Implementation of Synthetic Pathways to Foster Microbe-Based Production of Non-Naturally Occurring Carboxylic Acids and Derivatives
Abstract
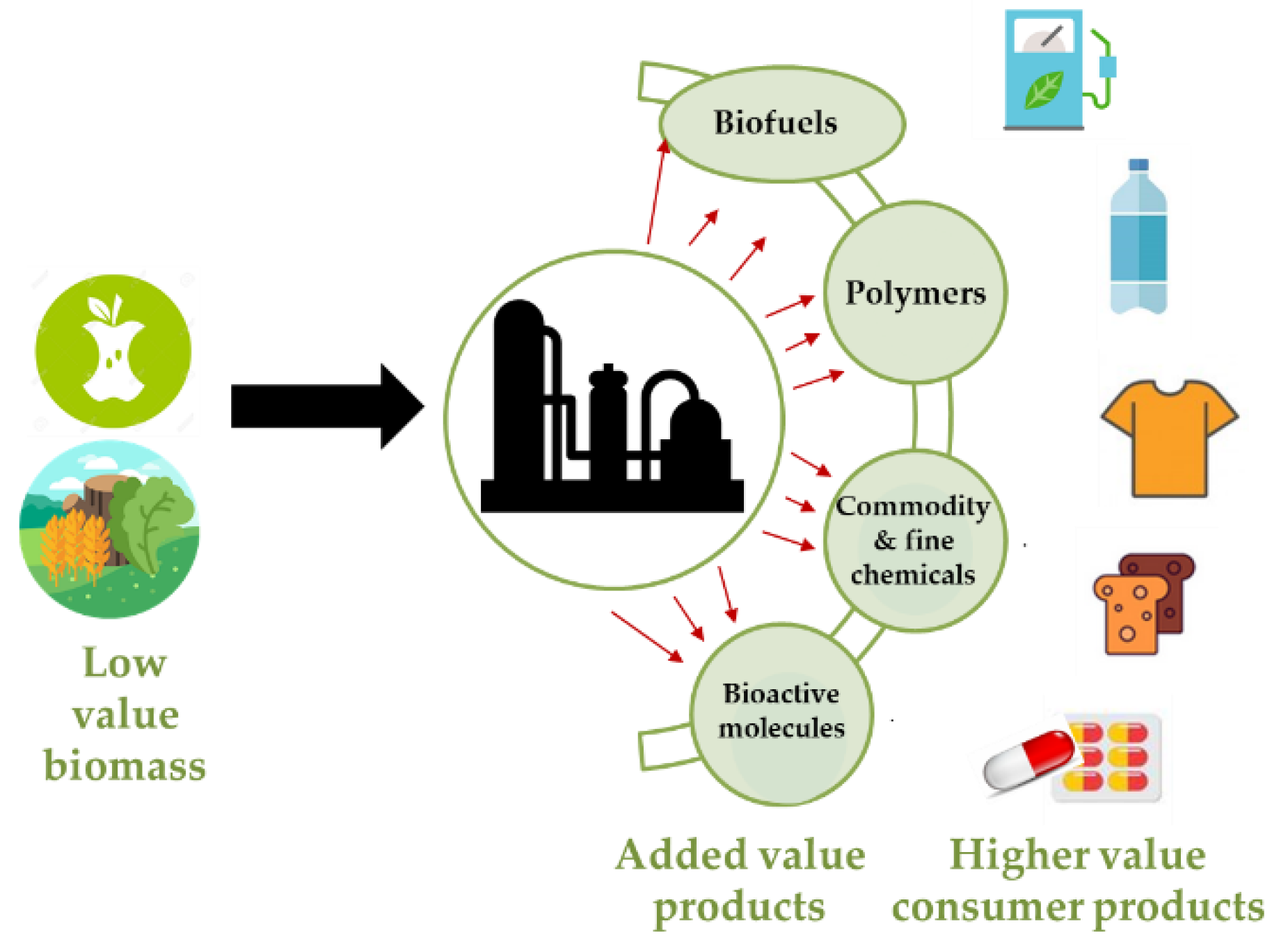
:1. Overview
2. Carboxylic Acids and Their Derivatives as “Green Building Blocks”
3. Implementation of New-to-Nature Pathways to Enable Microbe-Based Production of Naturally Occurring CAs
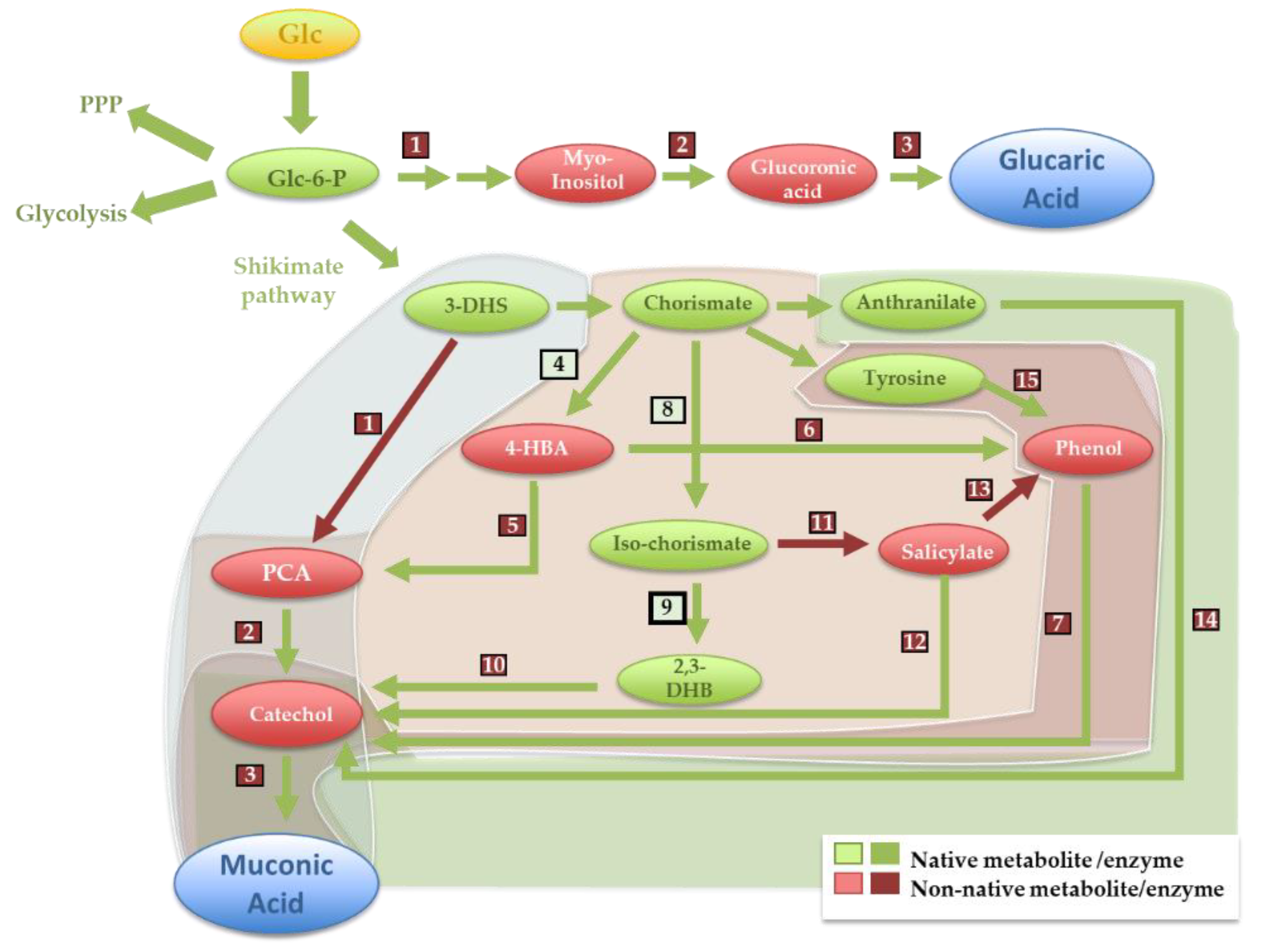
3.1. Glucaric Acid
3.2. Muconic Acid
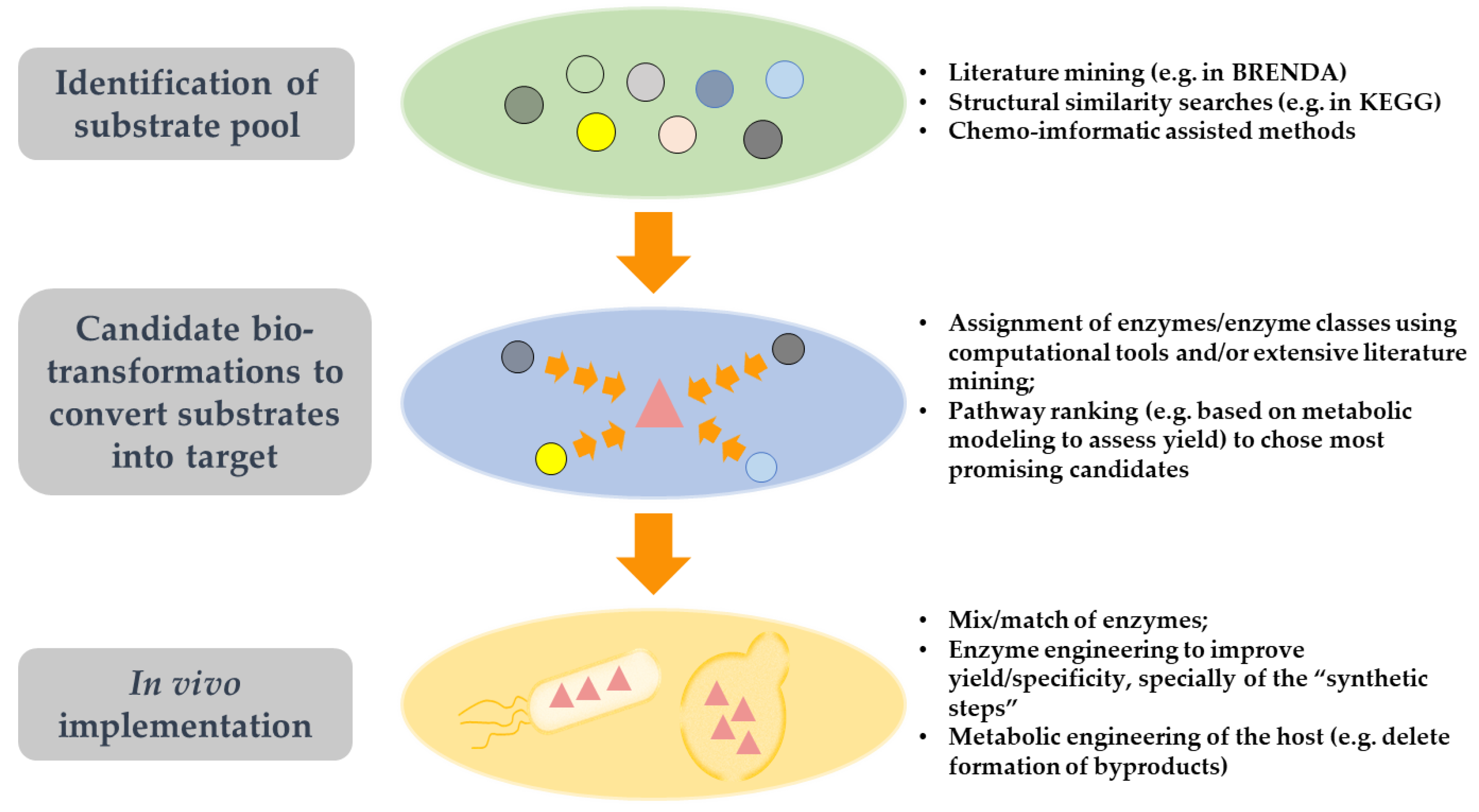
4. A General View on the Implementation of Synthetic Pathways for the Production of Non-Naturally Occurring Compounds
5. Implementation of New-to-Nature Synthetic Pathways to Enable Microbe-Based Production of Adipic, Acrylic and Levulinic Acids
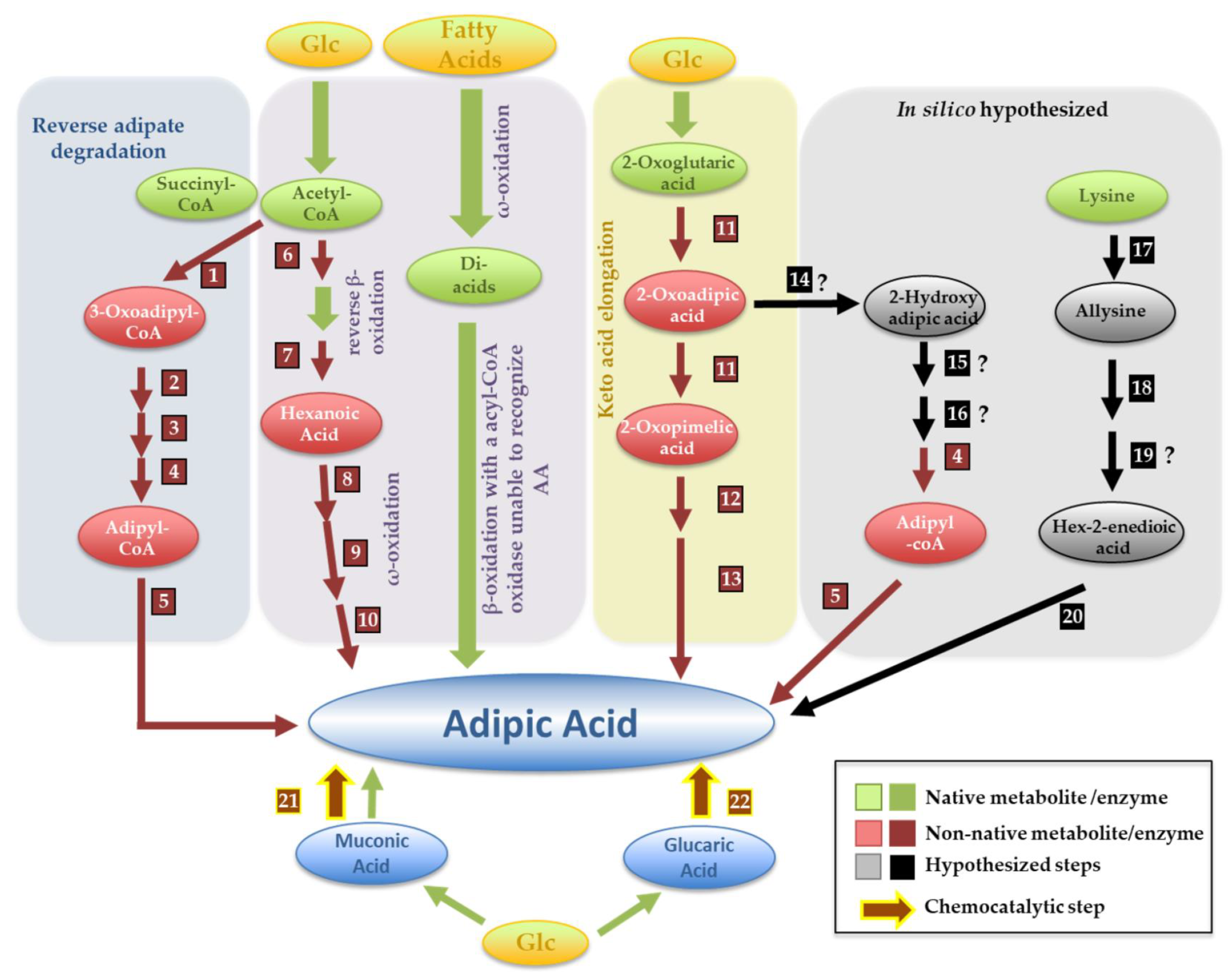
5.1. Adipic Acid
5.2. Acrylic Acid
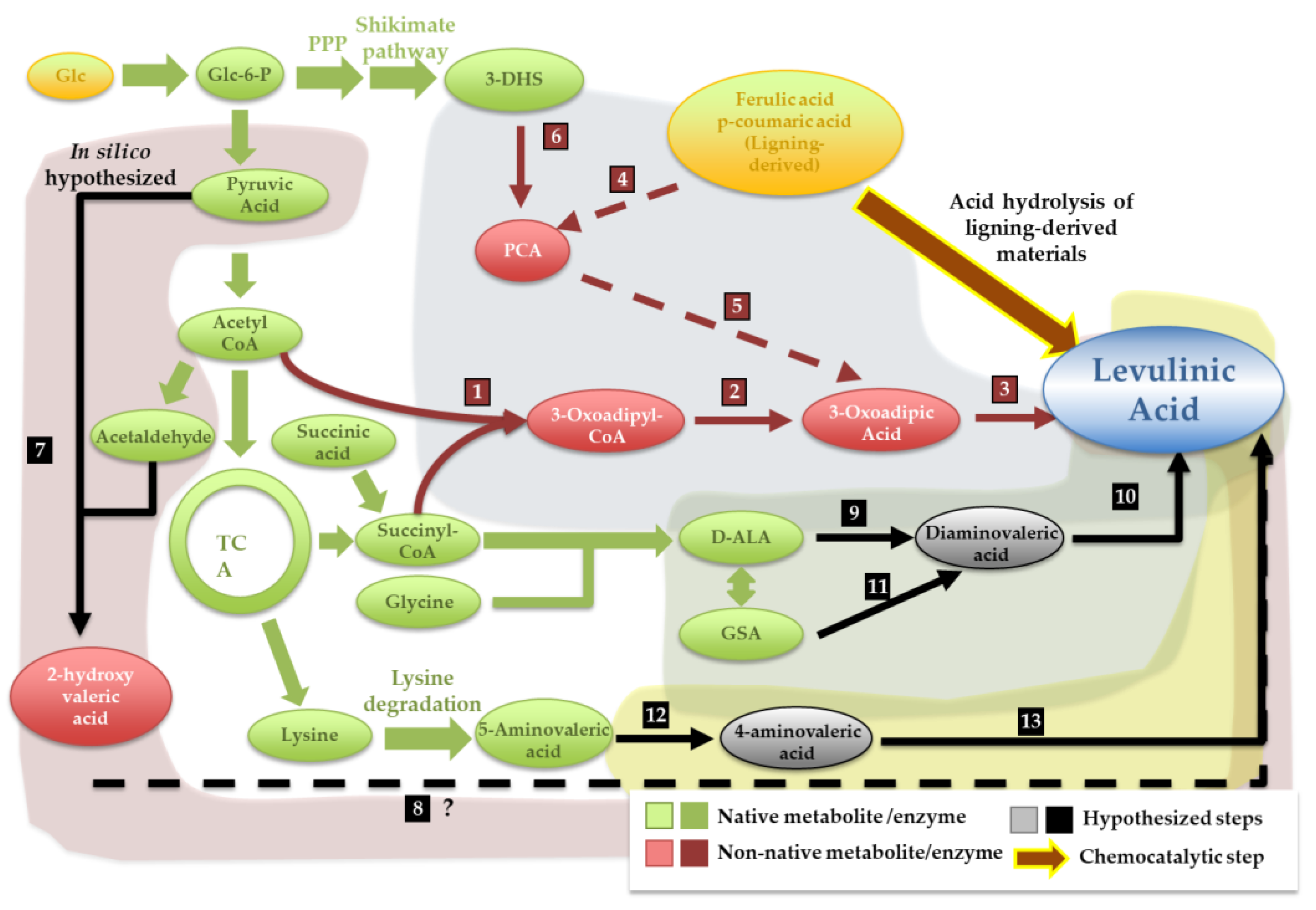
5.3. Levulinic Acid
6. Bridging the Gap between CAs and Their More Economically Relevant Derivatives through the Assembly of Synthetic Pathways
6.1. Poly-Lactate Polymers
6.2. Methacrylic and Methyl-Methacrylic Acids
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Willke, T.; Vorlop, K.D. Industrial bioconversion of renewable resources as an alternative to conventional chemistry. Appl. Microbiol. Biotechnol. 2004, 66, 131–142. [Google Scholar] [CrossRef]
- Cherubini, F. The biorefinery concept: Using biomass instead of oil for producing energy and chemicals. Energy Convers. Manag. 2010, 51, 1412–1421. [Google Scholar] [CrossRef]
- Tseng, H.-C.; Prather, K.L.J. Controlled biosynthesis of odd-chain fuels and chemicals via engineered modular metabolic pathways. Proc. Natl. Acad. Sci. USA 2012, 109, 17925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yim, H.; Haselbeck, R.; Niu, W.; Pujol-Baxley, C.; Burgard, A.; Boldt, J.; Khandurina, J.; Trawick, J.D.; Osterhout, R.E.; Stephen, R.; et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat. Chem. Biol. 2011, 7, 445–452. [Google Scholar] [CrossRef]
- Sheppard, M.J.; Kunjapur, A.M.; Wenck, S.J.; Prather, K.L.J. Retro-biosynthetic screening of a modular pathway design achieves selective route for microbial synthesis of 4-methyl-pentanol. Nat. Commun. 2014, 5, 5031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galanie, S.; Thodey, K.; Trenchard, I.J.; Filsinger Interrante, M.; Smolke, C.D. Complete biosynthesis of opioids in yeast. Science 2015, 349, 1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbott, D.A.; Zelle, R.M.; Pronk, J.T.; van Maris, A.J.A. Metabolic engineering of Saccharomyces cerevisiae for production of carboxylic acids: Current status and challenges. FEMS Yeast Res. 2009, 9, 1123–1136. [Google Scholar] [CrossRef] [Green Version]
- Consortia, B. BREW: Medium and Long-Term Opportunities and Risks of the Biotechnological Production of Bulk Chemicals from Renewable Resources—The Potential of White Biotechnology; Utrecht University: Utrecht, The Netherlands, 2006. [Google Scholar]
- Bozell, J.J.; Petersen, G.R. Technology development for the production of biobased products from biorefinery carbohydrates—The US Department of Energy’s “Top 10” revisited. Green Chem. 2010, 12, 539. [Google Scholar] [CrossRef]
- Murali, N.; Srinivas, K.; Ahring, B.K. Biochemical Production and Separation of Carboxylic Acids for Biorefinery Applications. Fermentation 2017, 3, 22. [Google Scholar] [CrossRef] [Green Version]
