Incidence, Speciation, and Morpho-Genetic Diversity of Penicillium spp. Causing Blue Mold of Stored Pome Fruits in Serbia
Abstract
:1. Introduction
2. Materials and Methods
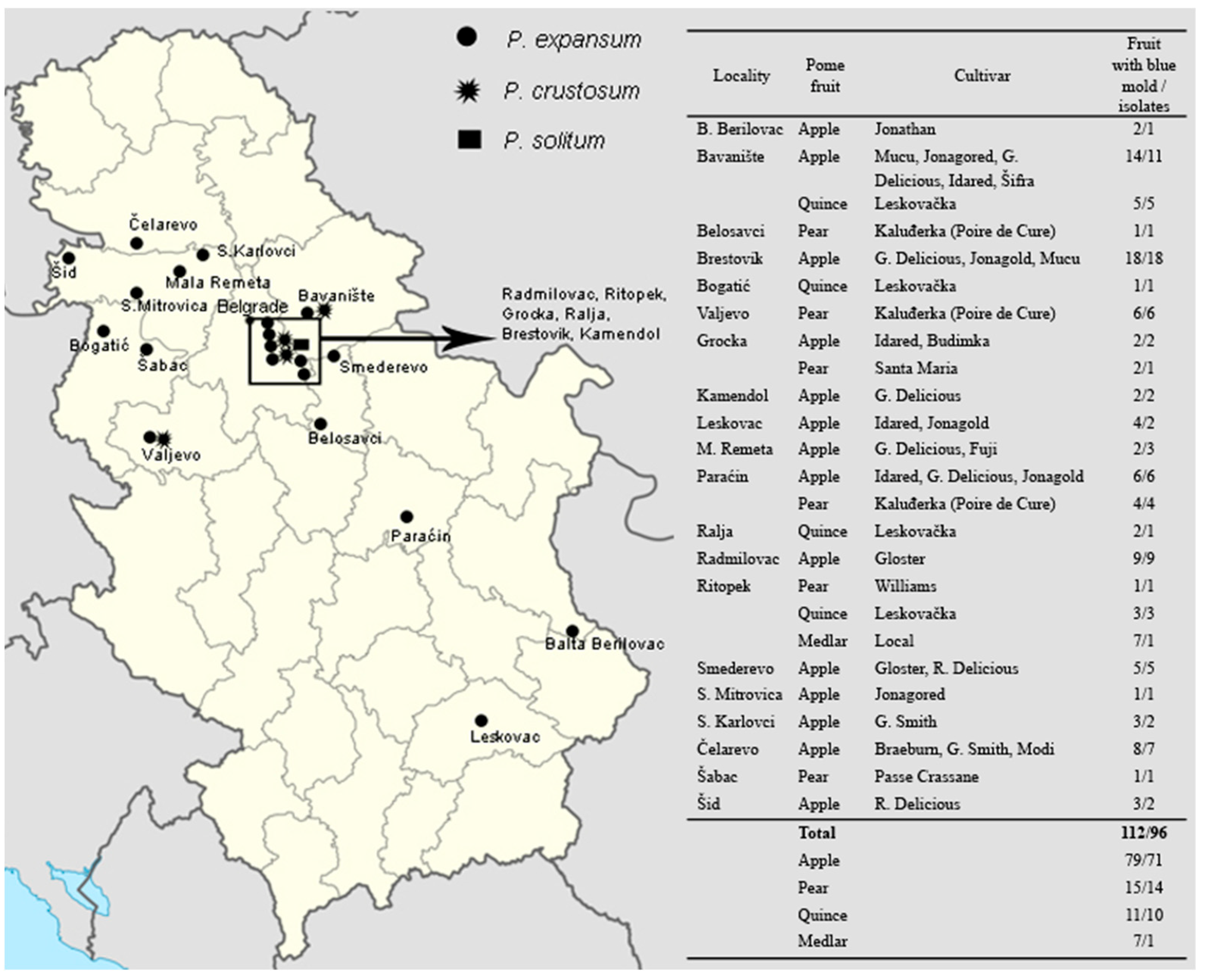
2.1. Sample Collection and Fungal Isolation
2.2. Pathogenicity Test
2.3. DNA Extraction, PCR Amplification, and Amplicon Sequencing
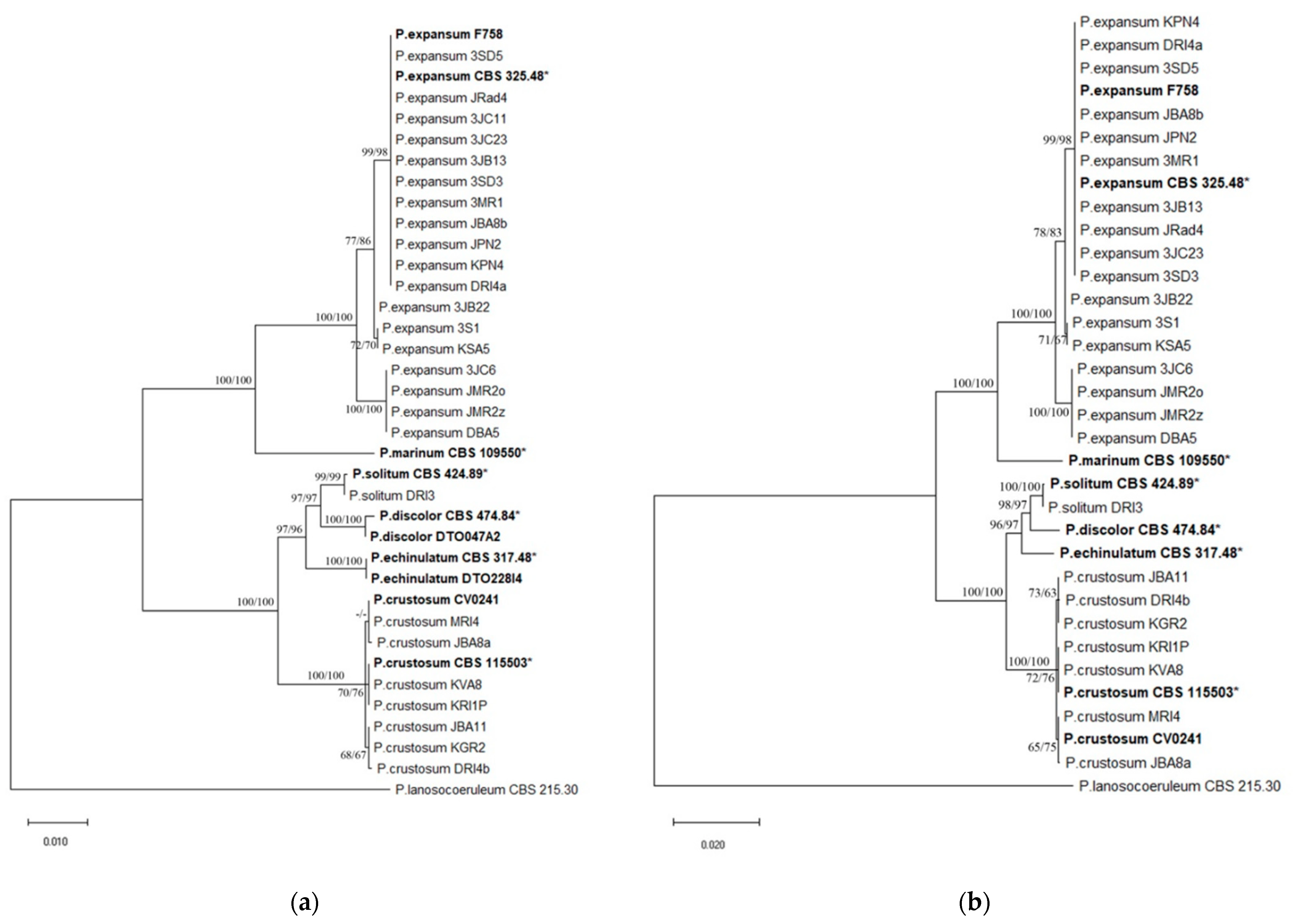
2.4. Multilocus Sequence Analysis and Phylogeny
2.5. Cultural Morphology
2.6. Ehrlich Test
2.7. Virulence Assessment in Apple Fruit
2.8. Statistical Analysis
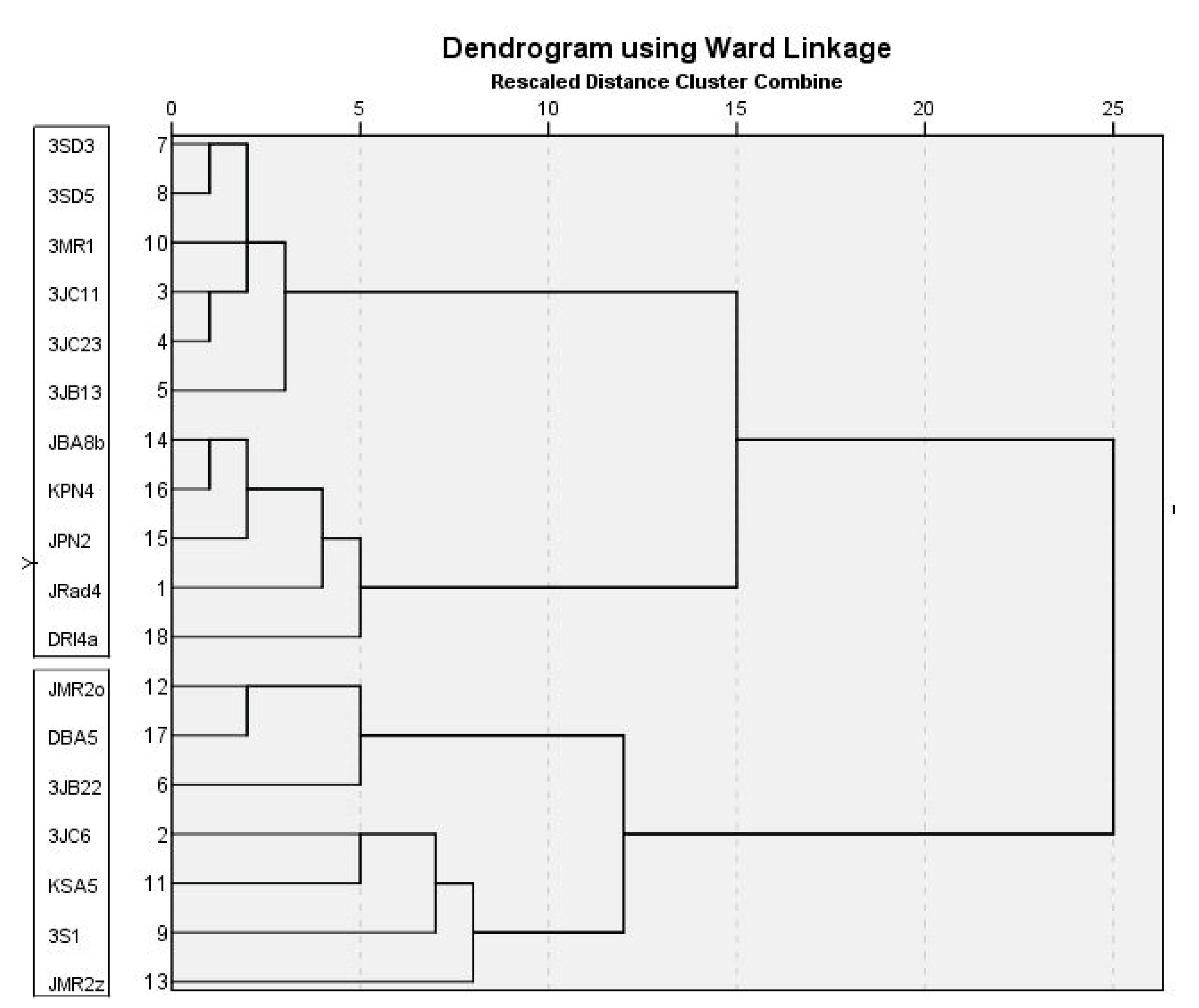
2.9. Cluster Analysis
2.10. Principle Component Analysis
| Isolate | Geographic Origin/Host cv. | Date | GenBank Accession Number | References | |||
|---|---|---|---|---|---|---|---|
| ITS | BenA | CaM | RPB2 | ||||
| Penicillium expansum | |||||||
| JRad4 | Serbia, Radmilovac/Apple ‘Gloster’ | December 2014 | MZ364021 | MZ364047 | MZ364097 | MZ364075 | This study |
| 3JC6 | Serbia, Čelarevo/Apple ‘Braeburn’ | January 2015 | MZ364023 | MZ364056 | MZ364098 | MZ364076 | This study |
| 3JC11 | Serbia, Čelarevo/Apple ’Granny Smith’ | April 2015 | OK432548 | MZ364048 | MZ364099 | MZ364077 | This study |
| 3JC23 | Serbia, Čelarevo/Apple ‘Modi’ | April 2015 | MZ364024 | MZ364049 | MZ364100 | MZ364078 | This study |
| 3JB13 | Serbia, Brestovik/Apple ’Jonagold’ | January 2015. | MZ364025 | MZ364050 | MZ364101 | MZ364079 | This study |
| 3JB22 | Serbia, Brestovik/ Apple ‘G. Delicious’ | January 2015 | MZ364026 | MZ364061 | MZ364102 | MZ364080 | This study |
| 3SD3 | Serbia, Smederevo/Apple ‘R. Delicious’ | January 2015 | MZ364027 | MZ364051 | MZ364103 | MZ364081 | This study |
| 3SD5 | Serbia, Smederevo/Apple ‘R. Delicious’ | January 2015 | MZ364028 | MZ364052 | MZ364110 | MZ364082 | This study |
| 3S1 | Serbia, Šid/Apple ‘R. Delicious’ | January 2015 | MZ364029 | MZ364057 | MZ364104 | MZ364083 | This study |
| 3MR1 | Serbia, Mala Remeta/Apple ‘G. Delicious’ | January 2015 | MZ364030 | MZ364053 | MZ364105 | MZ364084 | This study |
| KSA5 | Serbia, Šabac/Pear ‘Passe Crassane‘ | March 2015 | MZ364031 | MZ364045 | MZ364111 | MZ364071 | This study |
| JMR2o * | Serbia, Mala Remeta/Apple ‘Fuji’ | November 2015 | MZ364033 | MZ364058 | MZ364108 | MZ364085 | This study |
| JMR2z * | Serbia, Mala Remeta/Apple ‘Fuji’ | November 2015 | MZ364034 | MZ364059 | MZ364109 | MZ364086 | This study |
| JBA8b * | Serbia, Bavanište/Apple ‘Jonagored‘ | December 2015 | MZ364035 | MZ364054 | MZ364106 | MZ364087 | This study |
| JPN2 * | Serbia, Paraćin/Apple ‘Idared’ | December 2015 | MZ364036 | MZ364055 | MZ364107 | MZ364088 | This study |
| KPN4 * | Serbia, Paraćin/Pear ‘Poire de Cure‘ | December 2015 | MZ364032 | MZ364046 | MZ364112 | MZ364072 | This study |
| DBA5 * | Serbia, Bavanište/Quince ‘Leskovačka’ | December 2015 | MZ364022 | MZ364062 | MZ364114 | MZ364073 | This study |
| DRI4a * | Serbia, Ritopek/Quince ‘Leskovačka’ | December 2015 | MZ364020 | MZ364060 | MZ364113 | MZ364074 | This study |
| Penicillium crustosum | |||||||
| JBA8a * | Serbia, Bavanište/Apple ‘Jonagored’ | December 2015 | MZ364037 | MZ364067 | MZ389067 | MZ364089 | This study |
| JBA11 * | Serbia, Bavanište/Apple ‘Šifra’ | December 2015 | MZ364038 | MZ364068 | MZ389061 | MZ364090 | This study |
