Multilocus Phylogeography of the Tuber mesentericum Complex Unearths Three Highly Divergent Cryptic Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Specimens
2.2. Molecular Analyses
2.2.1. DNA Extraction, Amplification, and Sequencing of Voucher Specimens
2.2.2. ITS Amplification and Sequencing of Historical Voucher Specimens
2.2.3. Phylogenetic Analyses
2.3. Morphological Analyses
3. Results
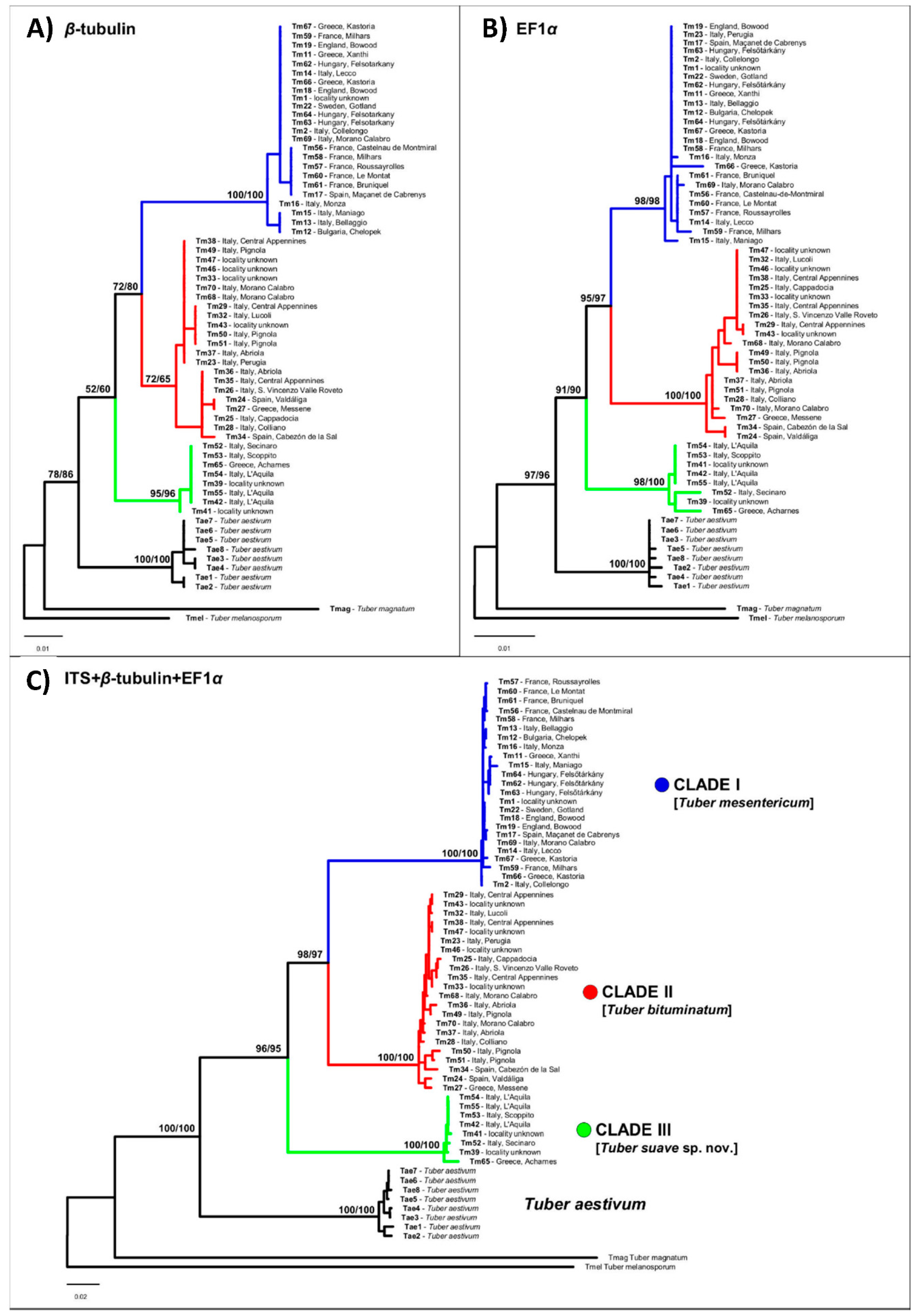
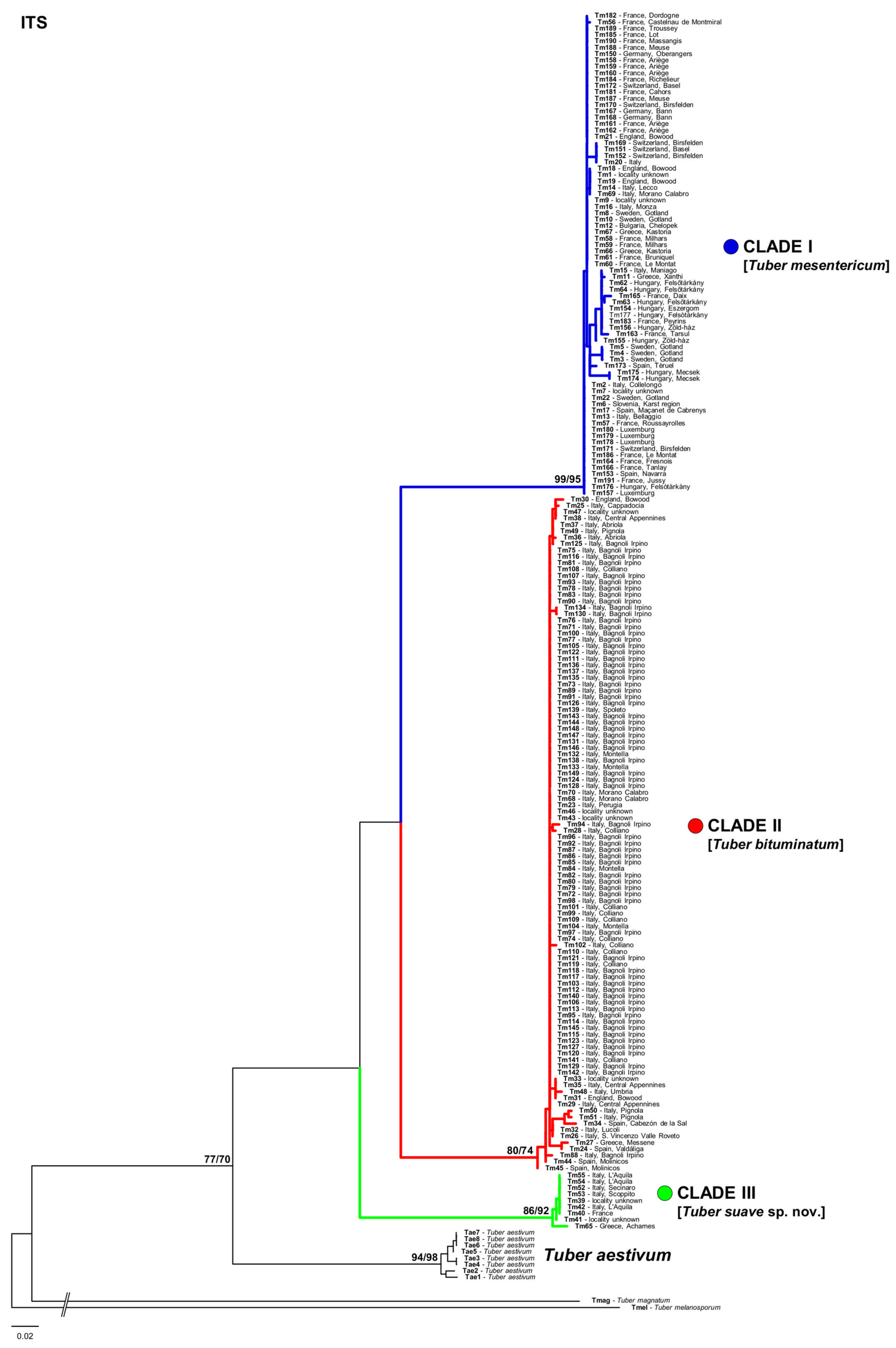
3.1. Phylogenetic Analyses
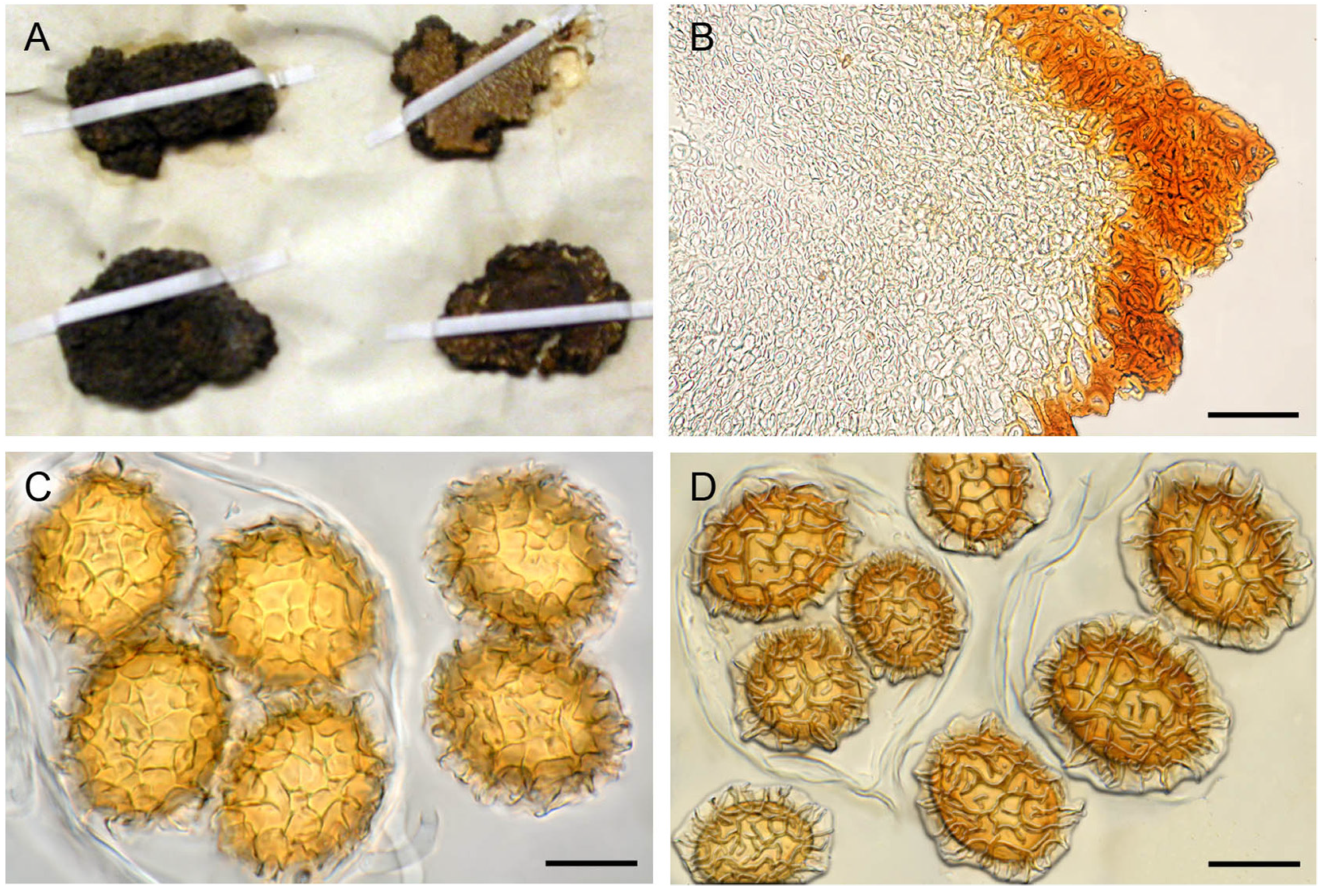
3.2. Ascoma Morphology
4. Discussion
5. Taxonomy
5.1. Tuber mesentericum Vittad.
5.2. Tuber bituminatum Berk. & Broome
5.3. Tuber suave Pacioni & M. Leonardi, sp. nov.
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gryndler, M. True truffle host diversity. In True Truffle (Tuber spp.) in the World, Soil Biology 47; Zambonelli, A., Iotti, M., Murat, C., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 267–281. [Google Scholar]
- Zambonelli, A.; Iotti, M.; Hall, I.R. Current status of truffle cultivation: Recent results and future perspectives. Ital. J. Mycol. 2015, 44, 31–40. [Google Scholar]
- Percudani, R.; Trevisi, A.; Zambonelli, A.; Ottonello, S. Molecular phylogeny of truffles (Pezizales: Terfeziaceae, Tuberaceae) derived from nuclear rDNA sequence analysis. Mol. Phylog. Evol. 1999, 13, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tan, Z.M.; Zhang, D.C.; Murat, C.; Jeandroz, S.; Le Tacon, F. Phylogenetic relationships between Tuber pseudoexcavatum, a Chinese truffle, and other Tuber species based on parsimony and distance analysis of four different gene sequences. FEMS Microbiol. Lett. 2006, 259, 269–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeandroz, S.; Murat, C.; Wang, Y.; Bonfante, P.; Le Tacon, F. Molecular phylogeny and historical biogeography of the genus Tuber, the ‘true truffles’. J. Biogeogr. 2008, 35, 815–829. [Google Scholar] [CrossRef]
- Kinoshita, A.; Sasaki, H.; Nara, K. Phylogeny and diversity of Japanese truffles (Tuber spp.) inferred from sequences of four nuclear loci. Mycologia 2011, 103, 779–794. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, M.; Iotti, M.; Pacioni, G.; Hall, I.R.; Zambonelli, A. Truffles: Biodiversity, ecological significances, and biotechnological applications. In Industrially Important Fungi for Sustainable Development; Abdel-Azeem, A.M., Yadav, A.N., Yadav, N., Usmani, Z., Eds.; Springer: Cham, Switzerland, 2021; pp. 107–146. [Google Scholar]