- Becker, J.; Wittmann, C. Advanced Biotechnology: Metabolically Engineered Cells for the Bio-Based Production of Chemicals and Fuels, Materials, and Health-Care Products. Angew. Chem. Int. Ed. 2015, 54, 3328–3350. [Google Scholar] [CrossRef]
- Iglesias, J.; Martínez-Salazar, I.; Maireles-Torres, P.; Martin Alonso, D.; Mariscal, R.; López Granados, M. Advances in catalytic routes for the production of carboxylic acids from biomass: A step forward for sustainable polymers. Chem. Soc. Rev. 2020, 49, 5704–5771. [Google Scholar] [CrossRef]
- Research, P.S.M. Bio-Based Organic Acids Market Size, Share; 2014; Available online: https://www.psmarketresearch.com/market-analysis/bio-based-organic-acids-market (accessed on 17 February 2020).
- Moon, T.S.; Yoon, S.H.; Lanza, A.M.; Roy-Mayhew, J.D.; Jones Prather, K.L. Production of glucaric acid from a synthetic pathway in recombinant Escherichia coli. Appl. Environ. Microbiol. 2009, 75, 589–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, T.S.; Dueber, J.E.; Shiue, E.; Prather, K.L.J. Use of modular, synthetic scaffolds for improved production of glucaric acid in engineered E. coli. Metab. Eng. 2010, 12, 298–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doong, S.J.; Gupta, A.; Prather, K.L.J. Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli. Proc. Natl. Acad. Sci. USA 2018, 115, 2964–2969. [Google Scholar] [CrossRef] [Green Version]
- Shiue, E.; Prather, K.L. Improving D-glucaric acid production from myo-inositol in E. coli by increasing MIOX stability and myo-inositol transport. Metab. Eng. 2014, 22, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Wang, J.; Zhao, Y.; Deng, Y. Metabolic engineering of Saccharomyces cerevisiae for efficient production of glucaric acid at high titer. Microb. Cell Factories 2018, 17, 67. [Google Scholar] [CrossRef] [Green Version]
- Prather, K.L.J.; Martin, C.H. De novo biosynthetic pathways: Rational design of microbial chemical factories. Curr. Opin. Biotechnol. 2008, 19, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Marques, W.L.; Anderson, L.A.; Sandoval, L.; Hicks, M.A.; Prather, K.L.J. Sequence-based bioprospecting of myo-inositol oxygenase (Miox) reveals new homologues that increase glucaric acid production in Saccharomyces cerevisiae. Enzym. Microb. Technol. 2020, 140, 109623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, C.; Liu, Y.; Wang, J.; Zhao, Y.; Deng, Y. Enhancement of glucaric acid production in Saccharomyces cerevisiae by expressing Vitreoscilla hemoglobin. Biotechnol. Lett. 2020, 42, 2169–2178. [Google Scholar] [CrossRef]
- Zeyaullah, M.; Abdelkafe, A.; Zabya, W.; Ali, A. Biodegradation of catechols by micro-organisms—A short review. Afr. J. Biotechnol. 2009, 8, 2916–2922. [Google Scholar]
- Becker, J.; Wittmann, C. A field of dreams: Lignin valorization into chemicals, materials, fuels, and health-care products. Biotechnol. Adv. 2019, 37, 107360. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Lee, H.-N.; Park, E.; Lee, S.-J.; Kim, E.-S. Recent Advances in Microbial Production of cis,cis-Muconic Acid. Biomolecules 2020, 10, 1238. [Google Scholar] [CrossRef]
