Spatio-Temporal Profiling of Metarhizium anisopliae—Responsive microRNAs Involved in Modulation of Plutella xylostella Immunity and Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Infection and Small RNA Sequencing
2.2. Bioinformatics Analysis of Small RNA Sequences
2.3. RT-qPCR Validation
2.4. In Vitro Luciferase Validation
2.5. Overexpression and Inhibition Treatment of miRNA In Vivo
2.6. Fungal Bioassay
3. Results
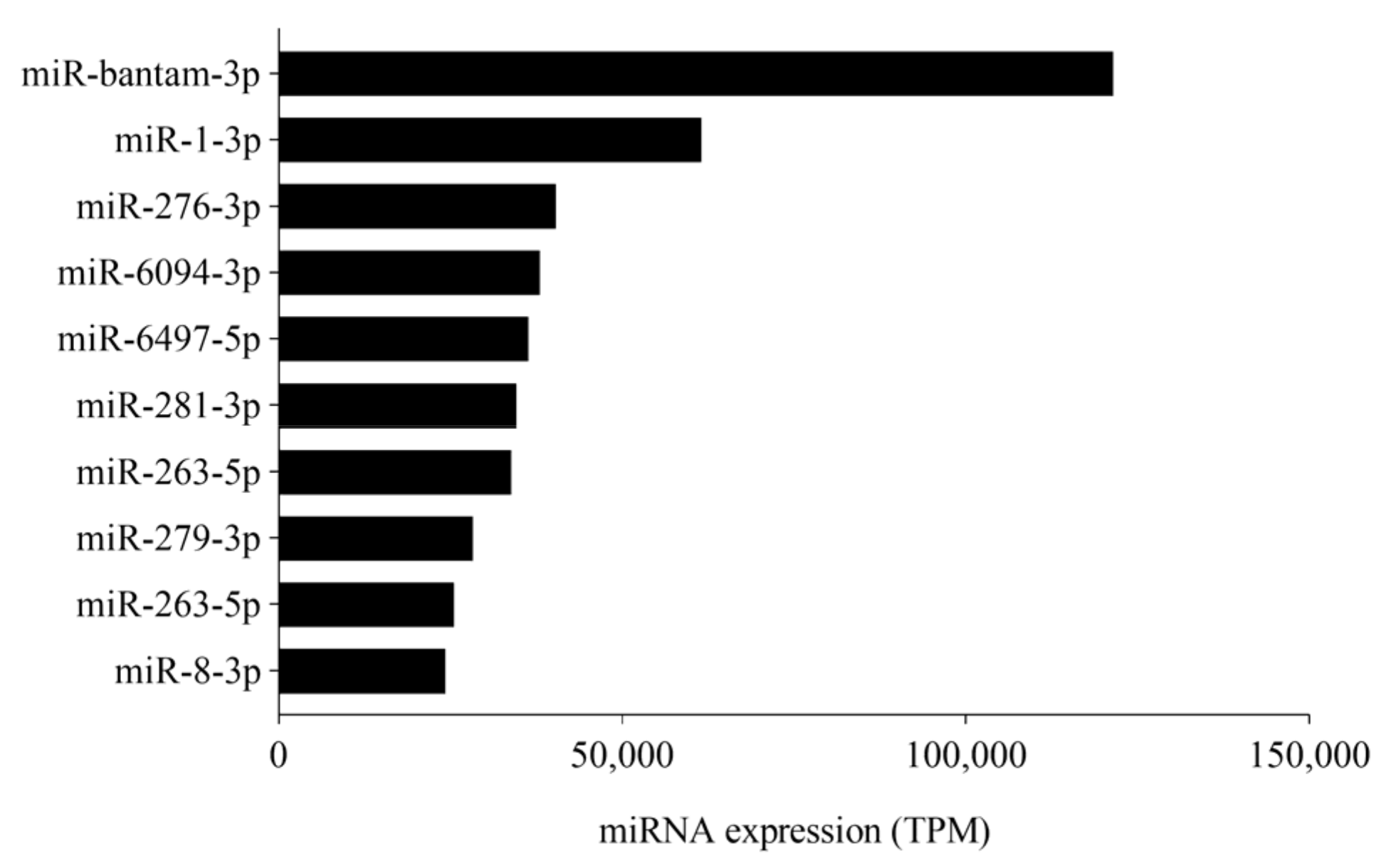
3.1. sRNA Sequencing Analysis
3.2. Identification of M. anisopliae Responsive Known and Novel miRNAs
3.3. Identification of Differentially Expressed miRNAs and Target Predictions
3.4. RT-qPCR Validation of miRNAs and Their Targets
3.5. Prediction and Annotation of miRNA Targets and Functional Classification
3.6. In Vitro Luciferase Validation
3.7. Overexpression and Inhibition Treatment of miRNA In Vivo
3.8. Impact on Development, Fecundity, and Survival
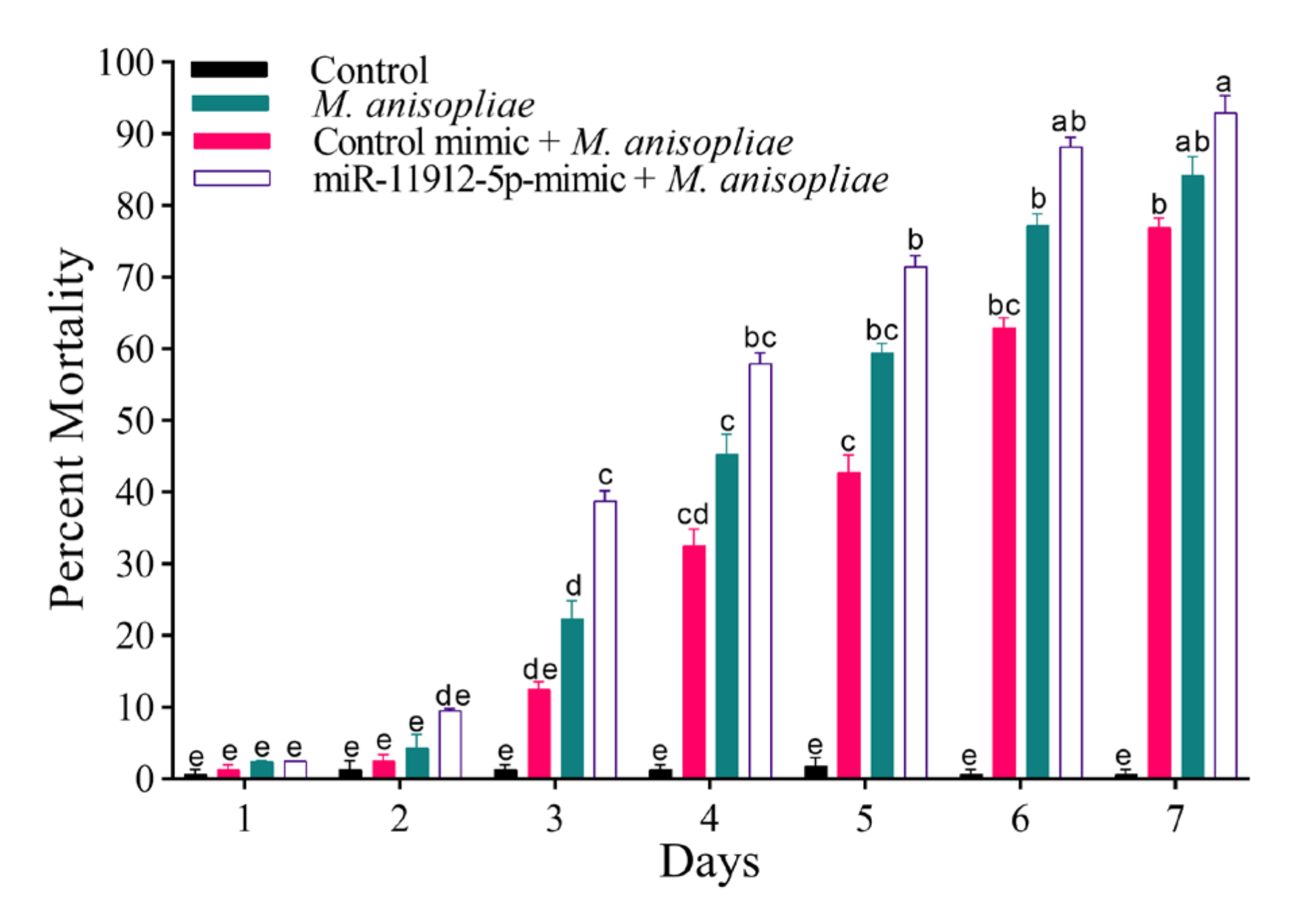
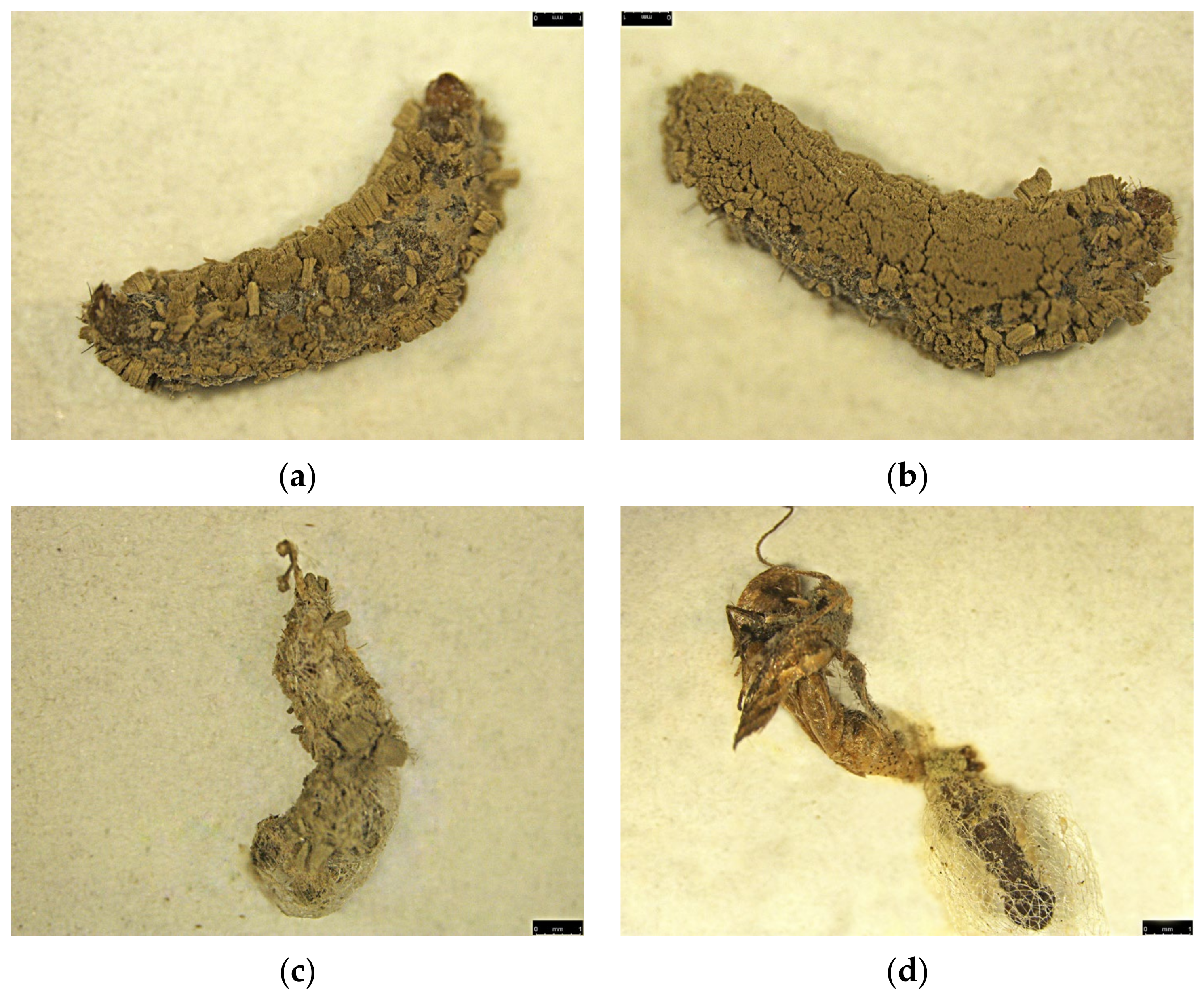
3.9. Fungal Susceptibility
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yusoff, N.; Abd Ghani, I.; Othman, N.W.; Aizat, W.M.; Hassan, M. Toxicity and sublethal effect of farnesyl acetate on diamondback moth, Plutella xylostella (L.) (Lepidoptera: Plutellidae). Insects 2021, 12, 109. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sun, H.; Tian, Z.; Li, Y.; Ye, X.; Li, R.; Li, X.; Zheng, S.; Liu, J.; Zhang, Y. Identification of key residues of carboxylesterase PxEst-6 involved in pyrethroid metabolism in Plutella xylostella (L.). J. Hazard. Mater. 2021, 407, 124612. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, Y. High levels of resistance to chlorantraniliprole evolved in field populations of Plutella xylostella. J. Econ. Entomol. 2012, 105, 1019–1023. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Li, Z.; Li, D.; Wang, R.; Zhang, S.; You, H.; Li, J. Biochemical mechanisms, cross-resistance and stability of resistance to metaflumizone in Plutella xylostella. Insects 2020, 11, 311. [Google Scholar] [CrossRef]
- Liu, Z.; Fu, S.; Ma, X.; Baxter, S.W.; Vasseur, L.; Xiong, L.; Huang, Y.; Yang, G.; You, S.; You, M. Resistance to Bacillus thuringiensis Cry1Ac toxin requires mutations in two Plutella xylostella ATP-binding cassette transporter paralogs. PLoS Pathog. 2020, 16, e1008697. [Google Scholar] [CrossRef]
- Alonso-Díaz, M.Á.; Fernández-Salas, A. Entomopathogenic fungi for tick control in cattle livestock from Mexico. Front. Fungal Biol. 2021, 2, 18. [Google Scholar] [CrossRef]