| KGR2 * | Serbia, Grocka/Pear ‘Santa Maria’ | October 2015 | MZ364039 | MZ364063 | MZ389062 | MZ364091 | This study |
| KRI1P * | Serbia, Ritopek/Pear ‘Williams’ | December 2015 | MZ364040 | MZ364064 | MZ389063 | MZ364092 | This study |
| KVA8 * | Serbia, Valjevo/Pear ‘Poire de Cure‘ | December 2015 | MZ364041 | MZ364065 | MZ389064 | MZ364093 | This study |
| DRI4b * | Serbia, Ritopek/Quince ‘Leskovačka’ | December 2015 | MZ364042 | MZ364066 | MZ389065 | MZ364094 | This study |
| MRI4 * | Serbia, Ritopek/Medlar local cv. | December 2015 | MZ364043 | MZ364069 | MZ389066 | MZ364095 | This study |
| Penicillium solitum | |||||||
| DRI3 * | Serbia, Ritopek/Quince ‘Leskovačka’ | December 2015 | MZ364044 | MZ364070 | MZ364115 | MZ364096 | This study |
| NCBI isolate | |||||||
| Penicillium expansum | |||||||
| CBS 325.48 ** | USA/Apple fruit | AY3739121 | AY6744002 | DQ9111342 | JF4174272 | 1[42]; 2[43] | |
| F758 | USA, Idaho/ Sugar beet | 2014 | MG714838 | MG714864 | MG714821 | MG714845 | [44] |
| PCAS | Italy/Chestnut | 2015 | MG821365 | MF100860 | MF100880 | / | [45] |
| P34 | Italy /Withered grape | / | KU554673 | / | / | [46] | |
| 4 | Greece/Kiwi fruit | 2017 | / | MH040784 | / | / | [47] |
| CV 2860 | South Africa/Fynbos biome | / | JX091539 | JX141580 | / | [48] | |
| CV 2861 | South Africa/Fynbos biome | / | JX091540 | JX141581 | / | [48] | |
| LUB | Serbia, Ub/Onion | 2015 | / | KY770971 | / | / | [49] |
| Penicillium marinum | |||||||
| CBS 109550 ** | Japan/Sandy soil | KJ834512 | AY674392 | KU896842 | KU904357 | [42] | |
| CBS 109547 | Tunisia/Sandy soil | / | AY674390 | / | / | [50] | |
| Penicillium crustosum | |||||||
| CBS 115503 ** | Scotland, Aberdeen/Lemon | MH8629851 | AY6743532 | DQ9111323 | MN9691143 | 1[51]; 2[50]; 3[42] | |
| CV 0241 | South Africa/Fynbos biome | JX091403 | JX091536 | JX141576 | MN149972 | [52] | |
| 5A | Italy/Chestnut | 2015 | MG821363 | MF100874 | MF100894 | / | [45] |
| N2AS | Serbia/Nectarine | 2021 | / | MT799805 | / | / | [53] |
| Penicillium echinulatum | |||||||
| CBS 317.48 ** | Canada, Culture contaminant | AF033473 | AY674341 | DQ911133 | KU904352 | [42] | |
| DTO228I4 | Unknown/Unknown | / | MN149925 | MN149944 | MN149964 | [52] | |
| Penicillium solitum | |||||||
| CBS 424.89 ** | Germany/Unknown | AY3739321 | AY6743542 | KU8968511 | KU9043631 | 1[42]; 2[50] | |
| XF | Italy/Chestnut | 2015 | MG821373 | MF100861 | MF100881 | / | [45] |
| Penicillium discolor | |||||||
| CBS 474.84 ** | Israel/Raphanus sativus | AJ004816 | AY674348 | KU896834 | KU904351 | [42] | |
| DTO047A2 | Unknown/Unknown | / | MN149922 | MN149941 | MN149961 | [52] | |
| Penicillium lanosocoeruleum | |||||||
| CBS 215.30 | USA/Culture contaminant | NR_1635411 | KU8968172 | JX9969672 | JX9967123 | 1[51]; 2[42]; 3[54] | |
3. Results
3.1. Blue Mold Symptoms and Koch’s Postulates
3.2. Molecular Identification and Single Nucleotide Polymorphism
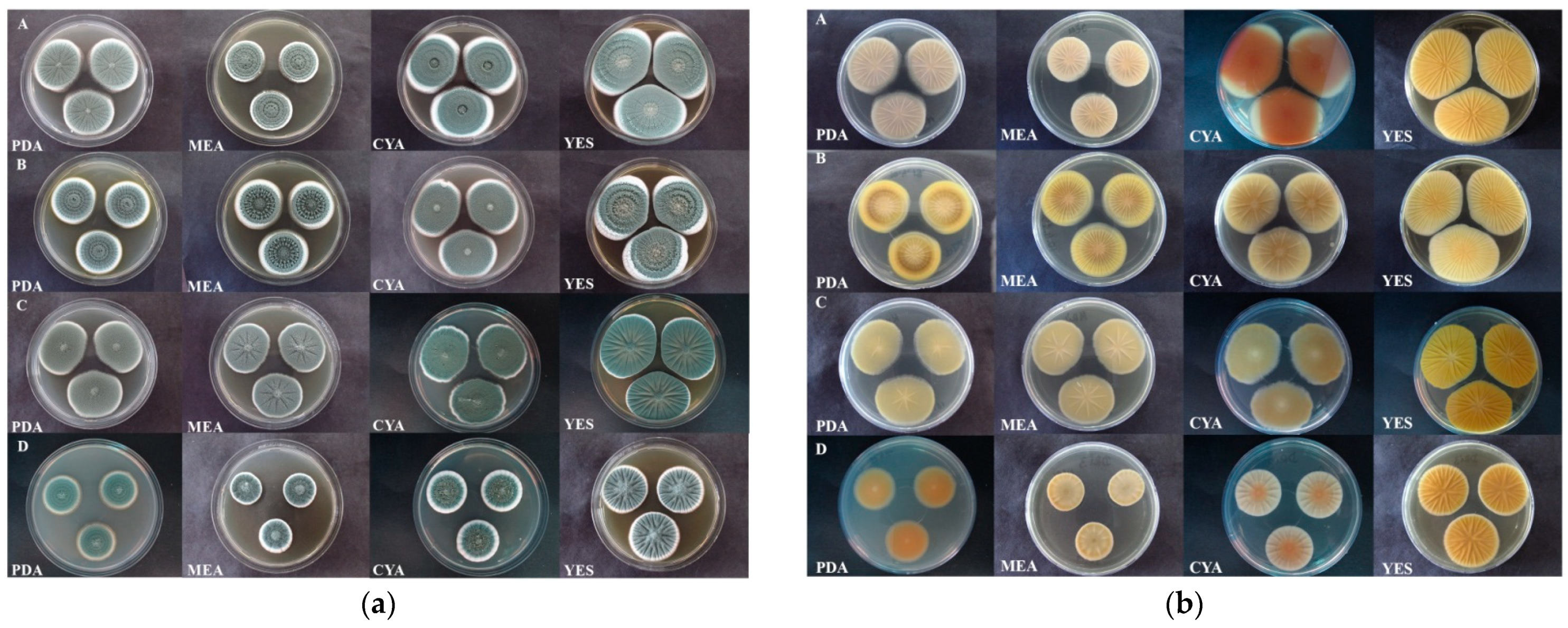
3.3. Macromorphology
3.4. Colony Growth
3.5. Micromorphology
3.6. Ehrlich Test
3.7. Penicillium Multilocus Phylogeny
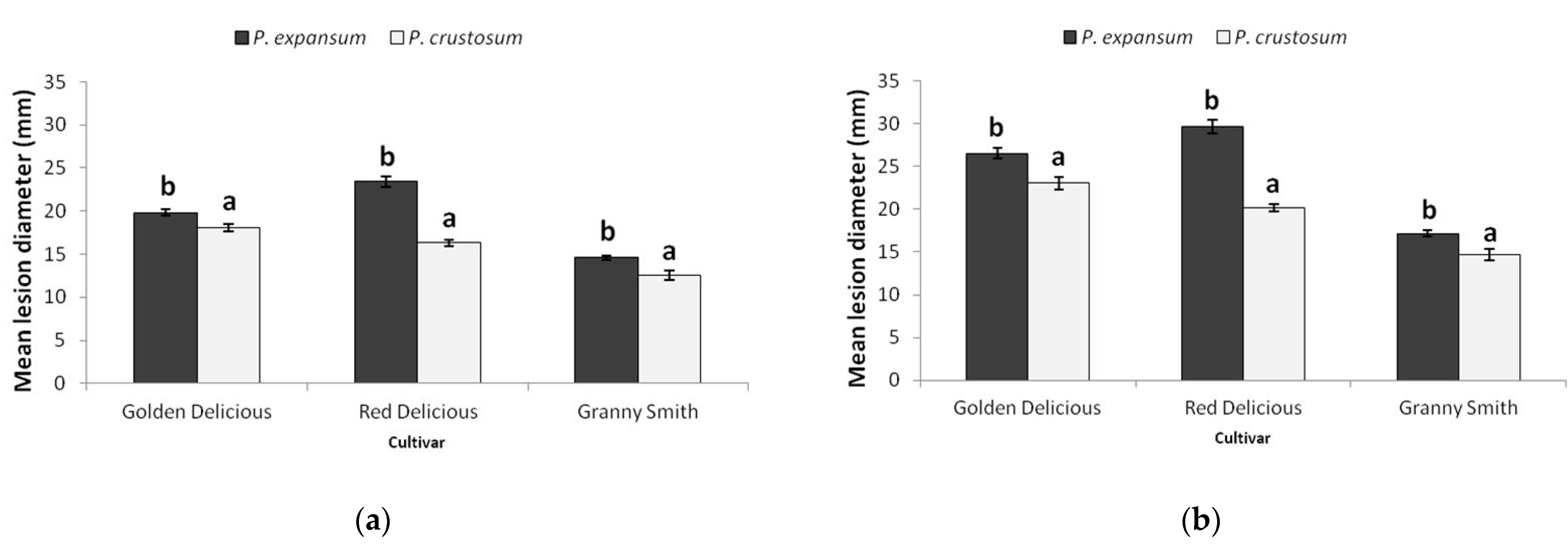
3.8. Virulence Phenotypes in Apple Fruit
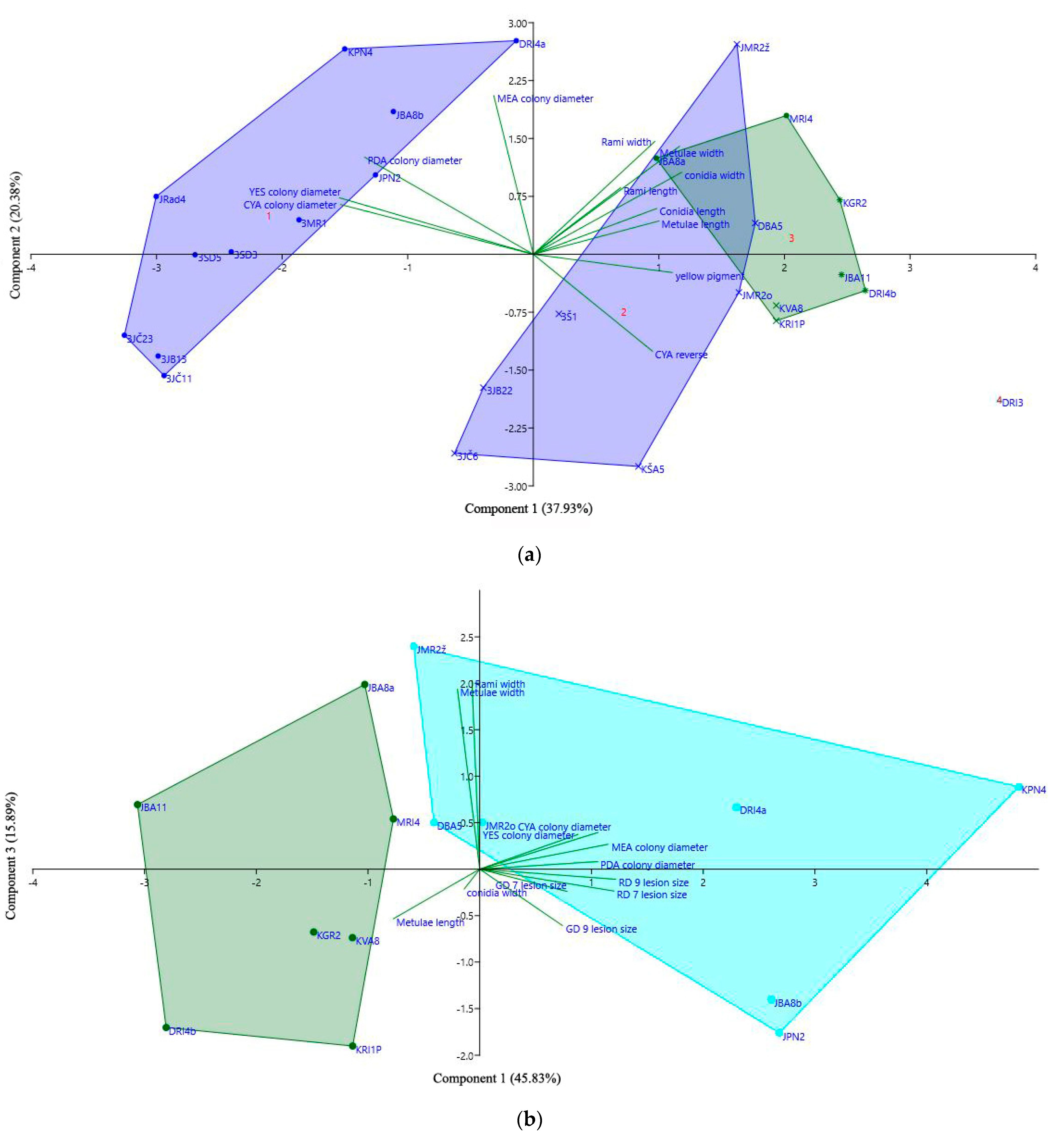
3.9. Principle Component Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Knee, M. Pome fruits. In Biochemistry of Fruit Ripening; Seymour, G.B., Taylor, J.E., Tucker, G.A., Eds.; Springer: Dordrecht, The Netherlands, 1993; p. 325. [Google Scholar]