- Zhang, H.; Pereira, B.; Li, Z.; Stephanopoulos, G. Engineering Escherichia coli coculture systems for the production of biochemical products. Proc. Natl. Acad. Sci. USA 2015, 112, 8266–8271. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Li, Z.; Pereira, B.; Stephanopoulos, G. Engineering E. coli–E. coli cocultures for production of muconic acid from glycerol. Microb. Cell Factories 2015, 14, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyne Michael, E.; Narcross, L.; Melgar, M.; Kevvai, K.; Mookerjee, S.; Leite Gustavo, B.; Martin Vincent, J.J.; Elliot Marie, A. An Engineered Aro1 Protein Degradation Approach for Increased cis,cis-Muconic Acid Biosynthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2018, 84, 84. [Google Scholar] [CrossRef] [Green Version]
- Brückner, C.; Oreb, M.; Kunze, G.; Boles, E.; Tripp, J. An expanded enzyme toolbox for production of cis, cis-muconic acid and other shikimate pathway derivatives in Saccharomyces cerevisiae. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, W.; Draths, K.M.; Frost, J.W. Benzene-Free Synthesis of Adipic Acid. Biotechnol. Prog. 2002, 18, 201–211. [Google Scholar] [CrossRef]
- Lin, Y.; Sun, X.; Yuan, Q.; Yan, Y. Extending shikimate pathway for the production of muconic acid and its precursor salicylic acid in Escherichia coli. Metab. Eng. 2014, 23, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.; Pugh, S.; Machas, M.; Nielsen, D.R. Muconic Acid Production via Alternative Pathways and a Synthetic “metabolic Funnel”. ACS Synth. Biol. 2018, 7, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Y.; Wang, J.; Zhao, Y.; Deng, Y. Biosynthesis of adipic acid in metabolically engineered Saccharomyces cerevisiae. J. Microbiol. 2020, 58, 1065–1075. [Google Scholar] [CrossRef]
- Clomburg, J.M.; Blankschien, M.D.; Vick, J.E.; Chou, A.; Kim, S.; Gonzalez, R. Integrated engineering of β -oxidation reversal and ω -oxidation pathways for the synthesis of medium chain ω -functionalized carboxylic acids. Metab. Eng. 2015, 28, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Turk, S.C.H.J.; Kloosterman, W.P.; Ninaber, D.K.; Kolen, K.P.A.M.; Knutova, J.; Suir, E.; Schürmann, M.; Raemakers-Franken, P.C.; Müller, M.; De Wildeman, S.M.A.; et al. Metabolic Engineering toward Sustainable Production of Nylon-6. ACS Synth. Biol. 2016, 5, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Raj, K.; Partow, S.; Correia, K.; Khusnutdinova, A.N.; Yakunin, A.F.; Mahadevan, R. Biocatalytic production of adipic acid from glucose using engineered Saccharomyces cerevisiae. Metab. Eng. Commun. 2018, 6, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.S.; Ahn, J.-H.; Yun, J.; Choi, I.S.; Nam, T.-W.; Cho, K.M. Direct fermentation route for the production of acrylic acid. Metab. Eng. 2015, 32, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Cheong, S.; Clomburg, J.M.; Gonzalez, R. Energy- and carbon-efficient synthesis of functionalized small molecules in bacteria using non-decarboxylative Claisen condensation reactions. Nat. Biotechnol. 2016, 34, 556–561. [Google Scholar] [CrossRef]
- Oh, M.-K.; Kim, B.S.; Chang, M.W.; Lo, T.-M.; Hwang, I.Y.; Cho, H.-S.; Fedora, R.E.; Chng, S.H.; Choi, W.J. Biosynthesis of Commodity Chemicals from Oil Palm Empty Fruit Bunch Lignin. Front. Microbiol. 2021. [Google Scholar] [CrossRef]