- Shoukat, R.F.; Zafar, J.; Shakeel, M.; Zhang, Y.; Freed, S.; Xu, X.; Jin, F. Assessment of lethal, sublethal, and transgenerational effects of Beauveria bassiana on the demography of Aedes albopictus (Culicidae: Diptera). Insects 2020, 11, 178. [Google Scholar] [CrossRef] [Green Version]
- Shin, T.Y.; Lee, M.R.; Park, S.E.; Lee, S.J.; Kim, W.J.; Kim, J.S. Pathogenesis-related genes of entomopathogenic fungi. Arch. Insect Biochem. Physiol. 2020, 105, e21747. [Google Scholar] [CrossRef]
- Shoukat, R.F.; Hassan, B.; Shakeel, M.; Zafar, J.; Li, S.; Freed, S.; Xu, X.; Jin, F. Pathogenicity and transgenerational effects of Metarhizium anisopliae on the demographic parameters of Aedes albopictus (Culicidae: Diptera). J. Med. Entomol. 2020, 57, 677–685. [Google Scholar] [CrossRef]
- Zafar, J.; Freed, S.; Khan, B.A.; Farooq, M. Effectiveness of Beauveria bassiana against cotton whitefly, Bemisia tabaci (Gennadius) (Aleyrodidae: Homoptera) on different host plants. Pak. J. Zool. 2016, 48, 91–99. [Google Scholar]
- Shoukat, R.F.; Freed, S.; Kanwar, W.A. Assessment of binary mixtures of entomopathogenic fungi and chemical insecticides on biological parameters of Culex pipiens (Diptera: Culicidae) under laboratory and field conditions. Pak. J. Zool. 2018, 50, 299–309. [Google Scholar] [CrossRef]
- Mwamburi, L.A. Endophytic fungi, Beauveria bassiana and Metarhizium anisopliae, confer control of the fall armyworm, Spodoptera frugiperda (JE Smith) (Lepidoptera: Noctuidae), in two tomato varieties. Egypt. J. Biol Pest. Control. 2021, 31, 1–6. [Google Scholar] [CrossRef]
- Evans, J.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [Green Version]
- Zafar, J.; Shoukat, R.F.; Zhang, Y.; Freed, S.; Xu, X.; Jin, F. Metarhizium anisopliae challenges immunity and demography of Plutella xylostella. Insects 2020, 11, 694. [Google Scholar] [CrossRef]
- O‘Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Lucas, K.J.; Zhao, B.; Liu, S.; Raikhel, A.S. Regulation of physiological processes by microRNAs in insects. Curr. Opin. Insect Sci. 2015, 11, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Ylla, G.; Piulachs, M.-D.; Belles, X. Comparative analysis of miRNA expression during the development of insects of different metamorphosis modes and germ-band types. BMC Genom. 2017, 18, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Guo, L.; Zhou, X.; Gao, X.; Liang, P. miRNAs regulated overexpression of ryanodine receptor is involved in chlorantraniliprole resistance in Plutella xylostella (L.). Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Wu, H.; Liu, H.; Zheng, J.; Lin, Y.; Chen, H. The overexpression of insect endogenous small RNAs in transgenic rice inhibits growth and delays pupation of striped stem borer (Chilo suppressalis). Pest. Manag. Sci. 2017, 73, 1453–1461. [Google Scholar] [CrossRef]