- Errampalli, D. Penicillium expansum (Blue Mould). In Postharvest Decay, Control Strategies; Bautista-Banos, S., Ed.; Elsevier: London, UK, 2004; pp. 190–191. [Google Scholar]
- Kim, Y.K.; Xiao, C.L. Distribution and incidence of Sphaeropsis rot in apple in Washington State. Plant Dis. 2008, 92, 940–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenberger, D.A.; Engle, C.A.; Meyer, F.W.; Watkins, C.B. Penicillium expansum invades apples through stems during controlled atmosphere storage. Plant Health Prog. 2006, 7, 1. [Google Scholar] [CrossRef]
- Amiri, A.; Bompeix, B. Diversity and population dynamics of Penicillium spp. on apples in pre- and postharvest environments: Consequences for decay development. Plant Pathol. 2005, 54, 74–81. [Google Scholar] [CrossRef]
- Louw, J.P.; Korsten, L. Pathogenic Penicillium spp. on apple and pear. Plant Dis. 2014, 98, 590–598. [Google Scholar] [CrossRef] [Green Version]
- Scholtz, I.; Korsten, L. Profile of Penicillium species in the pear supply chain. Plant Pathol. 2016, 65, 1126–1132. [Google Scholar] [CrossRef] [Green Version]
- Sutton, T.B.; Aldwinckle, A.; Agnello, A.M.; Walgenbach, J.F. Compendium of Apple and Pear Diseases and Pests, 2nd ed.; APS Press: St. Paul, MN, USA, 2014. [Google Scholar]
- Jurick II, W.M.; Cox, K.D. Pre and postharvest fungal apple diseases. In Achieving Sustainable Apple Production; Evans, K., Ed.; Burleigh Dodds Science Publishing: Cambridge, UK, 2016. [Google Scholar]
- Xiao, C.L.; Boal, R.J. Residual activity of fludioxonil and pyrimethanil against Penicillium expansum on apple fruit. Plant Dis. 2009, 93, 1003–1008. [Google Scholar] [CrossRef] [Green Version]
- Rosenberger, D.A. Blue mold. In Compendium of Apple and Pear Diseases; Sutton, T.B., Aldwinkle, H.S., Angello, A.M., Walgenbach, J.F., Eds.; APS Press: St. Paul, MN, USA, 2014; p. 76. [Google Scholar]
- van der Walt, L.; Spotts, R.A.; Visagie, C.M.; Jacobs, K.; Smit, F.J.; McLeod, A. Penicillium species associated with preharvest wet core rot in South Africa and their pathogenicity on apple. Plant Dis. 2010, 94, 666–675. [Google Scholar] [CrossRef] [Green Version]
- Pianzzola, M.J.; Moscatelli, M.; Vero, S. Characterization of Penicillium isolates associated with blue mold on apple in Uruguay. Plant Dis. 2004, 88, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Sanderson, P.G.; Spotts, R.A. Postharvest decay of winter pear and apple fruit caused by species of Penicillium. Phytopathology 1995, 85, 103–110. [Google Scholar] [CrossRef]
- Sholberg, P.L.; Haag, P.D. Incidence of postharvest pathogens of stored apples in British Columbia. Can. J. Plant Pathol. 1996, 18, 81–85. [Google Scholar] [CrossRef]
- Sharma, Y.P.; Sumbali, G. Unrecorded post-harvest fungal rot of quince fruits from India. Natl. Acad. Sci. Lett. 1997, 20, 35–37. [Google Scholar]
- Jurick II, W.M.; II Vico, I.; Gaskins, V.L.; Garrett, W.M.; Whitaker, B.D.; Janisiewicz, W.J.; Conway, W.S. Purification and biochemical characterization of polygalacturonase produced by Penicillium expansum during postharvest decay of ‘Anjou’ pear. Phytopathology 2010, 100, 42–48. [Google Scholar] [CrossRef] [Green Version]
- Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud. Mycol. 2004, 49, 1–174. [Google Scholar]