- Eastham, D.W.J.; Ronald, G.; Poliakoff, M.; Huddle, T.A. A Process for the Production of Methacrylic Acid and Its Derivatives and Polymers Produced Therefrom. Patent No. EP2643283A1, 12 August 2015. [Google Scholar]
- Nam, H.; Lewis, N.E.; Lerman, J.A.; Lee, D.H.; Chang, R.L.; Kim, D.; Palsson, B.O. Network context and selection in the evolution to enzyme specificity. Science 2012, 337, 1101–1104. [Google Scholar] [CrossRef] [Green Version]
- Lin, G.-M.; Warden-Rothman, R.; Voigt, C.A. Retrosynthetic design of metabolic pathways to chemicals not found in nature. Curr. Opin. Syst. Biol. 2019, 14, 82–107. [Google Scholar] [CrossRef]
- Walther, T.; Topham, C.M.; Irague, R.; Auriol, C.; Baylac, A.; Cordier, H.; Dressaire, C.; Lozano-Huguet, L.; Tarrat, N.; Martineau, N.; et al. Construction of a synthetic metabolic pathway for biosynthesis of the non-natural methionine precursor 2,4-dihydroxybutyric acid. Nat. Commun. 2017. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.; Calvayrac, F.; Malbert, Y.; Alkim, C.; Dressaire, C.; Cordier, H.; François, J.M. Construction of a synthetic metabolic pathway for the production of 2,4-dihydroxybutyric acid from homoserine. Metab. Eng. 2018, 45, 237–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, M.; Duigou, T.; Faulon, J.L. Reinforcement Learning for Bioretrosynthesis. ACS Synth. Biol. 2020. [Google Scholar] [CrossRef]
- Li, C.; Henry, C.S.; Jankowski, M.D.; Ionita, J.A.; Hatzimanikatis, V.; Broadbelt, L.J. Computational discovery of biochemical routes to specialty chemicals. Chem. Eng. Sci. 2004, 59, 5051–5060. [Google Scholar] [CrossRef]
- Hatzimanikatis, V.; Li, C.; Ionita, J.A.; Henry, C.S.; Jankowski, M.D.; Broadbelt, L.J. Exploring the diversity of complex metabolic networks. Bioinformatics 2005, 21, 1603–1609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriya, Y.; Shigemizu, D.; Hattori, M.; Tokimatsu, T.; Kotera, M.; Goto, S.; Kanehisa, M. PathPred: An enzyme-catalyzed metabolic pathway prediction server. Nucleic Acids Res. 2010, 38, W138–W143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, B.K.; Ellis, L.B.; Wackett, L.P. Encoding microbial metabolic logic: Predicting biodegradation. J. Ind. Microbiol. Biotechnol. 2004, 31, 261–272. [Google Scholar] [CrossRef]
- Hadadi, N.; Hafner, J.; Shajkofci, A.; Zisaki, A.; Hatzimanikatis, V. ATLAS of Biochemistry: A Repository of All Possible Biochemical Reactions for Synthetic Biology and Metabolic Engineering Studies. ACS Synth. Biol. 2016, 5, 1155–1166. [Google Scholar] [CrossRef]
- Sivakumar, T.V.; Giri, V.; Park, J.H.; Kim, T.Y.; Bhaduri, A. ReactPRED: A tool to predict and analyze biochemical reactions. Bioinformatics 2016, 32, 3522–3524. [Google Scholar] [CrossRef]
- Campodonico, M.A.; Andrews, B.A.; Asenjo, J.A.; Palsson, B.O.; Feist, A.M. Generation of an atlas for commodity chemical production in Escherichia coli and a novel pathway prediction algorithm, GEM-Path. Metab. Eng. 2014, 25, 140–158. [Google Scholar] [CrossRef]
- Bodor, Z.; Tompos, L.; Nechifor, A.C.; Bodor, K. In silico Analysis of 1,4-butanediol Heterologous Pathway Impact on Escherichia coli Metabolism. Rev. Chim. 2019, 70, 3448–3455. [Google Scholar] [CrossRef]
- Averesch, N.J.H.; Martínez, V.S.; Nielsen, L.K.; Krömer, J.O. Toward Synthetic Biology Strategies for Adipic Acid Production: An in Silico Tool for Combined Thermodynamics and Stoichiometric Analysis of Metabolic Networks. ACS Synth. Biol. 2018, 7, 490–509. [Google Scholar] [CrossRef] [PubMed]
