Effects of Laccaria bicolor on Gene Expression of Populus trichocarpa Root under Poplar Canker Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant and Fungal Materials
2.2. L. bicolor–P. trichocarpa–B. dothidea Coculturing in Two Sandwich Culture Systems
2.3. Estimation of Peroxidase (POD) and L-phenylalanine Ammonia-Lyase (PAL)
2.4. Content of Malondialdehyde (MDA)
2.5. RNA Extraction, Transcriptome Sequencing, and Bioinformatics Analysis
2.6. The Quantitative Real-Time PCR (qRT-PCR)
2.7. Statistical Analysis
3. Results
3.1. Enzyme Activity Analysis
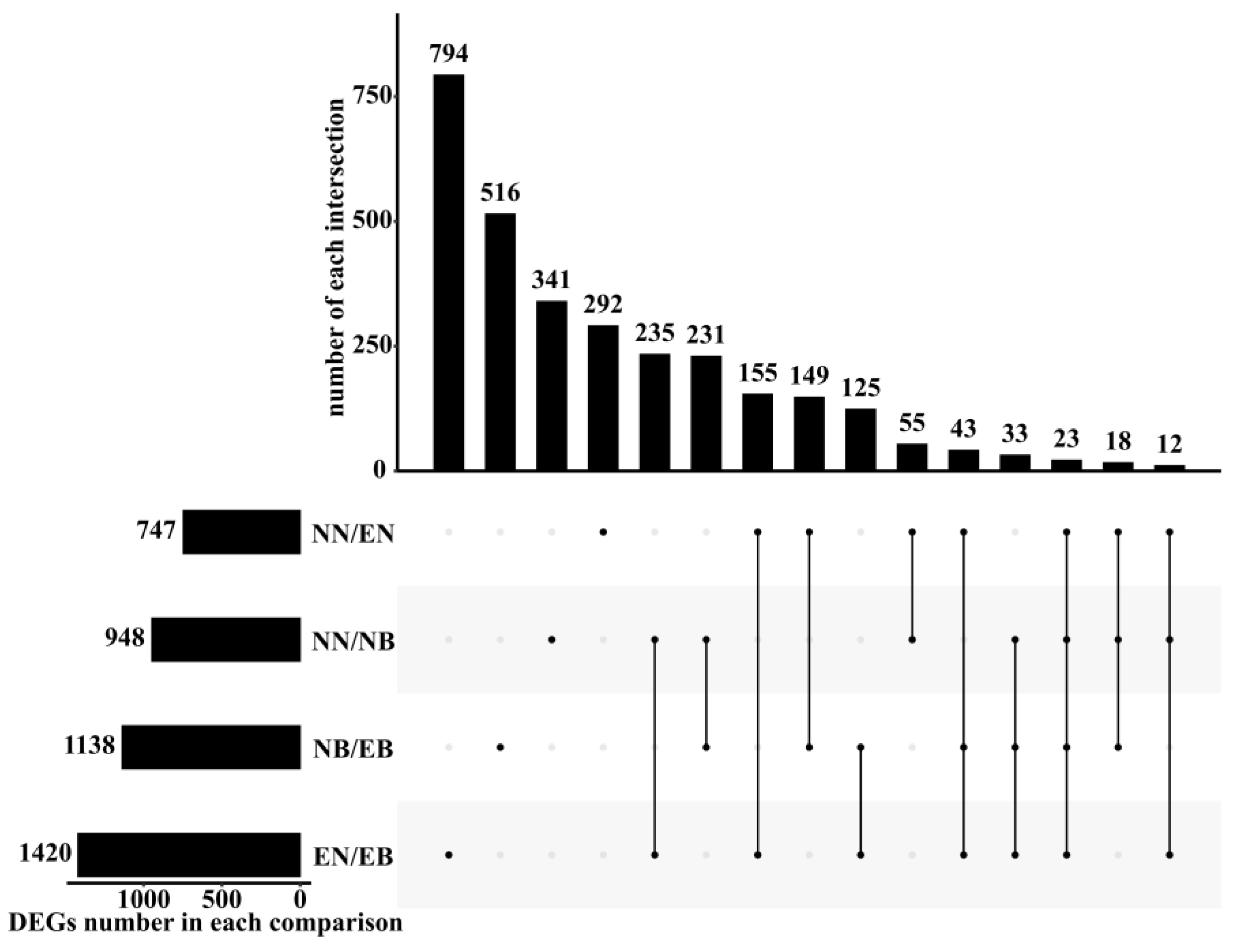
3.2. Analysis of Differentially Expressed Genes between Different Treatments
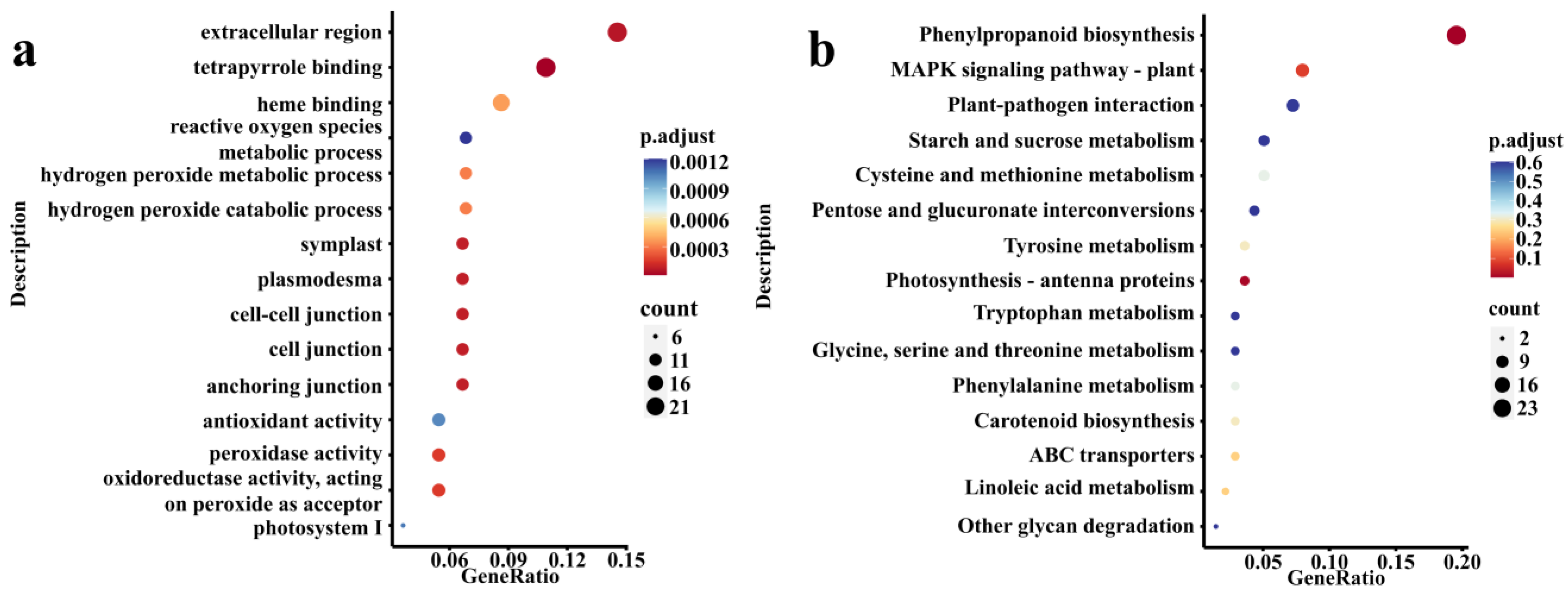
3.3. DEGs Enrichment Analysis
3.4. Analysis of Gene Expression Patterns Related to Signal Transduction Induced by L. bicolor
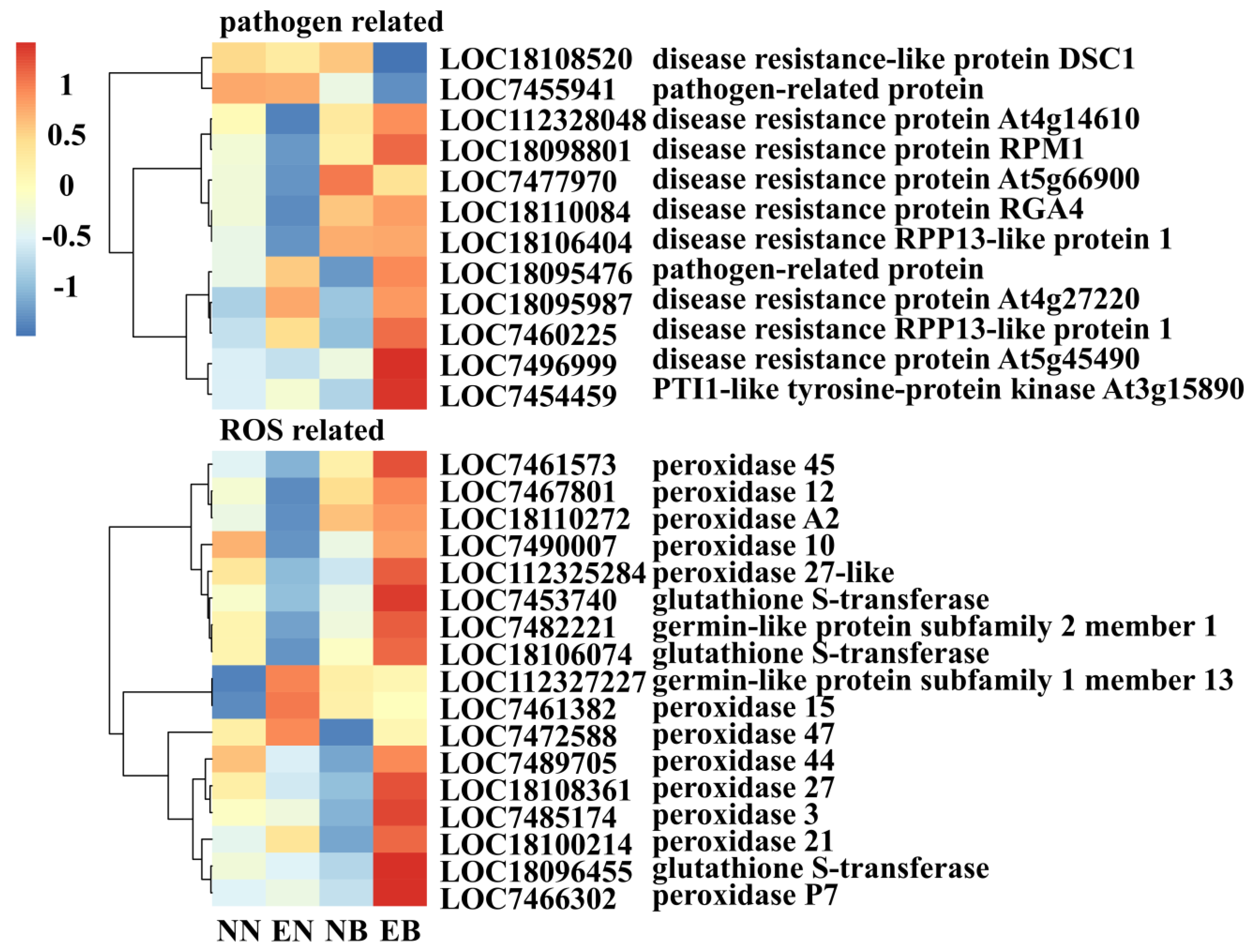
3.5. Analysis of the Expression Pattern of Disease Resistance-Related and Antioxidant-Related DEGs Induced by L. bicolor
3.6. The qRT-PCR Verification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xing, J.; Li, P.; Zhang, Y.; Li, J.; Liu, Y.; Lachenbruch, B.; Su, X.; Zhao, J. Fungal pathogens of canker disease trigger canopy dieback in poplar saplings by inducing functional failure of the phloem and cambium and carbon starvation in the xylem. Physiol. Mol. Plant Pathol. 2020, 112, 101523. [Google Scholar] [CrossRef]
- Abelleira, A.; Moura, L.; Aguín, O.; Salinero, C. First Report of Lonsdalea populi Causing Bark Canker Disease on Poplar in Portugal. Plant Dis. 2019, 103, 2121. [Google Scholar] [CrossRef]
- Niemczyk, M.; Thomas, B.R. Growth parameters and resistance to Sphaerulina musiva-induced canker are more important than wood density for increasing genetic gain from selection of Populus spp. hybrids for northern climates. Ann. For. Sci. 2020, 77, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Tabima, J.F.; Sondreli, K.L.; Kerio, S.; Feau, N.; Sakalidis, M.L.; Hamelin, R.C.; LeBoldus, J.M. Population Genomic Analyses Reveal Connectivity via Human-Mediated Transport across Populus Plantations in North America and an Undescribed Subpopulation of Sphaerulina musiva. Mol. Plant-Microbe Interact. 2020, 33, 189–199. [Google Scholar] [CrossRef]
- Zhong, Z.; Gao, Y. A brief report on the resistance of different poplar varieties to poplar vesicular canker. For. Sci. Technol. 1981, 1, 25–26. [Google Scholar] [CrossRef]