- Food and Agriculture Organization. FAOstat. World Agriculture Statistics Data Base. 2021. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 24 April 2021).
- Vico, I.; Duduk, N.; Vasić, M.; Nikolić, M. Identification of Penicillium expansum causing postharvest blue mold decay of apple fruit. Pestic. Fitomed. 2014, 29, 257–266. [Google Scholar] [CrossRef]
- Vico, I.; Gaskins, V.; Duduk, N.; Vasić, M.; Yu, J.J.; Peter, K.A.; Jurick II, W.M. First report of Penicillium crustosum causing blue mold on stored apple fruit in Serbia. Plant Dis. 2014, 98, 1430. [Google Scholar] [CrossRef] [PubMed]
- Babović, M.; Perišić, M.; Marković, S.; Stojanović, S.; Pantelić, Z. Investigation on rot of apple fruits in cold storehouse. Zaštita Bilja 1979, 147, 83–87. [Google Scholar]
- Perišić, M.M. Penicillium expansum, the cause of soft rot of apple fruits. Jugosl. Voćarstvo 1972, 21/22, 843–846. [Google Scholar]
- Stošić, S.; Ristić, D.; Savković, Ž.; Ljaljević-Grbić, M.; Vukojević, J.; Živković, S. Penicillium and Talaromyces species as postharvest pathogens of pear fruit (Pyrus communis L.) in Serbia. Plant Dis. 2021. [Google Scholar] [CrossRef]
- Jurick, W.M.; Vico, I.; McEvoy, J.L.; Whitaker, B.D.; Janisiewicz, W.; Conway, W.S. Isolation, purification, and characterization of a polygalacturonase produced in Penicillium solitum-decayed ‘Golden Delicious’ apple fruit. Phytopathology 2009, 99, 636–641. [Google Scholar] [CrossRef] [Green Version]
- Quaglia, M.; Ederli, L.; Pasqualini, S.; Zazzerini, A. Biological control agents and chemical inducers of resistance for postharvest control of Penicillium expansum Link. on apple fruit. Posthar. Biol. Technol. 2011, 59, 307–315. [Google Scholar] [CrossRef]
- Day, J.P.; Shattock, R.C. Aggressiveness and other factors relating to displacement of populations of Phytophthora infestans in England and Wales. Eur. J. Plant Pathol. 1997, 103, 379–391. [Google Scholar] [CrossRef]
- Marek, P.; Annamalai, Y.; Venkitanarayanan, K. Detection of Penicillium expansum by polymerase chain reaction. Int. J. Food Microbiol. 2003, 89, 139–144. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of premier sets designed for use with the PCR to amplify conserved genes from filamentous Ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, S.-B.; Cho, H.-S.; Shin, H.-D.; Frisvad, J.C.; Samson, A. Novel Neosartorya species isolated from soil in Korea. Int. J. Syst. Evol. Microbiol. 2006, 56, 477–486. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.-B.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef] [Green Version]
- Staden, R.; Beal, K.F.; Bonfield, J.K. The Staden package, 1998. Methods Mol. Biol. 2000, 132, 115–130. [Google Scholar] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The Clustal X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGAX: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Lund, F. Differentiating Penicillium species by detection of indole metabolites using a filter paper method. Lett. App. Microbiol. 1995, 20, 228–231. [Google Scholar] [CrossRef]
- Tabachnick, B.G.; Fidell, L.S. Using Multivariate Statistic; Pearson Education: Boston, MA, USA, 2007. [Google Scholar]