- Etebari, K.; Asgari, S. Conserved microRNA miR-8 blocks activation of the toll pathway by upregulating serpin 27 transcripts. RNA Biol. 2013, 10, 1356–1364. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Wang, Y.; Jiang, F.; Song, T.; Wang, H.; Liu, Q.; Zhang, J.; Zhang, J.; Kang, L. miR-71 and miR-263 jointly regulate target genes chitin synthase and chitinase to control locust molting. PLoS Genet. 2016, 12, e1006257. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Ling, L.; Xu, J.; Zeng, B.; Huang, Y.; Shang, P.; Tan, A. MicroRNA-14 regulates larval development time in Bombyx mori. Insect Biochem. Mol. Biol. 2018, 93, 57–65. [Google Scholar] [CrossRef]
- Zhang, X.; Zheng, Y.; Cao, X.; Ren, R.; Yu, X.-Q.; Jiang, H. Identification and profiling of Manduca sexta microRNAs and their possible roles in regulating specific transcripts in fat body, hemocytes, and midgut. Insect Biochem. Mol. Biol. 2015, 62, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Vilcinskas, A. Evolutionary ecology of parasitic fungi and their host insects. Fungal Ecol. 2019, 38, 12–20. [Google Scholar] [CrossRef]
- Roy, S.; Saha, T.T.; Zou, Z.; Raikhel, A.S. Regulatory pathways controlling female insect reproduction. Annu. Rev. Entomol. 2018, 63, 489–511. [Google Scholar] [CrossRef]
- Lin, J.; Yu, X.-Q.; Wang, Q.; Tao, X.; Li, J.; Zhang, S.; Xia, X.; You, M. Immune responses to Bacillus thuringiensis in the midgut of the diamondback moth, Plutella xylostella. Dev. Comp. Immunol. 2020, 107, 103661. [Google Scholar] [CrossRef]
- Santos, C.G.; Humann, F.C.; Hartfelder, K. Juvenile hormone signaling in insect oogenesis. Curr. Opin. Insect Sci. 2019, 31, 43–48. [Google Scholar] [CrossRef]
- Takeda, M. Structures and functions of insect midgut: The regulatory mechanisms by peptides, proteins and related compounds. In Hemolymph Proteins and Functional Peptides: Recent Advances in Insects and Other Arthropods; Tufail, M., Takeda, M., Eds.; Bentham Science Publishers: Sharjah, United Arab Emirates, 2012; Volume 1, pp. 94–110. [Google Scholar] [CrossRef] [Green Version]