- Babu, S.; Bidyarani, N.; Chopra, P.; Monga, D.; Kumar, R.; Prasanna, R.; Kranthi, S.; Saxena, A.K. Evaluating microbe-plant interactions and varietal differences for enhancing biocontrol efficacy in root rot disease challenged cotton crop. Eur. J. Plant Pathol. 2015, 142, 345–362. [Google Scholar] [CrossRef]
- Bahram, M.; Põlme, S.; Kõljalg, U.; Tedersoo, L. A single European aspen (Populus tremula) tree individual may potentially harbour dozens of Cenococcum geophilum ITS genotypes and hundreds of species of ectomycorrhizal fungi. FEMS Microbiol. Ecol. 2011, 75, 313–320. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Li, Z.; Wu, F.; Tang, M. Comparative photochemistry activity and antioxidant responses in male and female Populus cathayana cuttings inoculated with arbuscular mycorrhizal fungi under salt. Sci. Rep. 2016, 6, 37663. [Google Scholar] [CrossRef]
- Cui, J.Q.; Sun, H.B.; Sun, M.B.; Liang, R.T.; Jie, W.G.; Cai, B.Y. Effects of Funneliformis mosseae on Root Metabolites and Rhizosphere Soil Properties to Continuously-Cropped Soybean in the Potted-Experiments. Int. J. Mol. Sci. 2018, 19, 2160. [Google Scholar] [CrossRef] [Green Version]
- Leifheit, E.F.; Veresoglou, S.D.; Lehmann, A.; Morris, E.K.; Rillig, M.C. Multiple factors influence the role of arbuscular mycorrhizal fungi in soil aggregation—A meta-analysis. Plant Soil 2013, 374, 523–537. [Google Scholar] [CrossRef]
- Ortiz, N.; Armada, E.; Duque, E.; Roldan, A.; Azcon, R. Contribution of arbuscular mycorrhizal fungi and/or bacteria to enhancing plant drought tolerance under natural soil conditions: Effectiveness of autochthonous or allochthonous strains. J. Plant Physiol. 2015, 174, 87–96. [Google Scholar] [CrossRef]
- Ganugi, P.; Masoni, A.; Pietramellara, G.; Benedettelli, S. A Review of Studies from the Last Twenty Years on Plant–Arbuscular Mycorrhizal Fungi Associations and Their Uses for Wheat Crops. Agronomy 2019, 9, 840. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Feng, X.; Gao, P.; Li, Y.; Christensen, M.J.; Duan, T. Arbuscular mycorrhiza fungi increased the susceptibility of Astragalus adsurgens to powdery mildew caused by Erysiphe pisi. Mycology 2018, 9, 223–232. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, H.; Tang, M. Prior contact of Pinus tabulaeformis with ectomycorrhizal fungi increases plant growth and survival from damping-off. New For. 2017, 48, 855–866. [Google Scholar] [CrossRef]