- Stevens, J. Applied Multivariate Statistics for the Social Sciences; Lawrence Erlbaum: Mahwah, NJ, USA, 1996. [Google Scholar]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences; Routledge Academic: New York, NY, USA, 1988. [Google Scholar]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.-C.; Meijer, M.; Kraak, B.; Hubka, V.; Samson, R.A.; Frisvad, J.C. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef]
- Houbraken, J.; Wang, L.; Lee, H.B.; Frisvad, J.C. New sections in Penicillium containing novel species producing patulin, pyripyropens or other bioactive compounds. Persoonia 2016, 36, 299–314. [Google Scholar] [CrossRef] [Green Version]
- Strausbaugh, K. Incidence, distribution, and pathogenicity of fungi causing root rot in Idaho long-term sugar beet storage piles. Plant Dis. 2018, 102, 2296–2307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prencipe, S.; Siciliano, I.; Gatti, C.; Garibaldi, A.; Lodovica Gullino, M.; Botta, R.; Spadaro, D. Several species of Penicillium isolated from chestnut flour processing are pathogenic on fresh chestnuts and produce mycotoxins. Food Microbiol. 2018, 76, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Lorenzini, M.; Cappello, M.S.; Logrieco, A.; Zapparol, G. Polymorphism and phylogenetic species delimitation in filamentous fungi from predominant mycobiota in withered grapes. Int. J. Food Microbiol. 2016, 238, 56–62. [Google Scholar] [CrossRef]
- Prodromou, I.; Thomidis, T.; Zambounis, A. First report of Penicillium expansum (Link) Thom. causing postharvest fruit rot of kiwifruit in Northern Greece. Plant Dis. 2018, 102, 1851. [Google Scholar] [CrossRef]
- Visagie, C.M.; Houbraken, J.; Dijksterhuis, J.; Seifert, K.A.; Jacobs, K.; Samson, R.A. A taxonomic review of Penicillium species producing conidiophores with solitary phialides, classified in section Torulomyces. Persoonia 2016, 36, 134–155. [Google Scholar] [CrossRef] [Green Version]
- Duduk, N.; Lazarević, M.; Žebeljan, A.; Vasić, M.; Vico, I. Blue mold decay of stored onion bulbs caused by Penicillium polonicum, P. glabrum and P. expansum. J. Phytopathol. 2017, 165, 662–669. [Google Scholar] [CrossRef]
- Samson, R.A.; Seifert, K.A.; Kuijpers, A.F.A.; Houbraken, J.; Frisvad, J.C. Phylogenetic analysis of Penicillium subgenus Penicillium using partial β-tubulin sequences. Stud. Mycol. 2004, 49, 175–200. [Google Scholar]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Visagie, C.M.; Yilmaz, N.; Vanderwolf, K.; Renaud, J.B.; Sumarah, M.W.; Houbraken, J.; Assebgui, R.; Seifert, K.A.; Malloch, D. Penicillium diversity in Canadian bat caves, including a new species, P. speluncae. Fungal Systemat. Evol. 2020, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Duduk, N.; Bekčić, F.; Žebeljan, A.; Vučković, N.; Vico, I. First report of blue mold caused by Penicillium crustosum on nectarine fruit in Serbia. Plant Dis. 2020, 105, 487. [Google Scholar] [CrossRef]
- Houbraken, J.; Frisvad, J.C.; Seifert, K.A.; Overy, D.P.; Tuthill, D.M.; Valdez, J.G.; Samson, R.A. New penicillin-producing Penicillium species and an overview of section Chrysogena. Persoonia 2012, 29, 78–100. [Google Scholar] [CrossRef] [Green Version]
- Farr, D.F.; Rossman, A.Y. Fungal Databases. U.S. National Fungus Collections, Agricultural Research Service, U.S. Department of Agriculture. Available online: https://nt.ars-grin.gov/fungaldatabases/ (accessed on 19 November 2021).