- Sorokina, M.; Stam, M.; Médigue, C.; Lespinet, O.; Vallenet, D. Profiling the orphan enzymes. Biol. Direct 2014, 9, 10. [Google Scholar] [CrossRef] [Green Version]
- Delépine, B.; Duigou, T.; Carbonell, P.; Faulon, J.L. RetroPath2.0: A retrosynthesis workflow for metabolic engineers. Metab. Eng. 2018, 45, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Mao, Y. Production of adipic acid by the native-occurring pathway in Thermobifida fusca B6. J. Appl. Microbiol. 2015, 119, 1057–1063. [Google Scholar] [CrossRef] [Green Version]
- Kruyer, N.S.; Wauldron, N.; Bommarius, A.S.; Peralta-Yahya, P. Fully biological production of adipic acid analogs from branched catechols. Sci. Rep. 2020, 10, 13367. [Google Scholar] [CrossRef]
- Burgard, A.; Pharkya, P.; Osterhout, R.E. Microrganisms for the Production of Adipic Acid and Other Compounds. Patent No. US7799545B2, 21 September 2010. [Google Scholar]
- Picataggio, S.; Beardslee, T. Biological Methods for Preparing Adipic Acid. US20120021474A1, 14 August 2012. [Google Scholar]
- Skoog, E.; Shin, J.H.; Saez-Jimenez, V.; Mapelli, V.; Olsson, L. Biobased adipic acid—The challenge of developing the production host. Biotechnol. Adv. 2018, 36, 2248–2263. [Google Scholar] [CrossRef]
- Straathof, A.J.; Sie, S.; Franco, T.T.; van der Wielen, L.A. Feasibility of acrylic acid production by fermentation. Appl. Microbiol. Biotechnol. 2005, 67, 727–734. [Google Scholar] [CrossRef]
- Ko, Y.-S.; Kim, J.W.; Chae, T.U.; Song, C.W.; Lee, S.Y. A Novel Biosynthetic Pathway for the Production of Acrylic Acid through β-Alanine Route in Escherichia coli. ACS Synth. Biol. 2020, 9, 1150–1159. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, T. Production of acrylic acid and propionic acid by constructing a portion of the 3-hydroxypropionate/4-hydroxybutyrate cycle from Metallosphaera sedula in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2016, 43, 1659–1670. [Google Scholar] [CrossRef] [PubMed]
- Bertoldi, M.; Carbone, V.; Borri Voltattorni, C. Ornithine and glutamate decarboxylases catalyse an oxidative deamination of their alpha-methyl substrates. Biochem. J. 1999, 342 Pt 3, 509–512. [Google Scholar] [CrossRef]
- Vila-Santa, A.; Islam, M.A.; Ferreira, F.C.; Prather, K.L.J.; Mira, N.P. Prospecting Biochemical Pathways to Implement Microbe-Based Production of the New-to-Nature Platform Chemical Levulinic Acid. ACS Synth. Biol. 2021, 10. [Google Scholar] [CrossRef]
- Pinto-Ibieta, F.; Cea, M.; Cabrera, F.; Abanto, M.; Felissia, F.E.; Area, M.C.; Ciudad, G. Strategy for biological co-production of levulinic acid and polyhydroxyalkanoates by using mixed microbial cultures fed with synthetic hemicellulose hydrolysate. Bioresour. Technol. 2020, 309, 123323. [Google Scholar] [CrossRef]
- Dishisha, T.; Pyo, S.-H.; Hatti-Kaul, R. Bio-based 3-hydroxypropionic- and acrylicacid production from biodiesel glycerol via integrated microbial and chemicalcatalysis. Microb. Cell Factories 2015, 14, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lingoes, J.; Collias, D. Catalytic Conversion of Lactic Acid to Acrylic Acid. Patent No. US9,926,256B2, 27 March 2018. [Google Scholar]