- Zhang, Y.; Ye, J.; Zhao, Y.; Ma, S. Fermentation conditions of Xerocomus chrysenteron and its control effect on poplar canker disease. J. For. Environ. 2016, 36, 397–403. [Google Scholar] [CrossRef]
- Lu, C.C.; Guo, N.; Yang, C.; Sun, H.B.; Cai, B.Y. Transcriptome and metabolite profiling reveals the effects of Funneliformis mosseae on the roots of continuously cropped soybeans. BMC Plant Biol. 2020, 20, 479. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Y.; Chen, Q.; Huang, X.; Huang, X. Cloning, Characterization and Expression of a Phenylalanine Ammonialyase Gene (M-PAL) from Plantain (Musa ABB cv. Dongguandajiao). J. Trop. Subtrop. Bot. 2007, 15, 421–427. [Google Scholar] [CrossRef]
- Li, D.; Chen, Z.; Nie, Y. Antifungal substances producted by a high-yielding mutant of Bs−916 and their effects inducing-resistance on rice plant. Acta Phytopathol. Sin. 2008, 38, 192–198. [Google Scholar] [CrossRef]
- Kilic-Ekici, O.; Yuen, G.Y. Induced resistance as a mechanism of biological control by Lysobacter enzymogenes strain C3. Phytopathology 2003, 93, 1103–1110. [Google Scholar] [CrossRef] [Green Version]
- Zhan, W.; Liu, H.; Tang, M. Physiological and Biochemical Mechanism of Mycorrhizal Fungi Improving the Resistance of Poplar to Canker Disease. Acta Bot. Boreali-Occident. Sin. 2010, 30, 2437–2443. [Google Scholar]
- Morcillo, R.J.; Zhao, A.; Tamayo-Navarrete, M.I.; García-Garrido, J.M.; Macho, A.P. Tomato Root Transformation Followed by Inoculation with Ralstonia Solanacearum for Straightforward Genetic Analysis of Bacterial Wilt Disease. J. Vis. Exp. 2020, 157, e60302. [Google Scholar] [CrossRef]
- Liu, J.; Maldonado-Mendoza, I.; Lopez-Meyer, M.; Cheung, F.; Town, C.D.; Harrison, M.J. Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J. 2007, 50, 529–544. [Google Scholar] [CrossRef]
- Campos-Soriano, L.; Garcia-Martinez, J.; Segundo, B.S. The arbuscular mycorrhizal symbiosis promotes the systemic induction of regulatory defence-related genes in rice leaves and confers resistance to pathogen infection. Mol. Plant Pathol. 2012, 13, 579–592. [Google Scholar] [CrossRef]
- Grant, M.; Lamb, C. Systemic immunity. Curr. Opin. Plant Biol. 2006, 9, 414–420. [Google Scholar] [CrossRef]
- Lee, S.; Rojas, C.M.; Ishiga, Y.; Pandey, S.; Mysore, K.S. Arabidopsis heterotrimeric G-proteins play a critical role in host and nonhost resistance against Pseudomonas syringae pathogens. PLoS ONE 2013, 8, e82445. [Google Scholar] [CrossRef] [Green Version]