- Bonito, G.; Smith, M.E.; Nowak, M.; Healy, R.A.; Guevara, G.; Cázares, E.; Kinoshita, A.; Eduardo, R.; Nouhra, E.R.; Domínguez, L.S.; et al. Historical biogeography and diversification of truffles in the Tuberaceae and their newly identified southern hemisphere sister lineage. PLoS ONE 2013, 8, e52765. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Benucci, G.M.N.; Xie, X.D.; Bonito, G.; Leisola, M.; Liu, P.G.; Shamekh, S. Morphological, mycorrhizal and molecular characterization of Finnish truffles belonging to the Tuber anniae species-complex. Fungal. Ecol. 2013, 6, 269–280. [Google Scholar] [CrossRef]
- Bonuso, E.; Zambonelli, A.; Bergemann, S.E.; Iotti, M.; Garbelotto, M. Multilocus phylogenetic and coalescent analyses identify two cryptic species in the Italian bianchetto truffle, Tuber borchii Vittad. Conserv. Genet. 2010, 11, 1453–1466. [Google Scholar] [CrossRef]
- Merényi, Z.; Varga, T.; Hubai, A.G.; Pitlik, P.; Erős, A.; Trappe, J.M.; Bratek, Z. Challenges in the delimitation of morphologically similar species: A case study of Tuber brumale agg. (Ascomycota, Pezizales). Mycol. Prog. 2017, 16, 613–624. [Google Scholar] [CrossRef]
- Puliga, F.; Illice, M.; Iotti, M.; Baldo, D.; Zambonelli, A. Tuber iranicum, sp. nov., a truffle species belonging to the Excavatum clade. Mycologia 2020, 112, 932–940. [Google Scholar] [CrossRef]
- Belfiori, B.; Riccioni, C.; Paolocci, F.; Rubini, A. Mating type locus of Chinese black truffles reveals heterothallism and the presence of cryptic species within the T. indicum species complex. PLoS ONE 2013, 8, e82353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, P.; Tian, W.; Liu, P.; Yu, F.; Chen, J.; Deng, X.; Wan, S.; Wang, R.; Wang, Y.; Guo, H. Phylogeography and population genetic analyses reveal the speciation of the Tuber indicum complex. Fungal. Genet. Biol. 2018, 113, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Healy, R.; Bonito, G.M.; Smith, M.E. A brief overview of the systematics, taxonomy, and ecology of the Tuber rufum clade. In True Truffle (Tuber spp.) in the World, Soil Biology 47; Zambonelli, A., Iotti, M., Murat, C., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 125–136. [Google Scholar]