- Biswas, T.; Joop, G.; Rafaluk-Mohr, C. Cross-resistance: A consequence of bi-partite host-parasite coevolution. Insects 2018, 9, 28. [Google Scholar] [CrossRef] [Green Version]
- Batta, Y.A. Efficacy of two species of entomopathogenic fungi against the stored-grain pest, Sitophilus granarius L. (Curculionidae: Coleoptera), via oral ingestion. Egypt. J. Biol. Pest. Control 2018, 28, 44. [Google Scholar] [CrossRef] [Green Version]
- Wei, G.; Lai, Y.; Wang, G.; Chen, H.; Li, F.; Wang, S. Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. Proc. Natl. Acad. Sci. USA 2017, 114, 5994–5999. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, S.A.M.; Taha, A.M.; Salem, H.H.A. Fungal infection causes serious effects on cuticle and midgut of the greater wax moth, Galleria mellonella. Egypt. Acad. J. Biol. Sci. 2019, 12, 1–8. [Google Scholar] [CrossRef]
- Ibrahim, A.A.; Haroun, B.M.; El-Fekky, F.A.; Bekhiet, H.K. Effect of secondary metabolites from three entomopathogenic fungi on the cotton leaf worm, Spodoptera littoralis (Boisd.). Egypt. J. Agric. Res. 2012, 90, 575–587. [Google Scholar] [CrossRef]
- Intodia, A.; Prasad, A.; Veerwal, B. Histopathology of Beauveria bassiana (balsamo) vuillemin, an entomopathogenic fungus, infection in the midgut of termite, Odontotermes obesus (r.) Worker. Int. J. Recent Sci. Res. 2019, 10, 34326–34330. [Google Scholar] [CrossRef]
- Chavan, J.; Gaikwad, Y.; Chougale, A.; Bhawane, G. Curative effect of ethanolic plant extractives against Beauveria bassiana infection in silkworm, Bombyx mori l.: Histopathological observations on midgut. Int. J. Anim. Biol. 2015, 1, 266–272. [Google Scholar]
- Zhou, Q.; Yang, Y.-C.; Shen, C.; He, C.-T.; Yuan, J.-G.; Yang, Z.-Y. Comparative analysis between low-and high-cadmium-accumulating cultivars of Brassica parachinensis to identify difference of cadmium-induced microRNA and their targets. Plant. Soil 2017, 420, 223–237. [Google Scholar] [CrossRef]
- Shakeel, M.; Rodriguez, A.; Tahir, U.B.; Jin, F. Gene expression studies of reference genes for quantitative real-time PCR: An overview in insects. Biotechnol. Lett. 2018, 40, 227–236. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Etebari, K.; Afrad, M.; Tang, B.; Silva, R.; Furlong, M.; Asgari, S. Involvement of microRNA miR-2b-3p in regulation of metabolic resistance to insecticides in Plutella xylostella. Insect Mol. Biol. 2018, 27, 478–491. [Google Scholar] [CrossRef]
- Asgari, S.; Zareie, R.; Zhang, G.; Schmidt, O. Isolation and characterization of a novel venom protein from an endoparasitoid, Cotesia rubecula (Hym: Braconidae). Arch. Insect Biochem. 2003, 53, 92–100. [Google Scholar] [CrossRef]