- Bundo, M.; Coca, M. Enhancing blast disease resistance by overexpression of the calcium-dependent protein kinase OsCPK4 in rice. Plant Biotechnol. J. 2016, 14, 1357–1367. [Google Scholar] [CrossRef] [Green Version]
- Martos, G.G.; Teran Mdel, M.; Diaz Ricci, J.C. The defence elicitor AsES causes a rapid and transient membrane depolarization, a triphasic oxidative burst and the accumulation of nitric oxide. Plant Physiol. Biochem. 2015, 97, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, D.; Zhang, H.; Hong, Y.; Huang, L.; Liu, S.; Li, X.; Ouyang, Z.; Song, F. Tomato histone H2B monoubiquitination enzymes SlHUB1 and SlHUB2 contribute to disease resistance against Botrytis cinerea through modulating the balance between SA- and JA/ET-mediated signaling pathways. BMC Plant Biol. 2015, 15, 252. [Google Scholar] [CrossRef] [Green Version]
- Liao, W.; Ji, L.; Wang, J.; Chen, Z.; Ye, M.; Ma, H.; An, X. Identification of glutathione S-transferase genes responding to pathogen infestation in Populus tomentosa. Funct. Integr. Genom. 2014, 14, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yang, F.; Feng, J.; Wang, Y.; Lachenbruch, B.; Wang, J.; Wan, X. Genome-Wide Constitutively Expressed Gene Analysis and New Reference Gene Selection Based on Transcriptome Data: A Case Study from Poplar/Canker Disease Interaction. Front. Plant Sci. 2017, 8, 1876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Block, M. Factors Influencing the Tissue Culture and the Agrobacterium tumefaciens-Mediated Transformation of Hybrid Aspen and Poplar Clones. Plant Physiol. 1990, 93, 1110–1116. [Google Scholar] [CrossRef] [Green Version]
- Felten, J.; Kohler, A.; Morin, E.; Bhalerao, R.P.; Palme, K.; Martin, F.; Ditengou, F.A.; Legué, V.r. The Ectomycorrhizal Fungus Laccaria bicolor Stimulates Lateral Root Formation in Poplar and Arabidopsis through Auxin Transport and Signaling. Plant Physiol. 2009, 151, 1991–2005. [Google Scholar] [CrossRef] [Green Version]
- Wanwaen, S.; Youpensuk, S. Cultivation of Amanita princeps and Gyrodon suthepensis for Mycorrhizations with Castanopsis acuminatissima and their Effects on the Host Plants. Int. J. Agric. Biol. 2019, 22, 195–200. [Google Scholar] [CrossRef]