- Luciano-Rosario, D.; Keller, N.P.; Jurick, W.M. Penicillium spp.: Biology, omics, and management tools for a global postharvest pathogen causing blue mold of pome fruit. Mol. Plant Pathol. 2020, 21, 1391–1404. [Google Scholar] [CrossRef] [PubMed]
- Morales, H.; Marín, S.; Obea, L.; Patiño, B.; Doménech, M.; Ramos, A.J.; Sanchis, V. Ecophysiological characterization of Penicillium expansum population in lleida (Spain). Int. J. Food Microbiol. 2008, 122, 243–252. [Google Scholar] [CrossRef]
- Lichtner, F.J.; Gaskins, V.L.; Cox, K.D.; Jurick, W.M. Global transcriptomic responses orchestrate difenoconazole resistance in Penicillium spp. causing blue mold of stored apple fruit. BMC Genom. 2020, 21, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Žebeljan, A.; Vico, I.; Duduk, N.; Žiberna, B.; Urbanek Krajnc, A. Dynamic changes in common metabolites and antioxidants during Penicillium expansum-apple fruit interactions. Physiol. Mol. Plant Pathol. 2019, 106, 166–174. [Google Scholar] [CrossRef]
- Žebeljan, A.; Vico, I.; Duduk, N.; Žiberna, B.; Urbanek Krajnc, A. Profiling changes in primary metabolites and antioxidants during apple fruit decay caused by Penicillium crustosum. Physiol. Mol. Plant Pathol. 2021, 113, 101586. [Google Scholar] [CrossRef]
- Sun, J.; Janisiewicz, W.J.; Nichols, B.; Jurick, W.M.; Chen, P. Composition of phenolic compounds in wild apple with multiple resistance mechanisms against postharvest blue mold decay. Posthar. Biol. Technol. 2017, 127, 68–75. [Google Scholar] [CrossRef] [Green Version]
- Ahmadi-Afzadi, M.; Nybom, H.; Ekholm, A.; Tahir, I.; Rumpunen, K. Biochemical contents of apple peel and flesh affect level of partial resistance to blue mold. Posthar. Biol. Technol. 2015, 110, 173–182. [Google Scholar] [CrossRef]
- Schovankova, J.; Opatova, H. Changes in phenols composition and activity of phenylalanine-ammonia lyase in apples. Hortic. Sci. 2011, 38, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Čebulj, A.; Cunja, V.; Mikulič-Petkovšek, M.; Veberič, R. Importance of metabolite distribution in apple fruit. Hortic. Sci. 2017, 214, 214–220. [Google Scholar] [CrossRef]
- Lata, B.; Trampczynska, A.; Paczesna, J. Cultivar variation in apple peel and whole fruit phenolic composition. Hortic. Sci. 2009, 121, 176–181. [Google Scholar] [CrossRef]
- Vilanova, L.; Teixido, N.; Torres, R.; Usall, J.; Vinas, I. The infection capacity of P. expansum and P. digitatum on apples and histochemical analysis of host response. Int. J. Food Microbiol. 2012, 157, 360–367. [Google Scholar] [PubMed]
- Buron-Moles, G.; Torres, R.; Teixido, N.; Usall, J.; Vilanova, L.; Vinas, I. Characterisation of H2O2 production to study compatible and non-host pathogen interactions in orange and apple fruit at different maturity stages. Posthar. Biol. Technol. 2015, 99, 27–36. [Google Scholar] [CrossRef]
- Su, J.; Tu, K.; Cheng, L.; Tu, S.; Wang, M.; Xu, H.; Zhan, G. Wound-induced H2O2 and resistance to Botrytis cinerea decline with the ripening of apple fruit. Posthar. Biol. Technol. 2011, 62, 64–70. [Google Scholar] [CrossRef]
- Pitt, J.I.; Hocking, A.D. Fungi and Food Spoilage; Springer: New York, NY, USA, 2009. [Google Scholar]
- Barreto, M.C.; Houbraken, J.; Samson, R.A. Taxonomic studies of the Penicillium glabrum complex and the description of a new species P. subericola. Fungal Divers. 2011, 49, 23–33. [Google Scholar] [CrossRef] [Green Version]
- Sanzani, S.M.; Montemurro, C.; Di Rienzo, V.; Solfrizzo, M.; Ippolito, A. Genetic structure and natural variation associated with host of origin in Penicillium expansum strains causing blue mould. Int. J. Food Microbiol. 2013, 165, 111–120. [Google Scholar] [CrossRef]
- Julca, I.; Droby, S.; Sela, N.; Marcet-Houben, M.; Gabaldon, T. Contrasting genomic diversity in two closely related postharvest pathogens: Penicillium digitatum and Penicillium expansum. Genome Biol. Evol. 2015, 8, 218–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Sequence | Position | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Penicillium expansum | |||||||||||||||
| BenA | 232 | 260 | |||||||||||||
| I variant | T | C | |||||||||||||
| II variant | G | T | |||||||||||||
| CaM | 79 | 154 | 356 | ||||||||||||
| I variant | T | C | A | ||||||||||||
| II variant | C | T | G | ||||||||||||
| RPB2 | 139 | 151 | 325 | 340 | 373 | 454 | 520 | 760 | 841 | 854 | 889 | 943 | 958 | 979 | 1036 |