- Bomgardner, M. Cargill gives biobased acrylic acid one more go. Chemical & Engineering News, 20 May 2020. [Google Scholar]
- Riaz, S.; Fatima, N.; Rasheed, A.; Riaz, M.; Anwar, F.; Khatoon, Y. Metabolic Engineered Biocatalyst: A Solution for PLA Based Problems. Int. J. Biomater. 2018, 2018, 1963024. [Google Scholar] [CrossRef]
- Singhvi, M.S.; Zinjarde, S.S.; Gokhale, D.V. Polylactic acid: Synthesis and biomedical applications. J. Appl. Microbiol. 2019, 127, 1612–1626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.H.; Kim, T.W.; Kang, H.O.; Lee, S.H.; Lee, E.J.; Lim, S.C.; Oh, S.O.; Song, A.J.; Park, S.J.; Lee, S.Y. Biosynthesis of polylactic acid and its copolymers using evolved propionate CoA transferase and PHA synthase. Biotechnol. Bioeng. 2010, 105, 150–160. [Google Scholar] [CrossRef]
- Jung, Y.K.; Kim, T.Y.; Park, S.J.; Lee, S.Y. Metabolic engineering of Escherichia coli for the production of polylactic acid and its copolymers. Biotechnol. Bioeng. 2010, 105, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Lajus, S.; Dusséaux, S.; Verbeke, J.; Rigouin, C.; Guo, Z.; Fatarova, M.; Bellvert, F.; Borsenberger, V.; Bressy, M.; Nicaud, J.M.; et al. Engineering the Yeast Yarrowia lipolytica for Production of Polylactic Acid Homopolymer. Front. Bioeng. Biotechnol. 2020, 8, 954. [Google Scholar] [CrossRef]
- Tran, T.T.; Charles, T.C. Lactic acid containing polymers produced in engineered Sinorhizobium meliloti and Pseudomonas putida. PLoS ONE 2020, 15, e0218302. [Google Scholar] [CrossRef] [Green Version]
- De Carvalho, J.C.; Magalhães, A.I.; Soccol, C.R. Biobased itaconic acid market and research trends-is it really a promising chemical? Chem. Today 2018, 36, 56–58. [Google Scholar]
- Verified Market Research. Global Itaconic Acid Market Size by Derivative, by Application, by Geographic Scope and Forecast; Verified Market Research: Boonton, NJ, USA, 2020. [Google Scholar]
- Bauer, W. Methacrylic Acid and Derivatives. In Ullmann’s Encyclopedia of Industrial Chemistry; Wiley: Hoboken, NJ, USA, 2011. [Google Scholar] [CrossRef]
- Ali, U.; Karim, K.J.B.A.; Buang, N.A. A Review of the Properties and Applications of Poly (Methyl Methacrylate) (PMMA). Polym. Rev. 2015, 55, 678–705. [Google Scholar] [CrossRef]
- Tomić, S.L.; Suljovrujić, E.H.; Filipović, J.M. Biocompatible and bioadhesive hydrogels based on 2-hydroxyethyl methacrylate, monofunctional poly(alkylene glycol)s and itaconic acid. Polym. Bull. 2006, 57, 691–702. [Google Scholar] [CrossRef] [Green Version]
- Darabi Mahboub, M.J.; Dubois, J.L.; Cavani, F.; Rostamizadeh, M.; Patience, G.S. Catalysis for the synthesis of methacrylic acid and methyl methacrylate. Chem. Soc. Rev. 2018, 47, 7703–7738. [Google Scholar] [CrossRef]
- Nagai, K. New developments in the production of methyl methacrylate. Appl. Catal. A Gen. 2001, 221, 367–377. [Google Scholar] [CrossRef]
- Tullo, A.H. In with the new. Chemical & Engineering News, 19 October 2009; 22–23. [Google Scholar]
- Wang, X.L.; Tooze, R.P.; Whiston, K.; Eastham, G.R. Process for the Carbonylation of Ethylene and Catalyst Systems for Use therein. Patent No. US6348621 19 February 2002. [Google Scholar]
- Bacteria Like the Taste of Syngas. Available online: https://corporate.evonik.com/en/pages/article.aspx?articleId=103071 (accessed on 17 February 2020).