- Li, Y.; Feng, Y.; Lu, Q.; Yan, D.; Liu, Z.; Zhang, X. Comparative Proteomic Analysis of Plant–Pathogen Interactions in Resistant and Susceptible Poplar Ecotypes Infected with Botryosphaeria dothidea. Phytopathology 2019, 109, 2009–2021. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Kao, C.H. Enhanced peroxidase activity in rice leaves in response to excess iron, copper and zinc. Plant Sci. 2000, 158, 71–76. [Google Scholar] [CrossRef]
- Sreelakshmi, Y.; Sharma, R. Differential regulation of phenylalanine ammonia lyase activity and protein level by light in tomato seedlings. Plant Physiol. Biochem. 2008, 46, 444–451. [Google Scholar] [CrossRef]
- Kramer, G.F.; Norman, H.A.; Krizek, D.T.; Mirecki, R.M. Influence of UV-B radiation on polyamines, lipid peroxidation and membrane lipids in cucumber. Phytochemistry 1991, 30, 2101–2108. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wixon, J.; Kell, D. The Kyoto Encyclopedia of Genes and Genomes—KEGG. Yeast 2000, 17, 48–55. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Zhang, H.J.; Zhang, Y.X.; Liu, Y.Q.; Zhang, H.Q.; Tang, M. Arbuscular mycorrhizal fungi alter carbohydrate distribution and amino acid accumulation in Medicago truncatula under lead stress. Environ. Exp. Bot. 2020, 171, 103950. [Google Scholar] [CrossRef]
- Jiang, X. The Spatial and Temporal Expression of Thaumatin-Like Protein Coding Genes Induced by Tress Stem Canker Pathogen in Populus trichocarpa. Master’s Thesis, Hebei Agricultural University, Baoding, China, 2012. [Google Scholar]
- Su, X.; Fan, B.; Yuan, L.; Cui, X.; Lu, S. Selection and Validation of Reference Genes for Quantitative RT-PCR Analysis of Gene Expression in Populus trichocarpa. Chin. Bull. Bot. 2013, 48, 507–518. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Andersen, E.J.; Ali, S.; Byamukama, E.; Yen, Y.; Nepal, M.P. Disease Resistance Mechanisms in Plants. Genes 2018, 9, 339. [Google Scholar] [CrossRef] [Green Version]
- Mekapogu, M.; Jung, J.A.; Kwon, O.K.; Ahn, M.S.; Song, H.Y.; Jang, S. Recent Progress in Enhancing Fungal Disease Resistance in Ornamental Plants. Int. J. Mol. Sci. 2021, 22, 7956. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen Recognition and Inflammatory Signaling in Innate Immune Defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef] [Green Version]
- Bernaola, L.; Cosme, M.; Schneider, R.W.; Stout, M. Belowground Inoculation With Arbuscular Mycorrhizal Fungi Increases Local and Systemic Susceptibility of Rice Plants to Different Pest Organisms. Front. Plant Sci. 2018, 9, 747. [Google Scholar] [CrossRef]
