, 43, 430–451. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raja, H.A.; Baker, T.R.; Little, J.G.; Oberlies, N.H. DNA barcoding for identification of consumer-relevant mushrooms: A partial solution for product certification? Food Chem. 2017, 214, 383–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Wahab, M.A.; Pang, K.L.; Nagahama, T.; Abdel-Aziz, F.A.; Jones, E.B.G. Phylogenetic evaluation of anamorphic species of Cirrenalia and Cumulospora with the description of eight new genera and four new species. Mycol. Prog. 2010, 9, 537–558. [Google Scholar] [CrossRef]
- Azevedo, E.; Barata, M.; Marques, M.I.; Caeiro, M.F. Lulworthia atlantica: A new species supported by molecular phylogeny and morphological analysis. Mycologia 2017, 109, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Poli, A.; Bovio, E.; Perugini, I.; Varese, G.C.; Prigione, V. Corollospora mediterranea: A Novel Species Complex in the Mediterranean Sea. Appl. Sci. 2021, 11, 5452. [Google Scholar] [CrossRef]
- Poli, A.; Vizzini, A.; Prigione, V.; Varese, G.C. Basidiomycota isolated from the Mediterranean Sea—Phylogeny and putative ecological roles. Fungal Ecol. 2018, 36, 51–62. [Google Scholar] [CrossRef]
- Garzoli, L.; Poli, A.; Prigione, V.; Gnavi, G.; Varese, G.C. Peacock’s tail with a fungal cocktail: First assessment of the mycobiota associated with the brown alga Padina pavonica. Fungal Ecol. 2018, 35, 87–97. [Google Scholar] [CrossRef]
- Gnavi, G.; Garzoli, L.; Poli, A.; Prigione, V.; Burgaud, G.; Varese, G.C. The culturable mycobiota of Flabellia petiolata: First survey of marine fungi associated to a Mediterranean green alga. PLoS ONE 2017, 12, e0175941. [Google Scholar] [CrossRef] [PubMed]
- Zuccaro, A.; Schulz, B.; Mitchell, J.A. Molecular detection of ascomycetes associated with Fucus serratus. Mycol. Res. 2003, 107, 1451–1466. [Google Scholar] [CrossRef] [PubMed]
- Zuccaro, A.; Schoch, C.L.; Spatafora, J.W.; Kohlmeyer, J.; Draeger, S.; Mitchell, J.A. Detection and identification of fungi intimately associated with the brown seaweed Fucus serratus. Appl. Environ. Microbiol. 2008, 74, 931–941. [Google Scholar] [CrossRef] [Green Version]
- Bennett, R.J.; Turgeon, B.G. Fungal Sex: The Ascomycota. Microbiol. Spectr. 2016, 4, 4–5. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sorensen, J.L.; Hansen, F.; Arvas, M.; Syed, M.F.; Hassan, L.; Benz, J.P.; Record, E.; Henrissat, B.; Poggeler, S.; et al. Genome sequencing and analyses of two marine fungi from the North Sea unraveled a plethora of novel biosynthetic gene clusters. Sci. Rep. 2018, 8, 10187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, S.H.; Kim, H.K.; Lee, T.; Turgeon, B.G. Self-fertility in Chromocrea spinulosa is a consequence of direct repeat-mediated loss of MAT1-2, subsequent imbalance of nuclei differing in mating type, and recognition between unlike nuclei in a common cytoplasm. Plos Genet. 2017, 13, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, D.; Lane, F.A.; Steenkamp, E.T.; Wingfield, B.D.; Wilken, P.M. Unidirectional mating-type switching confers self-fertility to Thielaviopsis cerberus, the only homothallic species in the genus. Fungal Biol. 2021, 125, 427–434. [Google Scholar] [CrossRef]