- Haas, T.; Schiemann, Y.; Przybylski, D.; Rohwerder, T.; Mueller, R.H.; Harms, H. Biotechnological 2-hydroxyisobutyric Acid Production. Patent No. WO2013186340A1 24 December.
- Carlsson, M.; Habenicht, C.; Kam, L.C.; Antal, M.J.; Bian, N.; Cunningham, R.J.; Jones, M. Study of the Sequential Conversion of Citric to Itaconic to Methacrylic Acid in Near-critical and Supercritical Water. Ind. Eng. Chem. Res. 1994, 33, 1989–1996. [Google Scholar] [CrossRef]
- Lebeau, J.; Efromson, J.P.; Lynch, M.D. A Review of the Biotechnological Production of Methacrylic Acid. Front. Bioeng. Biotechnol. 2020, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Burk, M.J.; Burgard, A.P.; Osterhout, R.E.; Sun, J.; Pharkya, P. Microorganisms for Producing Methacrylic Acid and Methacrylate Esters and Methods Related thereto. Patent No. US9133487B2 15 September.
- Eastham, G.R.; Stephens, G.; Yiakoumetti, A. Process for the Biological Production of Methacrylic Acid and Derivatives thereof. Patent No. US 2018171368A1, 21 June 2018. [Google Scholar]
- Asano, Y.; Sato, E.; Yu, F.; Mizunashi, W. Method for producing methacrylic acid ester, and novel methacrylic acid ester synthase. Patent No. EP3115460A1 11 January.

| CA | Applications | Important Derivatives | Main Industrial Production Method | Microbe-Based Production Alternative |
|---|---|---|---|---|
| Malic | Detergents, food additives, pharmaceuticals, polyesters, solvents | Butanediol, THF, γ-butyrolactone | Hydration of maleic anhydride | Fermentation from renewable resources (Novozymes) using Aspergillus oryzae |
| Succinic | Hydrogenation of maleic anhydride, maleic or fumaric | Fermentation at pilot scale using E. coli (BioAmber and Myrant), S. cerevisiae (Reverdia) and Basfia succiniciproducens (Succinity) | ||
| Fumaric | Synthesis from butane-derived maleic | No industrial process established; production using fermentation with filamentous Fungi reported | ||
| 3-Hydroxypropionic | Superabsorbent, adhesives, surface coatings and paintings | Acrylates and 1,3-propanediol | Hydrolysis of 3-hydroxypropionitrile, hydrolysis of β-propiolactone and oxidation of 1,3- propanediol | No industrial process established; Fermentation from glucose or glycerol reported, mainly using E. coli and Klebsiella sp. |
| Aspartic | Nutritional supplement in food and animal feed, sweeteners | Polyaspartic, aspartic anhydride, amine butanediol, amine THF, amine butyrolactone | Amination of fumaric acid (enzymatic or with immobilized cells), fermentation with E. coli or Coreynebacterium glutamicum | - |
| Glucaric | Nylons and polyesters | Lactone, polyglucaric esters and amides | Chemical oxidation with nitric acid | Glucose fermentation using E. coli (Kalion) |
| Glutamic | Food additive, potential new polymers | 1,5-propanediol, 1,5-propanediacid, 5-amino, 1-butanol | Glucose fermentation using C. glutamicum | - |
| Itaconic | Rubber, solvents, acrylates, detergents, superabsorbents, drug delivery polymers, dental materials | MAA, MMA, polyesters, poly-itaconic acid polymers and styrene-butadiene | Glucose fermentation using Aspergillus terreus | - |
| Levulinic | Solvents, polymers, acrylates, herbicides, photodynamic therapy | 2-methyl-THF, levulinate esters, 1,4-pentanediol, β-acetoacrylate, lactones, δ-aminolevulinic, diphenolic acid | Acid hydrolysis of crystalline sugars or lignocellulosic residues | No industrial process established; reported production from glucose using E. coli or an undefined microbial consortia |
| Lactic | Biodegradable fibers in clothing, furniture and biomaterials | Lactate esters, propylene glycol, acrylates, poly-lactic acid | Fermentation of glucose from corn, cassava and sugarcane using Lactobacillus sp. | - |
| Acetic | Food additive, solvent, fibers, filters, cellulose plastics and resins (used in paints, adhesives, coatings and textiles) | Vinyl acetate, acetic anhydride, acetate esters, monochloroacetic acid | Methanol carbonylation; liquid-phase oxidation of aliphatic hydrocarbons; fermentation using acetic acid bacterial (mainly in the vinegar industry) | - |
| Citric | Acidulant, preservative, emulsifier, flavoring additive, sequestrant and buffering agent | - | Starch or glucose fermentation using A. niger | - |
| Gluconic | Cleaning and construction industries, food additives including prebiotics | Glucono-lactone, sodium gluconate | Oxidation of glucose; Glucose fermentation using A. niger | - |
| Adipic | Nylons and polyesters, plasticizers and lubricants | Esters for polymerization (PVC) | Synthesis from benzene | Fermentation of fatty acid rich-feedstocks (Verdezyne) or glucose (BioAmber) using yeasts (at a pilot scale) |
| Acrylic | Various coatings (decorative, industrial, drug tablets, clothes), adhesives, polishes, carpet backing compounds | Methyl acrylate, ethyl acrylate, butyl acrylate and 2-ethylhexyl acrylate, polyacrylates | Oxidation of propene | Fermentation from renewable feedstocks (Arkema); Fermentation of dextrose and sucrose-based feedstocks (OPXBio and Dow) using E. coli (at pilot-scale) |
| Glycolic | Tanning and dyeing agent for textiles, packaging materials | Polyglycolate, polyglicoside, butyl-glycolate | Catalysis from CO2 and formaldehyde and hydrolysis of chloroacetic acid | No industrial process established; reported production using E. coli (natural producer) or the yeasts S. cerevisiae and Kluveromyces lactis (engineered producers) |