- Zhang, R.Q.; Tang, M.; Chen, H.; Tian, Z.Q. Effects of ectomycorrhizal fungi on damping-off and induction of pathogenesis-related proteins in Pinus tabulaeformis seedlings inoculated with Amanita vaginata. For. Pathol. 2011, 41, 262–269. [Google Scholar] [CrossRef]
- Morales, J.; Kadota, Y.; Zipfel, C.; Molina, A.; Torres, M.A. The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J. Exp. Bot. 2016, 67, 1663–1676. [Google Scholar] [CrossRef] [Green Version]
- Emma, W.G.; Olusola, O.S.; Simeon, O.K. The molecular initiation and subsequent acquisition of disease resistance in plants. Afr. J. Biotechnol. 2003, 2, 26–32. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.; Yang, G. Research Advances in the Mechanism and Signal Transduction of Plant Disease Resistance. Biotechnol. Bull. 2016, 32, 109–117. [Google Scholar] [CrossRef]
- Zaretsky, M.; Sitrit, Y.; Mills, D.; Roth-Bejerano, N.; Kagan-Zur, V. Differential expression of fungal genes at preinfection and mycorrhiza establishment between Terfezia boudieri isolates and Cistus incanus hairy root clones. New Phytol. 2006, 171, 837–846. [Google Scholar] [CrossRef]
- Sitrit, Y.; Roth-Bejerano, N.; Kagan-Zur, V.; Turgeman, T. Pre-symbiotic interactions between the desert truffle Terfezia boudieri and its host plant Helianthemum sessiliflorum. In Desert Truffles; Springer: Berlin/Heidelberg, Germany, 2014; Volume 38, pp. 81–92. [Google Scholar]
- Turgeman, T.; Lubinsky, O.; Roth-Bejerano, N.; Kagan-Zur, V.; Kapulnik, Y.; Koltai, H.; Zaady, E.; Ben-Shabat, S.; Guy, O.; Lewinsohn, E.; et al. The role of pre-symbiotic auxin signaling in ectendomycorrhiza formation between the desert truffle Terfezia boudieri and Helianthemum sessiliflorum. Mycorrhiza 2016, 26, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, M.; Neve, J.; Kepinski, S. Defining auxin response contexts in plant development. Curr. Opin. Plant Biol. 2010, 13, 12–20. [Google Scholar] [CrossRef]
- Swarup, R.; Peret, B. AUX/LAX family of auxin influx carriers—An overview. Front. Plant Sci. 2012, 3, 225. [Google Scholar] [CrossRef] [Green Version]
- Kazan, K.; Manners, J.M. Linking development to defense: Auxin in plant-pathogen interactions. Trends Plant Sci. 2009, 14, 373–382. [Google Scholar] [CrossRef]
- Zhong, T. Cloning and Resistance Mechanism of Genes for Grey Leaf Spot and Stalk Rot Resistance in Maize. Ph.D. Thesis, China Agricultural University, Beijing, China, 2019. [Google Scholar]