- Calabrese, J.M.; Seila, A.C.; Yeo, G.W.; Sharp, P.A. RNA sequence analysis defines Dicer‘s role in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 18097–18102. [Google Scholar] [CrossRef] [Green Version]
- Etebari, K.; Asgari, S. Revised annotation of Plutella xylostella microRNAs and their genome-wide target identification. Insect Mol. Biol. 2016, 25, 788–799. [Google Scholar] [CrossRef]
- Shakeel, M.; Xu, X.; Xu, J.; Li, S.; Yu, J.; Zhou, X.; Xu, X.; Hu, Q.; Yu, X.; Jin, F. Genome-wide identification of Destruxin A-responsive immunity-related microRNAs in diamondback moth, Plutella xylostella. Front. Immunol. 2018, 9, 185. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Xu, X.; Li, S.; Wang, S.; Xu, X.; Zhou, X.; Yu, J.; Yu, X.; Shakeel, M.; Jin, F. Genome-wide profiling of Plutella xylostella immunity-related miRNAs after Isaria fumosorosea infection. Front. Physiol. 2017, 8, 1054. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Xu, X.; Zheng, Z.; Zheng, J.; Shakeel, M.; Jin, F. MicroRNA expression profiling of Plutella xylostella after challenge with B. thuringiensis. Dev. Comp. Immunol. 2019, 93, 115–124. [Google Scholar] [CrossRef]
- Hemmati, S.A.; Mehrabadi, M. Structural ensemble-based computational analysis of trypsin enzyme genes discovered highly conserved peptide motifs in insects. Arch. Phytopathol. 2020, 53, 335–354. [Google Scholar] [CrossRef]
- Macedo, M.L.R.; Durigan, R.A.; da Silva, D.S.; Marangoni, S.; Freire, M.d.G.M.; Parra, J.R.P. Adenanthera pavonina trypsin inhibitor retard growth of Anagasta kuehniella (Lepidoptera: Pyralidae). Arch. Insect Biochem. Physiol. 2010, 73, 213–231. [Google Scholar] [CrossRef]
- Farida, B.; Sonia, H.; Hakima, M.-K.; Fatma, B.; Fatma, H. Histological changes in the larvae of the domestic mosquito Culex pipiens treated with the entomopathogenic fungus Beauveria bassiana. Sci. Res. Essays 2018, 13, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Alves, S.; Pereira, R. Microbial control of insects. In Physiological Disorders Caused by Entomopathogens; Publisher of the Foundation for Agrarian Studies Luiz de Queiroz Piracicaba. FEALQ: Sao Dimas, Piracicaba, Brazil, 1998; pp. 39–53. [Google Scholar]
- Toledo, A.V.; Lenicov, A.M.M.d.R.; Lastra, C.C.L. Histopathology caused by the entomopathogenic fungi, Beauveria bassiana and Metarhizium anisopliae, in the adult planthopper, Peregrinus maidis, a maize virus vector. J. Insect Sci. 2010, 10, 35. [Google Scholar] [CrossRef] [Green Version]