- Bonito, G.M.; Smith, M.E. General systematic position of the truffles: Evolutionary theories. In True Truffle (Tuber spp.) in the World, Soil Biology 47; Zambonelli, A., Iotti, M., Murat, C., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 3–18. [Google Scholar]
- Molinier, V.; Peter, M.; Stobbe, U.; Egli, S. The Burgundy truffle (Tuber aestivum syn. uncinatum): A truffle species with a wide habitat range over Europe. In True Truffle (Tuber spp.) in the World, Soil Biology 47; Zambonelli, A., Iotti, M., Murat, C., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 33–47. [Google Scholar]
- Riccioni, C.; Rubini, A.; Belfiori, B.; Gregori, G.; Paolocci, F. Tuber magnatum: The special one. What makes it so different from the other Tuber spp.? In True Truffle (Tuber spp.) in the World, Soil Biology 47; Zambonelli, A., Iotti, M., Murat, C., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 87–103. [Google Scholar]
- Riccioni, C.; Rubini, A.; Türkoğlu, A.; Belfiori, B.; Paolocci, F. Ribosomal DNA polymorphisms reveal genetic structure and a phylogeographic pattern in the Burgundy truffle Tuber aestivum Vittad. Mycologia 2019, 111, 26–39. [Google Scholar] [CrossRef]
- Belfiori, B.; D’Angelo, V.; Riccioni, C.; Leonardi, M.; Paolocci, F.; Pacioni, G.; Rubini, A. Genetic structure and phylogeography of Tuber magnatum populations. Diversity 2020, 12, 44. [Google Scholar] [CrossRef] [Green Version]
- Hall, I.R.; Brown, G.T.; Zambonelli, A. Taming the Truffle: The History Lore and Science of the Ultimate Mushroom; Timber Press: Portland, Oregon, 2007. [Google Scholar]
- Pacioni, G.; Pomponi, G. Genotypic patterns of some Italian populations of the Tuber aestivum-T. mesentericum complex. Mycotaxon 1991, 42, 171–179. [Google Scholar]
- Pacioni, G.; Fantini, P. Tuber bellonae un tartufo mediterraneo del complesso Tuber aestivum-T. mesentericum. Micol. E Veg. Mediterr. 1997, 12, 15–20. [Google Scholar]
- Sica, M.; Gaudio, L.; Aceto, S. Genetic structure of Tuber mesentericum Vitt. based on polymorphisms at the ribosomal DNA ITS. Mycorrhiza 2007, 17, 405–414. [Google Scholar] [CrossRef] [Green Version]