| Muconic | Plastics industry (automotive and packaging applications), synthetic fibers for textiles or industry (mainly nylon) and food acidifying agent | Adipic, terphthalic acid and trimellitic acid, caprolactam | Catalytic oxidation of cyclohexanol or cyclohexanol/cyclohexanone mixtures | No industrial process established; reported production from degradation of benzene-like xenobiotics and from glucose fermentation using Pseudomonas spp. (natural producers) and E. coli and S. cerevisiae (engineered hosts) |
| Glucaric acid | Titer | Ref | |||||||||||||||||
| Myo-inositol synthase (1) 5.5.1.4 | Myo-inositol oxygenase (2) 1.13.99.1 | Glucuronic acid dehydrogenase (3); 1.1.1.305 | |||||||||||||||||
| E. coli | Scaffolded—S. cerevisiae Ino1 | Scaffolded—M. musculus MIOX | Scaffolded—P. syringae Udh | 2.5 g/L | [15] | ||||||||||||||
| S. cerevisiae | Endogenous Ino1 | Stabilized Arabidopsis thaliana MIOX | P. syringae Udh | 6 g/L | [18] | ||||||||||||||
| Muconic acid | Titer | Ref | |||||||||||||||||
| From Dihydroshikimate | E. coli | Dihydroshikimate Hydratase (1); 4.2.1.118 Klebsiella pneumoniae aroZ | Protocatechuate decarboxylase (2) 4.1.1.63 K. pneumoniae aroY | Catechol 1.2-dioxygenase (3) 1.13.11.1 Acinetobacter calcoaceticus CatA | 38.6 g/L ● | [29] | |||||||||||||
| S. cerevisiae | P. anserina aroZ | Talaromyces atroroseus GDC1 | A. radioresistens CatA | 1.24 g/L | [28] | ||||||||||||||
| From chorismate | E. coli | Isochorismate synthase (8); 5.4.4.2 Endogenous EntC | Isochorismate pyruvate lyase (11); 4.2.99.21 P. fluorescens PchB | Salicylate monoxygenase (12); 1.14.13.1 P. putida nahG | Catechol 1.2-dioxygenase (3) 1.13.11.1 P. putida CatA | 1.5 g/L | [30] | ||||||||||||
| From anthranilate | E. coli | Anthranilate 1.2-dioxygenase (14) 1.14.12.1 P. aeruginosa AntABC | Catechol 1.2-dioxygenase (3) 1.13.11.1 P. putida CatA | 389.96 mg/L | [30] | ||||||||||||||
| From tyrosine | E. coli | Tyrosine phenol lyase (15);4.1.99 Citrobacter brakii tutA | Phenol hydrolyase (7)1.14.13.7 P. steutzeri PhKLMOP | Catechol 1.2-dioxygenase (3) 1.13.11.1 P. putida CatA | 186 mg/L | [31] | |||||||||||||
| Adipic acid | Titer | Ref | |||||||||||||||||
| Reverse adipate route | S. cerevisiae | 3-Oxoadipyl-CoA thiolase (1); 2.3.1.174 T. fusca Tfu_0875 | 3-Hydroxyadipyl-CoA dehydrogenase (2) 1.1.1.35 T. fusca Tfu_2399 | 2,3-Dehydroadipyl-CoA hydratase (3); 4.2.1.17 T. fusca Tfu_0067 | Adipyl-CoA dehydrogenase (4);1.1.1.35 T. fusca Tfu_1647 | Adipyl-CoA thioesterase (5) T. fusca Tfu_2577 and 2576 | 3.83 mg/L | [32] | |||||||||||
| Reverse β-oxidation followed by ω-reduction * | E. coli | 3-ketoacyl-CoA thiolase (6)2.3.1.16 C. necator BktB | Trans-enoyl-CoA reductase (7) 1.3.1.44 E. gracilis Ter | ω-Hydroxylase (8) 1.14.15.3 P. putida AlkBGT | Alcohol dehydrogenase (9) * Acinetobacter spp. ChnD | Aldehyde dehydrogenase (10) * Acinetobacter spp. ChnE | - | [33] | |||||||||||
| 2-oxopimelic route | E. coli | 2-oxoglutaric elongation to 2-oxopimelic (11) A. vinelandii nifV (2.3.3.14) and M. aeolicus Nankai AksD (4.2.1.114), AksE (4.2.1.33), and AksF * | 2-Oxopimelic decarboxylase (12)4.1.1.72 Lactococcus lactis KdcA | Adipic semi-aldehyde oxidation (13) Unknown endogenous enzyme | 0.3 g/L | [34] | |||||||||||||
| From muconic acid | S. cerevisiae | DHS hydratase 4.2.1.118 P. anserina aroZ | Protocatechuate decarboxylase; 4.1.1.63 Enterobacter cloacae aroY | Catechol 1,2-dioxygenase1.13.11.1 C. albicans HQD2 | Enoate reductase(22)1.3.1.31 Bacillus coagulans MAR (MAR-BC) | 2.6 mg/L | [35] | ||||||||||||
| Acrylic acid | Titer | Ref | |||||||||||||||||
| From glycerol | E. coli | Glycerol-3-P phosphatase3.1.3.21 S. cerevisiae Gpp2 | Glycerol Dehydratase 4.2.1.30 K. pneumoniae DhaB | Aldehyde Dehydrogenase 1.2.1.16 C. necator GabD4 | CoA transferase 2.8.3.8 C. necator YdiF | CoA dehydratase* A. flavithermus Aflv_0566 | 0.12 g/L | [36] | |||||||||||
| Levulinic acid | Titer | Ref | |||||||||||||||||
| From 3-oxoadipic | E. coli | Succinyl-CoA transferase 2.8.3.18 C.kluyveri Cat1 | β-ketoadipyl-CoA thiolase (1) 2.3.1.174 Endogenous PaaJ | 3-Oxoadipyl-CoA transferase (2); 2.8.3.6 P. putida PcaIJ | 3-Oxoadipic acid decarboxylase (3);4.1.1.4 C. acetobutylicum Adc | 159 mg/L | [37] | ||||||||||||
| PCA synthesis (4) P. putida Fcs (6.2.1.34), Ech (4.1.2.61), Vdh (1.2.1.67), VanAb (1.14.13.82), PobA (1.14.13.2) | Dearomatization pathway (5) P. putida PcaGH (1.13.11.3), PcaB (5.5.1.2), PcaC (4.1.1.44), PcaD (3.1.1.24) | 3-Oxoadipic Acid decarboxylase (3); 4.1.1.4 C. acetobutylicum Adc | 455 mg/L | [38] | |||||||||||||||
| Methacrylic acid | Titer | Ref | |||||||||||||||||
| Through Isobutyryl-CoA | E. coli | Isobutyryl-CoA synthase (1) 6.2.1.3 P. chlorpraphis AcsA | Acyl-CoA oxidase (2); 1.3.3.6 A. thaliana ACX4 | Hydroxybenzoyl-CoA thioesterase (3); 3.1.2.23 Arthrobacter spp. 4HBT | ~250 µM | [39] | |||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vila-Santa, A.; Mendes, F.C.; Ferreira, F.C.; Prather, K.L.J.; Mira, N.P. Implementation of Synthetic Pathways to Foster Microbe-Based Production of Non-Naturally Occurring Carboxylic Acids and Derivatives. J. Fungi 2021, 7, 1020. https://doi.org/10.3390/jof7121020
Vila-Santa A, Mendes FC, Ferreira FC, Prather KLJ, Mira NP. Implementation of Synthetic Pathways to Foster Microbe-Based Production of Non-Naturally Occurring Carboxylic Acids and Derivatives. Journal of Fungi. 2021; 7(12):1020. https://doi.org/10.3390/jof7121020
Chicago/Turabian StyleVila-Santa, Ana, Fernão C. Mendes, Frederico C. Ferreira, Kristala L. J. Prather, and Nuno P. Mira. 2021. "Implementation of Synthetic Pathways to Foster Microbe-Based Production of Non-Naturally Occurring Carboxylic Acids and Derivatives" Journal of Fungi 7, no. 12: 1020. https://doi.org/10.3390/jof7121020