| I variant | C | A | G | T | G | C | T | C | T | C | T | G | A | C | C |
| II variant | T | G | A | C | A | T | C | T | C | C | C | A | C | T | T |
| III variant | C | A | G | T | G | C | T | C | T | T | T | G | A | C | C |
| Penicillium crustosum | |||||||||||||||
| CaM | 303 | 337 | 414 | ||||||||||||
| I variant | C | A | T | ||||||||||||
| II variant | - | G | - | ||||||||||||
| RPB2 | 175 | 724 | 940 | 979 | |||||||||||
| I variant | T | C | A | C | |||||||||||
| II variant | T | C | C | T | |||||||||||
| III variant | T | T | C | C | |||||||||||
| IV variant | C | T | C | C | |||||||||||
| Isolate | Colony Diameter on Different Culture Media (mm ± Standard Deviation) | |||
|---|---|---|---|---|
| PDA | MEA | CYA | YES | |
| Penicillium expansum | ||||
| JRad4 | 47.00 ± 0.97f * | 37.22 ± 0.73hi | 48.72 ± 1.25h | 51.31 ± 1.21d–f |
| 3JC11 | 42.69 ± 0.86d–f | 30.67 ± 1.08b–d | 44.18 ± 1.59d–g | 49.71 ± 1.78c–f |
| 3JC23 | 45.81 ± 1.02ef | 31.58 ± 0.69c–e | 43.09 ± 1.10c–f | 52.44 ± 0.63ef |
| 3JB13 | 40.58 ± 0.81b–e | 29.42 ± 1.48a–c | 46.13 ± 1.09e–h | 50.53 ± 0.85d–f |
| 3SD3 | 43.85 ± 1.26d–f | 31.06 ± 0.97c–e | 46.94 ± 1.57f–h | 51.69 ± 1.14d–f |
| 3SD5 | 43.92 ± 0.97d–f | 30.72 ± 0.89b–d | 46.34 ± 1.66e–h | 53.28 ± 1.17f |
| 3MR1 | 44.83 ± 1.20d–f | 32.08 ± 1.03de | 47.73 ± 1.02gh | 49.10 ± 1.54b–f |
| JBA8b | 43.47 ± 7.05d–f | 36.69 ± 2.12gh | 44.39 ± 6.26e–h | 49.64 ± 9.88c–f |
| JPN2 | 43.92 ± 7.85d–f | 37.89 ± 1.92hi | 42.42 ± 5.53b–e | 48.39 ± 8.64b–f |
| KPN4 | 44.56 ± 8.03d–f | 39.03 ± 3.58i | 44.50 ± 5.96e–h | 48.89 ± 8.77b–f |
| DRI4a | 44.91 ± 7.60d–f | 37.50 ± 2.81–i | 45.14 ± 6.31e–h | 50.21 ± 8.46d–f |
| 3JC6 | 36.77 ± 0.87a–c | 27.33 ± 0.49a | 46.22 ± 0.86e–h | 49.31 ± 1.01b–f |
| 3JB22 | 39.33 ± 1.42b–d | 28.64 ± 1.41ab | 38.69 ± 0.67a–c | 45.00 ± 2.61b–d |
| 3S1 | 32.92 ± 1.62a | 36.38 ± 1.50gh | 42.47 ± 0.63b–e | 48.03 ± 1.47b–f |
| KSA5 | 36.64 ± 0.70ab | 27.25 ± 0.46a | 38.36 ± 1.12ab | 42.50 ± 1.72ab |
| JMR2o | 35.03 ± 4.24ab | 32.06 ± 1.03de | 38.39 ± 3.21ab | 37.53 ± 2.17a |
| JMR2z | 42.41 ± 6.47c–f | 34.75 ± 2.06fg | 39.94 ± 2.43a–d | 45.72 ± 6.56b–e |
| DBA5 | 36.78 ± 4.56a–c | 33.17 ± 1.20ef | 36.28 ± 2.15a | 43.00 ± 5.54a–c |
| Avg. | 41.45 ± 5.76b ** | 32.94 ± 4.00b | 43.17 ± 4.75c | 48.03 ± 6.25c |
| Penicillium crustosum | ||||
| JBA8a | 39.58 ± 5.18a | 35.03 ± 2.27c | 43.81 ± 5.99b | 47.19 ± 8.38ab |
| JBA11 | 36.38 ± 3.09a | 30.39 ± 1.12a | 36.00 ± 2.54a | 41.61 ± 5.13ab |
| KGR2 | 38.36 ± 2.84a | 32.39 ± 1.36ab | 39.13 ± 6.09ab | 44.09 ± 6.19ab |
| KRI1P | 38.97 ± 2.61a | 31.34 ± 1.57ab | 38.69 ± 3.28ab | 40.92 ± 4.72a |
| KVA8 | 37.91 ± 3.37a | 31.64 ± 1.60ab | 38.92 ± 3.79ab | 41.58 ± 5.12ab |
| DRI4b | 37.71 ± 3.07a | 33.25 ± 1.40bc | 35.53 ± 1.69a | 41.44 ± 4.35ab |
| MRI4 | 39.71 ± 4.10a | 35.26 ± 2.48c | 40.53 ± 5.64ab | 48.47 ± 8.46b |
| Avg. | 38.39 ± 3.64b | 32.77 ± 2.45b | 38.90 ± 5.05b | 43.51 ± 6.67b |
| Penicillium solitum | ||||
| DRI3 | 28.02 ± 0.50a | 24.31 ± 0.68a | 29.75 ± 0.69a | 34.53 ± 1.22a |
| Isolate | Average Size (µm ± Standard Deviation) | |||||||
|---|---|---|---|---|---|---|---|---|
| Conidia | Phialides | Metulae | Rami | |||||
| Length | Width | Length | Width | Length | Width | Length | Width | |
| Penicillium expansum | ||||||||
| JRad4 | 3.25 ± 0.25c–e * | 2.72 ± 0.19bc | 9.74 ± 1.38ab | 2.84 ± 0.55a–d | 12.72 ± 2.41a–e | 3.35 ± 0.59a–e | 19.64 ± 4.20a–c | 3.67 ± 0.63ab |
| 3JC11 | 3.13 ± 0.30a–d | 2.56 ± 0.26ab | 10.06 ± 0.91a–d | 2.85 ± 0.34a–d | 11.63 ± 1.46a | 3.19 ± 0.38ab | 18.61 ± 2.10a–c | 3.58 ± 0.43a |
| 3JC23 | 3.13 ± 0.24a–d | 2.52 ± 0.18a | 9.09 ± 1.35a | 2.89 ± 0.53a–e | 11.99 ± 2.65a–c | 3.23 ± 0.68a–c | 16.67 ± 3.51a | 3.78 ± 0.77ab |
| 3JB13 | 3.00 ± 0.22ab | 2.52 ± 0.19a | 9.82 ± 0.77ab | 2.69 ± 0.28a–c | 12.94 ± 1.83a–e | 3.27 ± 0.36a–d | 19.58 ± 2.77a–c | 3.56 ± 0.39a |
| 3SD3 | 3.05 ± 0.22a–c | 2.54 ± 0.21ab | 9.65 ± 1.05ab | 2.74 ± 0.39a–d | 12.77 ± 3.19a–e | 3.56 ± 0.72a-g | 18.55 ± 4.15a–c | 4.15 ± 0.68a–c |
| 3SD5 | 2.96 ± 0.23a | 2.56 ± 0.20ab | 10.13 ± 1.21b–d | 2.97 ± 0.40b-e | 11.81 ± 2.09ab | 3.78 ± 0.76e-h | 17.83 ± 3.16a–c | 4.10 ± 0.64a–c |
| 3MR1 | 3.16 ± 0.29a-d | 2.56 ± 0.21ab | 10.48 ± 1.11b–d | 2.92 ± 0.37b-e | 13.51 ± 1.76c–e | 3.51 ± 0.37a-f | 24.65 ± 5.84d | 3.74 ± 0.39ab |
| JBA8b | 3.22 ± 0.24b-e | 2.98 ± 0.24de | 9.86 ± 1.11a–c | 3.03 ± 0.39d–f | 14.30 ± 2.26e | 3.64 ± 0.54b-g | 20.45 ± 4.33a-d | 4.01 ± 0.64a–c |