- Acuña, S.M.; Floeter-Winter, L.M.; Muxel, S.M. MicroRNAs: Biological regulators in pathogen–host interactions. Cells 2020, 9, 113. [Google Scholar] [CrossRef] [Green Version]
- Boulan, L.; Martín, D.; Milán, M. Bantam miRNA promotes systemic growth by connecting insulin signaling and ecdysone production. Curr. Biol. 2013, 23, 473–478. [Google Scholar] [CrossRef] [Green Version]
- Fullaondo, A.; Lee, S.Y. Identification of putative miRNA involved in Drosophila melanogaster immune response. Dev. Comp. Immunol. 2012, 36, 267–273. [Google Scholar] [CrossRef]
- Etebari, K.; Hussain, M.; Asgari, S. Identification of microRNAs from Plutella xylostella larvae associated with parasitization by Diadegma semiclausum. Insect Biochem. Mol. Biol. 2013, 43, 309–318. [Google Scholar] [CrossRef]
- Zhu, B.; Li, X.; Liu, Y.; Gao, X.; Liang, P. Global identification of microRNAs associated with chlorantraniliprole resistance in diamondback moth Plutella xylostella (L.). Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Asgari, S. MicroRNA functions in insects. Insect Biochem. Mol. Biol. 2013, 43, 388–397. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, Y.; Yan, H.; Li, Y.; Zhang, H.; Xu, J.; Puthiyakunnon, S.; Chen, X. miR-281, an abundant midgut-specific miRNA of the vector mosquito Aedes albopictus enhances dengue virus replication. Parasit. Vectors. 2014, 7, 488. [Google Scholar] [CrossRef]
- Luo, W.; Huang, L.-X.; Qin, S.-K.; Zhang, X.; Feng, Q.-L.; Gu, J.; Huang, L.-H. Multiple microRNAs control ecdysone signaling in the midgut of Spodoptera litura. Insect Sci. 2020, 27, 1208–1223. [Google Scholar] [CrossRef]
- Caygill, E.E.; Johnston, L.A. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr. Biol. 2008, 18, 943–950. [Google Scholar] [CrossRef] [Green Version]
- Garbuzov, A.; Tatar, M. Hormonal regulation of Drosophila microRNA let-7 and miR-125 that target innate immunity. Fly 2010, 4, 306–311. [Google Scholar] [CrossRef] [Green Version]
- Sunkar, R.; Jagadeeswaran, G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. 2008, 8, 37. [Google Scholar] [CrossRef] [Green Version]
- Vilcinskas, A. The role of epigenetics in host–parasite coevolution: Lessons from the model host insects Galleria mellonella and Tribolium castaneum. Zoology 2016, 119, 273–280. [Google Scholar] [CrossRef]