- Benucci, G.M.N.; Gógán Csorbai, A.; Baciarelli Falini, L.; Marozzi, G.; Suriano, E.; Sitta, N.; Donnini, D. Taxonomy, biology and ecology of Tuber macrosporum Vittad. and Tuber mesentericum Vittad. In True Truffle (Tuber spp.) in the World, Soil Biology 47; Zambonelli, A., Iotti, M., Murat, C., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 69–86. [Google Scholar]
- Marozzi, G.; Benucci, G.M.N.; Suriano, E.; Sitta, N.; Raggi, L.; Lancioni, H.; Baciarelli Falini, L.; Albertini, E.; Donnini, D. Tuber mesentericum and Tuber aestivum truffles: New insights based on morphological and phylogenetic analyses. Diversity 2020, 12, 349. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with specificity for Basidiomycetes: Application to the identification of mycorrhizae and rust. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microb. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paolocci, F.; Rubini, A.; Riccioni, C.; Topini, F.; Arcioni, S. Tuber aestivum and Tuber uncinatum: Two morphotypes or two species? FEMS Microbiol. Lett. 2004, 235, 109–115. [Google Scholar] [CrossRef]
- Zampieri, E.; Mello, A.; Bonfante, P.; Murat, C. PCR primers specific for the genus Tuber reveal the presence of several truffle species in a truffle-ground. FEMS Microbiol. Lett. 2009, 297, 67–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acid. S 1999, 41, 95–98. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Untergrasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Silvestro, D.; Michalak, I. raxmlGUI: A graphical front-end for RAxML. Org. Divers. Evol. 2012, 12, 335–337. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Leonardi, M.; Paz-Conde, A.; Guevara, G.; Salvi, D.; Pacioni, G. Two new species of Tuber previously reported as Tuber malacodermum. Mycologia 2019, 111, 676–689. [Google Scholar] [CrossRef]
- Rayner, R.W. A Mycological Colour Chart; Commonwealth Mycological Institute: Kew, UK, 1970. [Google Scholar]
- Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Austria, Vienna, 2019. [Google Scholar]
- Bonito, G.M.; Gryganskyi, A.P.; Trappe, J.M.; Vilgalys, R. A global meta-analysis of Tuber ITS rDNA sequences: Species diversity, host associations and long-distance dispersal. Mol. Ecol. 2010, 19, 4994–5008. [Google Scholar] [CrossRef]