- Ding, X.; Cao, Y.; Huang, L.; Zhao, J.; Xu, C.; Li, X.; Wang, S. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 2008, 20, 228–240. [Google Scholar] [CrossRef] [Green Version]
- Pons, S.; Fournier, S.; Chervin, C.; Becard, G.; Rochange, S.; Frei Dit Frey, N.; Puech Pages, V. Phytohormone production by the arbuscular mycorrhizal fungus Rhizophagus irregularis. PLoS ONE 2020, 15, e0240886. [Google Scholar] [CrossRef]
- Jiang, C. Transcriptome Analysis of the Auxin Key Genes in Populus davidiana × P. alba var. pyramidalis Response to Trichoderma asperellum and Alternaria alternata. Master’s Thesis, Northeast Forestry University, Harbin, China, 2017. [Google Scholar]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y. Identification of Resistance of Sesame Varieties to Stem Rot and Functional Analysis of SiPYL4 and SiTLP genes. Master’s Thesis, Zhengzhou University, Zhengzhou, China, 2019. [Google Scholar]
- Miller, G.; Shulaev, V.; Mittler, R. Reactive oxygen signaling and abiotic stress. Physiol. Plant 2008, 133, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B.; Sauter, M. Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. Plant Cell 2009, 21, 184–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halliwell, B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gechev, T.S.; Van Breusegem, F.; Stone, J.M.; Denev, I.; Laloi, C. Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 2006, 28, 1091–1101. [Google Scholar] [CrossRef]
- Neill, S.J.; Desikan, R.; Clarke, A.; Hurst, R.D.; Hancock, J.T. Hydrogen peroxide and nitric oxide as signalling molecules in plants. J. Exp. Bot. 2002, 53, 1237–1247. [Google Scholar] [CrossRef]
- Yoshioka, H.; Numata, N.; Nakajima, K.; Katou, S.; Kawakita, K.; Rowland, O.; Jones, J.D.; Doke, N. Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 2003, 15, 706–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, X.; Qi, J.; Dou, W.; Chen, S.; He, Y.; Li, Q. Identification of Rboh Family and the Response to Hormone and Citrus Bacterial Canker in Citrus. Sci. Agric. Sin. 2020, 53, 4189–4203. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Pati, P.K. Analysis of cis-acting regulatory elements of Respiratory burst oxidase homolog (Rboh) gene families in Arabidopsis and rice provides clues for their diverse functions. Comput. Biol. Chem. 2016, 62, 104–118. [Google Scholar] [CrossRef]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY Transcription Factor Family in Model Plants and Crops. Crit. Rev. Plant Sci. 2018, 36, 311–335. [Google Scholar] [CrossRef]