- Chethana, K.W.T.; Jayawardena, R.S.; Hyde, K.D. Hurdles in fungal taxonomy: Effectiveness of recent methods in discriminating taxa. Megataxa 2020, 1, 114–122. [Google Scholar]
- Jeewon, R.; Hyde, K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 2016, 7, 1669–1677. [Google Scholar] [CrossRef]
- Paco, A.; Duarte, K.; da Costa, J.P.; Santos, P.S.M.; Pereira, R.; Pereira, M.E.; Freitas, A.C.; Duarte, A.C.; Rocha-Santos, T.A.P. Biodegradation of polyethylene microplastics by the marine fungus Zalerion maritimum. Sci. Total Environ. 2017, 586, 10–15. [Google Scholar] [CrossRef]


| Species | Strain | Source | nrITS | nrSSU | nrLSU |
|---|---|---|---|---|---|
| Lulworthiales | |||||
| Lulworthiaceae | |||||
| Cumulospora marina Schmidt | MF46 | Submerged wood | – | GU252136 | GU252135 |
| GC53 | Submerged wood | – | GU256625 | GU256626 | |
| C. varia Chatmala and Somrithipol | GR78 | Submerged wood | – | EU848593 | EU848578 |
| IT 152 | Submerged wood | EU848579 | EU848579 | ||
| Halazoon mehlae Abdel-Aziz, Abdel-Wahab and Nagah. | MF819 T | Drift stems of Phragmites australis | – | GU252144 | GU252143 |
| H. fuscus (Schmidt) Abdel-Wahab, Pang, Nagah., Abdel-Aziz and Jones | NBRC 105256 | Driftwood | – | GU252148 | GU252147 |
| Hydea pigmaea (Kohlm) Pang and Jones | NBRC 33069 | Driftwood | – | GU252134 | GU252133 |
| IT081 | Driftwood | – | GU256632 | GU256633 | |
| Kohlmeyeriella crassa (Nakagiri) Kohlm., Volkm.–Kohlm., Campb., Spatafora and Gräfenhan | NBRC 32133 T | Sea foam | LC146741 | AY879005 | LC146742 |
| K. tubulata (Kohlm.) Jones, Johnson and Moss | PP115 | Marine environment | – | AY878998 | AF491265 |
| PP0989 | Marine environment | – | AY878997 | AF491264 | |
| Lindra marinera Meyers | JK 5091 | Marine environment | – | AY879000 | AY878958 |
| L. obtusa Nakagiri and Tubaki | NRBC 31317 T | Sea foam | LC146744 | AY879002 | AY878960 |
| AFTOL 5012 | Marine environment | – | FJ176847 | FJ176902 | |
| CBS 113030 | n.d. | AY879001 | AY878959 | ||
| L. thalassiae Orpurt, Meyers, Boral and Simms | JK 5090A | Marine environment | – | U46874 | U46891 |
| AFTOL 413 | Marine environment | DQ491508 | DQ470994 | DQ470947 | |
| JK 5090 | Marine environment | – | AF195634 | AF195635 | |
| JK 4322 | Thalassia testudinum leaves | – | AF195632 | AF195633 | |
| Lulwoana uniseptata (Nakagiri) Kohlmeyer et al. | NBRC 32137 T | Submerged wood | LC146746 | LC146746 | LC146746 |
| CBS 16760 | Driftwood | – | AY879034 | AY878991 | |
| Zalerion maritima (Linder) Anastasiou | FCUL280207CP1 | Sea water | KT347216 | KT347203 | JN886806 |
| FCUL010407SP2 | Sea water | KT347217 | KT347204 | JN886805 | |
| Lulworthia atlantica Azevedo, Caeiro and Barata | FCUL210208SP4 | Sea water | KT347205 | KT347193 | JN886843 |