- Bonito, G.M.; Trappe, J.M.; Rawlinson, P.; Vilgalys, R. Improved resolution of major clades within Tuber and taxonomy of species within the Tuber gibbosum complex. Mycologia 2010, 102, 1042–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Guo, S.X.; Liu, P.G. Species recognition and cryptic species in the Tuber indicum complex. PLoS ONE 2011, 6, e14625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vittadini, C. Monographia Tuberacearum; Mediolani: Milan, Italy, 1831. [Google Scholar]
- Lazzari, G. Storia della Micologia Italiana; Saturnia: Trento, Italy, 1973. [Google Scholar]
- Berkeley, M.J.; Broome, C.E. Notices of British Hypogeous Fungi. Ann. Mag Nat. Hist 1851, 7, 176–186. [Google Scholar]
- Fischer, E. Ascomyceten. Tuberaceen und Hemiasceen. In Kryptogamen–Flora von Deutschland, Oesterreich und der Schweiz Band, Pilze V, Lief 57; Rabenhorst, L., Ed.; E. Kummer: Leipzig, Germany, 1897; pp. 1–64. [Google Scholar]
- Hawker, L.E. British hypogeous fungi. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1954, 237, 429–546. [Google Scholar]
- Pegler, D.N.; Spooner, B.M.; Young, T.W.K. British Truffles. A Revision of British Hypogeous Fungi; Royal Botanic Gardens: Kew, UK, 1993. [Google Scholar]
- Rodríguez, A.; Navarro-Ródenas, A.; Arenas, F.; Guarnizo, A.L.; Morte, A. Fungal Planet 1107: Tuber alcaracense Ant. Rodr. & Morte, sp. nov. Persoonia 2020, 44, 301–459. [Google Scholar]
- Pacioni, G.; Bellina-Agostinone, C.; D’Antonio, M. On the odour of Tuber mesentericum. Mycol. Res. 1991, 94, 201–204. [Google Scholar] [CrossRef]
- Mauriello, G.; Marino, R.; D’Auria, M.; Cerone, G.; Rana, G.L. Determination of volatile organic compounds from truffles via SPME-GC-MS. J. Chromatogr. Sci. 2004, 42, 299–305. [Google Scholar] [CrossRef]
- Wedén, C.; Danell, E.; Tibell, L. Species recognition in the truffle genus Tuber– the synonyms Tuber aestivum and Tuber uncinatum. Environ. Microbiol. 2005, 7, 1535–1546. [Google Scholar] [CrossRef]
- Roux, C.; Séjalon-Delmas, N.; Martins, M.; Parguey-Leduc, A.; Dargent, R.; Bécard, G. Phylogenetic relationships between European and Chinese truffles based on parsimony and distance analysis of ITS sequences. FEMS Microbiol. Lett. 1999, 180, 147–155. [Google Scholar] [CrossRef]