- Bittner-Eddy, P.D.; Crute, I.R.; Holub, E.B.; Beynon, J.L. RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 2000, 21, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Fan, H.; Li, L.; Hu, B.; Liu, H.; Liu, Z. Genome-wide Identification and Expression Analyses of RPP13-like Genes in Barley. BioChip J. 2018, 12, 102–113. [Google Scholar] [CrossRef]
- Chao, J.; Jin, J.; Wang, D.; Han, R.; Zhu, R.; Zhu, Y.; Li, S. Cytological and transcriptional dynamics analysis of host plant revealed stage-specific biological processes related to compatible rice-Ustilaginoidea virens interaction. PLoS ONE 2014, 9, e91391. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Gao, H.; Liang, Y.; Cao, Y. Full-length transcriptome analysis of asparagus roots reveals the molecular mechanism of salt tolerance induced by arbuscular mycorrhizal fungi. Environ. Exp. Bot. 2021, 185, 104402. [Google Scholar] [CrossRef]


| Treatment | POD | PAL | MDA |
|---|---|---|---|
| NN | 1054.02 ± 325.66 c | 5678.41 ± 1428.19 c | 14.30 ± 3.90 d |
| EN | 3516.56 ± 484.89 ab | 16600.94 ± 779.80 ab | 35.49 ± 5.53 c |
| NB | 2330.89 ± 465.16 bc | 10399.50 ± 2842.16 bc | 128.66 ± 11.48 a |
| EB | 5252.68 ± 1370.74 a | 23236.47 ± 5403.70 a | 60.68 ± 8.61 b |
| L. bicolor | ** | ** | ** |
| B. dothidea | * | * | ** |
| L. bicolor & B. dothidea | ns | ns | ** |
| Comparisons (Control/Treatment) | All DEGs | Up Regulated DEGs | Down Regulated DEGs |
|---|---|---|---|
| NN/EN | 747 | 297 | 450 |
| NN/NB | 948 | 734 | 214 |
| EN/EB | 1420 | 736 | 684 |
| NB/EB | 1138 | 288 | 850 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, F.; Wang, Y.; Tang, M. Effects of Laccaria bicolor on Gene Expression of Populus trichocarpa Root under Poplar Canker Stress. J. Fungi 2021, 7, 1024. https://doi.org/10.3390/jof7121024
Dong F, Wang Y, Tang M. Effects of Laccaria bicolor on Gene Expression of Populus trichocarpa Root under Poplar Canker Stress. Journal of Fungi. 2021; 7(12):1024. https://doi.org/10.3390/jof7121024
Chicago/Turabian StyleDong, Fengxin, Yihan Wang, and Ming Tang. 2021. "Effects of Laccaria bicolor on Gene Expression of Populus trichocarpa Root under Poplar Canker Stress" Journal of Fungi 7, no. 12: 1024. https://doi.org/10.3390/jof7121024