- Skalsky, R.L.; Vanlandingham, D.L.; Scholle, F.; Higgs, S.; Cullen, B.R. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genom. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Huang, Y.; Zhang, Q.; Zhou, H.; Jin, P.; Ma, F. The miR-317 functions as a negative regulator of toll immune response and influences Drosophila survival. Dev. Comp. Immunol. 2019, 95, 19–27. [Google Scholar] [CrossRef]
- Yang, J.; Xu, X.; Lin, S.; Chen, S.; Lin, G.; Song, Q.; Bai, J.; You, M.; Xie, M. Profiling of microRNAs in midguts of Plutella xylostella provides novel insights into the Bacillus thuringiensis resistance. Front. Genet. 2021, 12, 1690. [Google Scholar] [CrossRef] [PubMed]
- Marco, A.; Hooks, K.; Griffiths-Jones, S. Evolution and function of the extended miR-2 microRNA family. RNA Biol. 2012, 9, 242–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozano, J.; Montañez, R.; Belles, X. MiR-2 family regulates insect metamorphosis by controlling the juvenile hormone signaling pathway. Proc. Natl. Acad. Sci. USA 2015, 112, 3740–3745. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Sun, Y.; Xiao, H.; Ge, C.; Li, F.; Han, Z. Multiple miRNAs jointly regulate the biosynthesis of ecdysteroid in the holometabolous insects, Chilo suppressalis. RNA 2017, 23, 1817–1833. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Ge, X.; Li, Z.; Wang, Y.; Song, Q.; Stanley, D.W.; Tan, A.; Huang, Y. MicroRNA-281 regulates the expression of ecdysone receptor (EcR) isoform B in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2013, 43, 692–700. [Google Scholar] [CrossRef]
- Zhu, Y.C.; Liu, X.; Maddur, A.A.; Oppert, B.; Chen, M.-S. Cloning and characterization of chymotrypsin-and trypsin-like cDNAs from the gut of the Hessian fly [Mayetiola destructor (Say)]. Insect Biochem. Mol. Biol. 2005, 35, 23–32. [Google Scholar] [CrossRef]
- Jayachandran, B.; Hussain, M.; Asgari, S. Regulation of Helicoverpa armigera ecdysone receptor by miR-14 and its potential link to baculovirus infection. J. Invertebr. Pathol. 2013, 114, 151–157. [Google Scholar] [CrossRef]
- Zotti, M.; Dos Santos, E.A.; Cagliari, D.; Christiaens, O.; Taning, C.N.T.; Smagghe, G. RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest. Manag. Sci. 2018, 74, 1239–1250. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Huang, Q.X.; Yin, G.H.; Lee, S.; Jia, R.Z.; Liu, Z.X.; Yu, N.T.; Pennerman, K.K.; Chen, X.; Guo, A.P. Identification of microRNAs by small RNA deep sequencing for synthetic microRNA mimics to control Spodoptera exigua. Gene 2015, 557, 215–221. [Google Scholar] [CrossRef]