- Stobbe, U.; Buntgen, U.; Sproll, L.; Tegel, W.; Egli, S.; Fink, S. Spatial distribution and ecological variation of re-discovered German truffle habitats. Fungal. Ecol. 2012, 5, 591–599. [Google Scholar] [CrossRef]
- Zhang, J.P.; Liu, P.G.; Chen, J. Tuber sinoaestivum sp. nov., and edible truffle from southwestern China. Mycotaxon 2012, 122, 73–82. [Google Scholar] [CrossRef]
- Paolocci, F.; Rubini, A.; Granetti, B.; Arcioni, S. Typing Tuber melanosporum and Chinese black truffle species by molecular markers. FEMS Microbiol. Lett. 1997, 153, 255–260. [Google Scholar] [CrossRef] [PubMed]



| ID | Clade | Locality | ITS | β-tub | EF-1-α |
|---|---|---|---|---|---|
| Tm2 | I | Italy—Collelongo | KP686246 | KT456266 | OL753348 |
| Tm11 | I | Greece—Xanthi | OL711588 | OL753394 | OL753349 |
| Tm12 | I | Bulgaria—Chelopek | OL711589 | OL753395 | OL753350 |
| Tm13 | I | Italy—Bellagio | OL711590 | OL753396 | OL753351 |
| Tm14 | I | Italy—Lecco | OL711591 | OL753397 | OL753352 |
| Tm15 | I | Italy—Maniago | OL711592 | OL753398 | OL753353 |
| Tm16 | I | Italy—Monza | OL711593 | OL753399 | OL753354 |
| Tm17 | I | Spain—Maçanet de Cabrenys | OL711594 | OL753400 | OL753355 |
| Tm18 | I | England—Bowood | OL711595 | OL753401 | OL753356 |
| Tm19 | I | England—Bowood | OL711596 | OL753402 | OL753357 |
| Tm20 | I | Italy | KT456267 | - | - |
| Tm21 | I | England—Bowood | OL711597 | - | - |
| Tm22 | I | Sweden—Gotland | AJ888043 | KT456265 | JX022595 |
| Tm56 | I | France—Castelnau-de-Montmiral | OL711598 | OL753418 | OL753379 |
| Tm57 | I | France—Roussayrolles | OL711599 | OL753419 | OL753380 |
| Tm58 | I | France—Milhars | OL711600 | OL753420 | OL753381 |
| Tm59 | I | France—Milhars | OL711601 | OL753421 | OL753382 |
| Tm60 | I | France—Le Montat | OL711602 | OL753422 | OL753383 |
| Tm61 | I | France—Bruniquel | OL711603 | OL753423 | OL753384 |
| Tm62 | I | Hungary—Felsőtárkány | OL711604 | OL753424 | OL753385 |
| Tm63 | I | Hungary—Felsőtárkány | OL711605 | OL753425 | OL753386 |
| Tm64 | I | Hungary—Felsőtárkány | OL711606 | OL753426 | OL753387 |
| Tm66 | I | Greece—Kastoria | OL711607 | OL753428 | OL753389 |
| Tm67 | I | Greece—Kastoria | OL711608 | OL753429 | OL753390 |