| JPN2 | 3.25 ± 0.31c–e | 2.80 ± 0.22cd | 10.36 ± 1.35b–d | 2.98 ± 0.36c–e | 13.50 ± 1.96b-e | 3.67 ± 0.43c–h | 19.39 ± 4.75a–c | 3.83 ± 0.63ab |
| KPN4 | 3.27 ± 0.37c–e | 2.98 ± 0.30de | 10.91 ± 1.46d | 3.20 ± 0.35ef | 14.01 ± 2.51e | 3.96 ± 0.53fgh | 22.02 ± 4.64cd | 4.17 ± 0.56a–c |
| DRI4a | 3.61 ± 0.39f | 3.22 ± 0.30f | 9.75 ± 1.38ab | 3.03 ± 0.44d–f | 13.62 ± 2.06c–e | 3.91 ± 0.65fgh | 18.91 ± 3.56a–c | 4.23 ± 0.45bc |
| 3JC6 | 3.56 ± 0.27f | 2.99 ± 0.29e | 10.27 ± 1.05b–d | 2.57 ± 0.27a | 11.29 ± 1.18a | 3.13 ± 0.47a | 17.89 ± 2.26a–c | 3.58 ± 0.54a |
| 3JB22 | 3.10 ± 0.24a–d | 2.47 ± 0.17a | 10.33 ± 1.02b–d | 2.64 ± 0.38ab | 13.84 ± 2.38de | 3.58 ± 0.77a–g | 19.91 ± 4.39a–c | 3.83 ± 0.81ab |
| 3S1 | 2.93 ± 0.29a | 2.48 ± 0.23a | 10.39 ± 1.79b–d | 2.97 ± 0.44b–e | 12.79 ± 2.09a–e | 3.69 ± 0.77d–h | 20.92 ± 3.60b–d | 4.15 ± 0.94a–c |
| KSA5 | 3.32 ± 0.29de | 2.63 ± 0.19ab | 10.24 ± 1.32b–d | 2.79 ± 0.32a–d | 12.63 ± 1.76a–e | 3.36 ± 0.44a–e | 19.75 ± 4.11a–c | 3.83 ± 0.48ab |
| JMR2o | 3.46 ± 0.39ef | 2.82 ± 0.31c–e | 9.78 ± 1.10ab | 3.32 ± 0.55f | 12.15 ± 1.45a–d | 4.10 ± 0.47h | 16.79 ± 3.22ab | 4.25 ± 0.65bc |
| JMR2z | 3.42 ± 0.36ef | 2.99 ± 0.25e | 10.34 ± 1.11b–d | 3.20 ± 0.53ef | 13.90 ± 2.45e | 4.11 ± 0.60h | 21.75 ± 5.48cd | 4.58 ± 0.53c |
| DBA5 | 3.45 ± 0.47ef | 2.81 ± 0.31c–e | 10.83 ± 1.47cd | 3.20 ± 0.52ef | 13.46 ± 2.28b–e | 4.01 ± 0.53gh | 20.61 ± 5.60a–d | 4.26 ± 0.53bc |
| Avg. | 3.24 ± 0.30a ** | 2.73 ± 0.24a | 10.14 ± 1.32a | 2.94 ± 0.47a | 12.94 ± 2.30a | 3.61 ± 0.65a | 19.66 ± 4.47ab | 3.96 ± 0.97a |
| Penicillium crustosum | ||||||||
| JBA8a | 3.10 ± 0.24a | 2.76 ± 0.23a | 10.67 ± 1.56d | 3.08 ± 0.49a | 14.37 ± 2.30ab | 4.10 ± 0.53b | 21.52 ± 4.38ab | 4.31 ± 0.69b |
| JBA11 | 3.27 ± 0.38a–c | 3.00 ± 0.27b | 10.59 ± 1.52cd | 3.23 ± 0.51a | 13.30 ± 11.86a | 4.01 ± 0.56ab | 20.37 ± 3.65ab | 4.22 ± 0.57ab |
| KGR2 | 3.61 ± 0.27d | 3.34 ± 0.22c | 9.67 ± 1.05a–c | 3.03 ± 0.32a | 13.71 ± 1.88ab | 3.71 ± 0.46a | 21.18 ± 3.87ab | 4.16 ± 0.44ab |
| KRI1P | 3.28 ± 0.23a–c | 3.06 ± 0.18b | 10.36 ± 1.57b–d | 3.09 ± 0.34a | 13.97 ± 2.79ab | 3.77 ± 0.53a | 20.01 ± 4.73a | 3.88 ± 0.48ab |
| KVA8 | 3.23 ± 0.21ab | 2.97 ± 0.20b | 9.63 ± 1.10ab | 3.06 ± 0.38a | 13.62 ± 3.05a | 3.83 ± 0.48ab | 19.87 ± 4.55a | 4.10 ± 0.66ab |
| DRI4b | 3.36 ± 0.33bc | 2.98 ± 0.27b | 9.36 ± 0.96a | 2.99 ± 0.39a | 15.04 ± 2.24b | 3.75 ± 0.54a | 23.37 ± 4.33b | 3.78 ± 0.59a |
| MRI4 | 3.45 ± 0.23c | 3.28 ± 0.23c | 10.07 ± 1.41a–d | 3.21 ± 0.45a | 14.34 ± 1.79ab | 3.86 ± 0.58ab | 22.26 ± 4.82ab | 4.28 ± 0.55b |
| Avg. | 3.33 ± 0.27a | 3.06 ± 0.23b | 10.05 ± 1.35a | 3.10 ± 0.41a | 14.02 ± 2.43b | 3.86 ± 0.54b | 21.23 ± 4.45b | 4.11 ± 0.60ab |
| Penicillium solitum | ||||||||
| DRI3 | 3.67 ± 0.17b | 3.42 ± 0.18c | 10.13 ± 1.92a | 3.11 ± 0.41a | 12.20 ± 2.03a | 4.21 ± 0.56c | 18.17 ± 4.09a | 4.41 ± 0.69b |
| Macromorphology (Cluster Analysis: Colony Diameter, PDA, MEA, CYA Reverse) | Phylogeny | ||
|---|---|---|---|
| BenA CaM | RPB2 Multilocus | ||
| I group | JRad4 * | JRad4 | JRad4 |
| 3JC11 | 3JC11 | 3JC11 | |
| 3JC23 | 3JC23 | 3JC23 | |
| 3JB13 | 3JB13 | 3JB13 | |
| 3SD3 | 3SD3 | 3SD3 | |
| 3SD5 | 3SD5 | 3SD5 | |
| 3MR1 | 3MR1 | 3MR1 | |
| JBA8b | JBA8b | JBA8b | |
| JPN2 | JPN2 | JPN2 | |
| KPN4 | KPN4 | KPN4 | |
| DRI4a | DRI4a | DRI4a | |
| 3JB22 | |||
| 3S1 | |||
| KSA5 | |||
| II group | 3JB22 | 3JB22 | |
| 3S1 | 3S1 | ||
| KSA5 | KSA5 | ||
| 3JC6 | 3JC6 | 3JC6 | |
| JMR2o | JMR2o | JMR2o | |
| JMR2z | JMR2z | JMR2z | |
| DBA5 | DBA5 | DBA5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Žebeljan, A.; Duduk, N.; Vučković, N.; Jurick, W.M., II; Vico, I. Incidence, Speciation, and Morpho-Genetic Diversity of Penicillium spp. Causing Blue Mold of Stored Pome Fruits in Serbia. J. Fungi 2021, 7, 1019. https://doi.org/10.3390/jof7121019
Žebeljan A, Duduk N, Vučković N, Jurick WM II, Vico I. Incidence, Speciation, and Morpho-Genetic Diversity of Penicillium spp. Causing Blue Mold of Stored Pome Fruits in Serbia. Journal of Fungi. 2021; 7(12):1019. https://doi.org/10.3390/jof7121019
Chicago/Turabian StyleŽebeljan, Aleksandra, Nataša Duduk, Nina Vučković, Wayne M. Jurick, II, and Ivana Vico. 2021. "Incidence, Speciation, and Morpho-Genetic Diversity of Penicillium spp. Causing Blue Mold of Stored Pome Fruits in Serbia" Journal of Fungi 7, no. 12: 1019. https://doi.org/10.3390/jof7121019