- Agrawal, A.; Rajamani, V.; Reddy, V.S.; Mukherjee, S.K.; Bhatnagar, R.K. Transgenic plants over-expressing insect-specific microRNA acquire insecticidal activity against Helicoverpa armigera: An alternative to Bt-toxin technology. Transgenic Res. 2015, 24, 791–801. [Google Scholar] [CrossRef]
- Xu, D.; Xue, Q.; McElroy, D.; Mawal, Y.; Hilder, V.A.; Wu, R. Constitutive expression of a cowpea trypsin inhibitor gene, CpTi, in transgenic rice plants confers resistance to two major rice insect pests. Mol. Breed. 1996, 2, 167–173. [Google Scholar] [CrossRef]
- Singh, D.; Kesavan, A.K.; Sohal, S.K. Exploration of anti-insect potential of trypsin inhibitor purified from seeds of Sapindus mukorossi against Bactrocera cucurbitae. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Mukherjee, K.; Vilcinskas, A. The entomopathogenic fungus Metarhizium robertsii communicates with the insect host Galleria mellonella during infection. Virulence 2018, 9, 402–413. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Ren, X.; Liu, Y.; Smagghe, G.; Liang, P.; Gao, X. MiR-189942 regulates fufenozide susceptibility by modulating ecdysone receptor isoform B in Plutella xylostella (L.). Pestic. Biochem. Physiol. 2020, 163, 235–240. [Google Scholar] [CrossRef]
- Zhenghao, W.; Miao, J.; Yanfeng, Z.; Guilin, D.; Guangchun, C.; Xiangqun, N.; Changzhong, L.; Guangjun, W.; Zehua, Z. Effects of different inhibitors on digestive enzyme and detoxification enzyme in midgut of Locusta migratoria manilensis during Metarhizium anisopliae infection. Chin. J. Biol. 2016, 32, 756–761. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zafar, J.; Zhang, Y.; Huang, J.; Freed, S.; Shoukat, R.F.; Xu, X.; Jin, F. Spatio-Temporal Profiling of Metarhizium anisopliae—Responsive microRNAs Involved in Modulation of Plutella xylostella Immunity and Development. J. Fungi 2021, 7, 942. https://doi.org/10.3390/jof7110942
Zafar J, Zhang Y, Huang J, Freed S, Shoukat RF, Xu X, Jin F. Spatio-Temporal Profiling of Metarhizium anisopliae—Responsive microRNAs Involved in Modulation of Plutella xylostella Immunity and Development. Journal of Fungi. 2021; 7(11):942. https://doi.org/10.3390/jof7110942
Chicago/Turabian StyleZafar, Junaid, Yuxin Zhang, Junlin Huang, Shoaib Freed, Rana Fartab Shoukat, Xiaoxia Xu, and Fengliang Jin. 2021. "Spatio-Temporal Profiling of Metarhizium anisopliae—Responsive microRNAs Involved in Modulation of Plutella xylostella Immunity and Development" Journal of Fungi 7, no. 11: 942. https://doi.org/10.3390/jof7110942