| Tm69 | I | Italy—Morano Calabro | OL711609 | OL753431 | OL753392 |
| Tm23 | II | Italy—Perugia | KP686243 | KT456263 | OL753358 |
| Tm24 | II | Spain—Valdáliga | OL711610 | OL753403 | OL753359 |
| Tm25 | II | Italy—Cappadocia | OL711611 | OL753404 | OL753360 |
| Tm26 | II | Italy—S. Vincenzo Valle Roveto | OL711612 | OL753405 | OL753361 |
| Tm27 | II | Greece—Messene | OL711613 | OL753406 | OL753362 |
| Tm28 | II | Italy—Colliano | OL711614 | OL753407 | OL753363 |
| Tm29 | II | Italy—Central Appennines | KP686240 | KT456261 | OL753364 |
| Tm30 | II | England—Bowood | OL711615 | - | - |
| Tm31 | II | England—Bowood | OL711616 | - | - |
| Tm32 | II | Italy—Lucoli | KP686239 | KT456260 | OL753365 |
| Tm34 | II | Spain—Cabezón de la Sal | OL711617 | OL753408 | OL753366 |
| Tm35 | II | Italy—Central Appennines | KP686242 | KX290123 | OL753367 |
| Tm36 | II | Italy—Abriola | KP686245 | KX290124 | OL753368 |
| Tm37 | II | Italy—Abriola | KP686244 | OL753409 | OL753369 |
| Tm38 | II | Italy—Central Appennines | KP686241 | KT456262 | OL753370 |
| Tm68 | II | Italy—Morano Calabro | OL711618 | OL753430 | OL753391 |
| Tm49 | II | Italy—Pignola | OL711619 | OL753411 | OL753372 |
| Tm50 | II | Italy—Pignola | OL711620 | OL753412 | OL753373 |
| Tm51 | II | Italy—Pignola | OL711621 | OL753413 | OL753374 |
| Tm70 | II | Italy—Morano Calabro | OL711622 | OL753432 | OL753393 |
| Tm52 | III | Italy—Secinaro | OL711623 | OL753414 | OL753375 |
| Tm53 | III | Italy—Scoppito | OL711624 | OL753414 | OL753376 |
| Tm54 | III | Italy—L’Aquila | OL711625 | OL753416 | OL753377 |
| Tm55 | III | Italy—L’Aquila | OL711626 | OL753417 | OL753378 |
| Tm65 | III | Greece—Acharnes | OL711627 | OL753427 | OL753388 |
| Tm42 | III | Italy—L’Aquila | OL711628 | OL753410 | OL753371 |
| Clade | Forward (5′→3′) | Reverse (5′→3′) | Amplicon Size (bp) |
|---|---|---|---|
| I | ATAACGAGAAGTCTGAACC | CTGCTCTACGCTTATCACA | 357 |
| II | CGTGAACACACTTTGGACA | CTGCCTTACGCTAATTACAAC | 372 |
| III | ACTTGGTAAACTGAAGCAGAC | CTTCACGCCGATTACAACG | 365 |
| Clade I | Clade II | Clade III | ||||
|---|---|---|---|---|---|---|
| Min–Max | Mean ± SD | Min–Max | Mean ± SD | Min–Max | Mean ± SD | |
| L1 | 18–53 | 27.84 ± 6.64 | 16–52 | 27.41 ± 5.98 | 16–52 | 27.64 ± 6.28 |