| FCUL190407CF4 | Sea water | KT347207 | KT347198 | JN886816 | |
| FCUL061107CP3 | Sea water | KT347208 | KT347196 | JN886825 | |
| L. fucicola Sutherl. | ATCC 64288 T | Intertidal wood | – | AY879007 | AY878965 |
| PP1249 | Marine environment | – | AY879008 | AY878966 | |
| L. grandispora Meyers | AFTOL 424 | Dead Rhizophora sp. branch | – | DQ522855 | DQ522856 |
| NTOU3841 | Driftwood | – | KY026044 | KY026048 | |
| NTOU3847 | Decayed mangrove wood | – | KY026046 | KY026049 | |
| NTOU3849 | Decayed mangrove wood | – | KY026047 | KY026050 | |
| Lulworthia lignoarenaria (Koch and Jones) Kohlm., Volkm.–Kohlm., Campb., Spatafora and Gräfenhan | AFTOL 5013 | Marine environment | – | FJ176848 | FJ176903 |
| L. medusa (Ellis and Everh.) Cribb and Cribb | JK 5581 T | Spartina | – | AF195636 | AF195637 |
| L. opaca (Linder) Cribb and J.W. Cribb | CBS 218.60 | Driftwood in seawater | – | AY879003 | AY87896 |
| L. cf. purpurea (Wilson) Johnson | FCUL170907CP5 | Sea water | KT347219 | KT347201 | JN886824 |
| FCUL280207CF9 | Sea water | KT347218 | KT347202 | JN886808 | |
| Matsusporium tropicale (Kohlm.) Jones and Pang | NBRC 32499 | Submerged wood | – | GU252142 | GU252141 |
| Moleospora maritima Abdel-Wahab, Abdel-Aziz and Nagah. | MF 836 T | Drift stems of Phragmites australis | – | GU252138 | GU252137 |
| Paralulworthia candida sp. nov. | MUT 5430 | P. oceanica | MZ357724 | MZ357767 | MZ357746 |
| Paralulworthia elbensis sp. nov. | MUT 377 | P. oceanica | MZ357710 | MZ357753 | MZ357732 |
| MUT 5422 | P. oceanica | MZ357723 | MZ357766 | MZ357745 | |
| MUT 5438 | P. oceanica | MZ357712 | MZ357755 | MZ357734 | |
| MUT 5461 | P. oceanica | MZ357725 | MZ357768 | MZ357747 | |
| Paralulworthia gigaspora Prigione, Poli, Bovio and Varese | MUT 435 T | P. oceanica | MN649242 | MN649246 | MN649250 |
| MUT 5413 | P. oceanica | MN649243 | MN649247 | MN649251 | |
| MUT 263 | Oil-contaminated sea water | MZ357729 | MZ357772 | MZ357751 | |
| MUT 465 | P. oceanica | MZ357726 | MZ357769 | MZ357748 | |
| MUT 1753 | Oil-contaminated sea water | MZ357730 | MZ357773 | MZ357752 | |
| MUT 5085 | P. oceanica | MZ357715 | MZ357758 | MZ357737 | |
| MUT 5086 | P. oceanica | MZ357716 | MZ357759 | MZ357738 | |
| MUT 5093 | P. oceanica | MZ357718 | MZ357761 | MZ357740 | |
| MUT 5094 | P. oceanica | MZ357719 | MZ357762 | MZ357741 | |
| Paralulworthia halima (Anastasiou) Gonçalves, Abreu and Alves | CMG 68 | Submerged wood | MT235736 | MT235712 | MT235753 |
| CMG 69 | Submerged wood | MT235737 | MT235713 | MT235754 | |
| MUT 1483 | Submerged wood | MZ357727 | MZ357770 | MZ357749 | |
| MUT 2919 | Submerged wood | MZ357713 | MZ357756 | MZ357735 | |
| MUT 3347 | Submerged wood | MZ357728 | MZ357771 | MZ357750 | |
| Paralulworthia posidoniae Poli, Prigione, Bovio and Varese | MUT 5261 T | P. oceanica | MN649245 | MN649249 | MN649253 |
| MUT 5092 | P. oceanica | MZ357717 | MZ357760 | MZ357739 | |
| MUT 5110 | P. oceanica | MZ357720 | MZ357763 | MZ357742 | |
| MUT 5419 | P. oceanica | MZ357722 | MZ35776 | MZ357744 | |
| Paralulworthia mediterranea sp. nov. | MUT 654 | P. oceanica | MZ357711 | MZ357754 | MZ357733 |
| MUT 5080 | P. oceanica | MZ357714 | MZ357757 | MZ357736 | |
| MUT 5417 T | P. oceanica | MZ357721 | MZ357764 | MZ357743 | |
| Pisorisporiales | |||||
| Pisorisporiaceae | |||||
| Achroceratosphaeria potamia Réblová, Fourn. and Hyde | JF 08139 T | Submerged wood of Platanus sp. | – | GQ996541 | GQ996538 |
| Pleosporales | |||||
| Melanommataceae | |||||
| Bimuria novae-zelandiae Hawksw., Chea and Sheridan | CBS 107.79 T | Soil | MH861181 | FJ190605 | MH872950 |
| Pleosporaceae | |||||
| Setosphaeria monoceras Alcorn | CBS 154.26 | n.d. | DQ337380 | DQ238603 | AY016368 |
| Dydimosphaeriaceae | |||||
| Letendraea helminthicola (Berk. and Broome) Weese ex Petch | CBS 884.85 | Yerba mate | MK404145 | AY016345 | AY016362 |
| Forward and Reverse Primers | Thermocycler Conditions | References | |
|---|---|---|---|
| ITS | ITS1–ITS4 | 95 °C: 5 min (95 °C: 40 s, 55 °C: 50 s, 72 °C: 50 s) × 35 cycles; 72 °C: 8 min; 4 °C: ∞ | [19] |
| LSU | LR0R–LR7 | 95 °C: 5 min (95 °C: 1 min, 50 °C: 1 min, 72 °C: 2 min) × 35 cycles; 72 °C: 10 min; 4 °C: ∞ | [20] |
| SSU | NS1–NS4 | 95 °C: 5 min (95 °C: 1 min, 50 °C: 1 min, 72 °C: 2 min) × 35 cycles; 72 °C: 10 min; 4 °C: ∞ | [19] |
| TEF-1α | EF-dF/EF-2218R | 95 °C: 5 min (95 °C: 1 min, 50 °C: 1 min; 72 °C: 2 min) × 40 cycles, 72 °C: 10 min; 4 °C: ∞ | [21] |
| βTUB | Bt2a–Bt2b | 94 °C: 4 min (94 °C: 35 s, 58 °C: 35 s, 72 °C: 50 s) × 35 cycles; 72 °C: 5 min; 4 °C: ∞ | [22] |
| RPB1 | RPB1Af–RPB1Cr | 96 °C: 5 min (94 °C: 30 s, 52 °C: 30 s, 72 °C: 1 min) × 40 cycles; 72 °C: 8 min; 4 °C: ∞ | [23] |
| RPB2 | fRPB2-5F/fPB2-7cR | 94 °C: 3 min (94 °C: 30 s; 55 °C: 30 s; 72 °C: 1 min) × 40 cycles, 72 °C: 10 min; 4 °C: ∞ | [24] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poli, A.; Prigione, V.; Bovio, E.; Perugini, I.; Varese, G.C. Insights on Lulworthiales Inhabiting the Mediterranean Sea and Description of Three Novel Species of the Genus Paralulworthia. J. Fungi 2021, 7, 940. https://doi.org/10.3390/jof7110940
Poli A, Prigione V, Bovio E, Perugini I, Varese GC. Insights on Lulworthiales Inhabiting the Mediterranean Sea and Description of Three Novel Species of the Genus Paralulworthia. Journal of Fungi. 2021; 7(11):940. https://doi.org/10.3390/jof7110940
Chicago/Turabian StylePoli, Anna, Valeria Prigione, Elena Bovio, Iolanda Perugini, and Giovanna Cristina Varese. 2021. "Insights on Lulworthiales Inhabiting the Mediterranean Sea and Description of Three Novel Species of the Genus Paralulworthia" Journal of Fungi 7, no. 11: 940. https://doi.org/10.3390/jof7110940
APA StylePoli, A., Prigione, V., Bovio, E., Perugini, I., & Varese, G. C. (2021). Insights on Lulworthiales Inhabiting the Mediterranean Sea and Description of Three Novel Species of the Genus Paralulworthia. Journal of Fungi, 7(11), 940. https://doi.org/10.3390/jof7110940