| L2 | 24–61 | 33.15 ± 7.12 | 22–61 | 33.06 ± 4.66 | 22–59 | 33.47 ± 5.91 |
| L3 | 30–70 | 41.71 ± 8.06 | 29–70 | 43.71 ± 7.24 | 29–70 | 43.24 ± 7.24 |
| W1 | 16–31 | 20.99 ± 3.49 | 14–33 | 21.41 ± 3.69 | 14–33 | 21.49 ± 3.69 |
| W2 | 20–41 | 26.84 ± 4.38 | 20–43 | 25.04 ± 3.63 | 20–39 | 27.11 ± 3.78 |
| W3 | 26–56 | 35.84 ± 5.61 | 26–54 | 37.13 ± 5.62 | 26–54 | 36.85 ± 5.62 |
| Q1 (L1/W1) | 1.00–1.83 | 1.32 ± 0.19 | 1.00–2.21 | 1.28 ± 0.19 | 1.00–1.93 | 1.38 ± 0.16 |
| Q2 (L2/W2) | 1.00–1.69 | 1.23 ± 0.14 | 1.00–2.09 | 1.23 ± 0.15 | 1.00–1.75 | 1.31 ± 0.16 |
| Q3 (L3/W3) | 0.92–1.45 | 1.17 ± 0.12 | 0.95–1.67 | 1.17 ± 0.12 | 1.00–1.54 | 1.22 ± 0.12 |
| L2–L1 | 3–10 | 5.43 ± 1.41 | 3–12 | 8.86 ± 2.41 | 2.0–10.0 | 4.98 ± 1.42 |
| L3–L2 | 4–16 | 8.57 ± 2.33 | 4–16 | 11.00 ± 2.31 | 4.0–14.0 | 8.02 ± 1.98 |
| W2–W1 | 2–8 | 5.85 ± 1.41 | 4–13 | 9.71 ± 1.80 | 3.0–10.0 | 5.03 ± 1.32 |
| W3–W2 | 5–15 | 8.74 ± 2.15 | 9–19 | 10.14 ± 1.46 | 4.0–14.0 | 8.44 ± 2.06 |
| Episp. Thickness 1 | 1.5–5.0 | 3.14 ± 0.95 | 1.5–6.5 | 2.93 ± 0.90 | 1.0–5.0 | 2.50 ± 0.68 |
| Exosp. Height 2 | 2.0–8.0 | 4.07 ± 1.13 | 2.0–9.5 | 4.95 ± 1.31 | 2.0–7.0 | 4.11 ± 1.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leonardi, M.; Salvi, D.; Iotti, M.; Rana, G.L.; Paz-Conde, A.; Pacioni, G. Multilocus Phylogeography of the Tuber mesentericum Complex Unearths Three Highly Divergent Cryptic Species. J. Fungi 2021, 7, 1090. https://doi.org/10.3390/jof7121090
Leonardi M, Salvi D, Iotti M, Rana GL, Paz-Conde A, Pacioni G. Multilocus Phylogeography of the Tuber mesentericum Complex Unearths Three Highly Divergent Cryptic Species. Journal of Fungi. 2021; 7(12):1090. https://doi.org/10.3390/jof7121090
Chicago/Turabian StyleLeonardi, Marco, Daniele Salvi, Mirco Iotti, Gian Luigi Rana, Aurelia Paz-Conde, and Giovanni Pacioni. 2021. "Multilocus Phylogeography of the Tuber mesentericum Complex Unearths Three Highly Divergent Cryptic Species" Journal of Fungi 7, no. 12: 1090. https://doi.org/10.3390/jof7121090
APA StyleLeonardi, M., Salvi, D., Iotti, M., Rana, G. L., Paz-Conde, A., & Pacioni, G. (2021). Multilocus Phylogeography of the Tuber mesentericum Complex Unearths Three Highly Divergent Cryptic Species. Journal of Fungi, 7(12), 1090. https://doi.org/10.3390/jof7121090






