Post-Translational Modifications Drive Success and Failure of Fungal–Host Interactions
Abstract
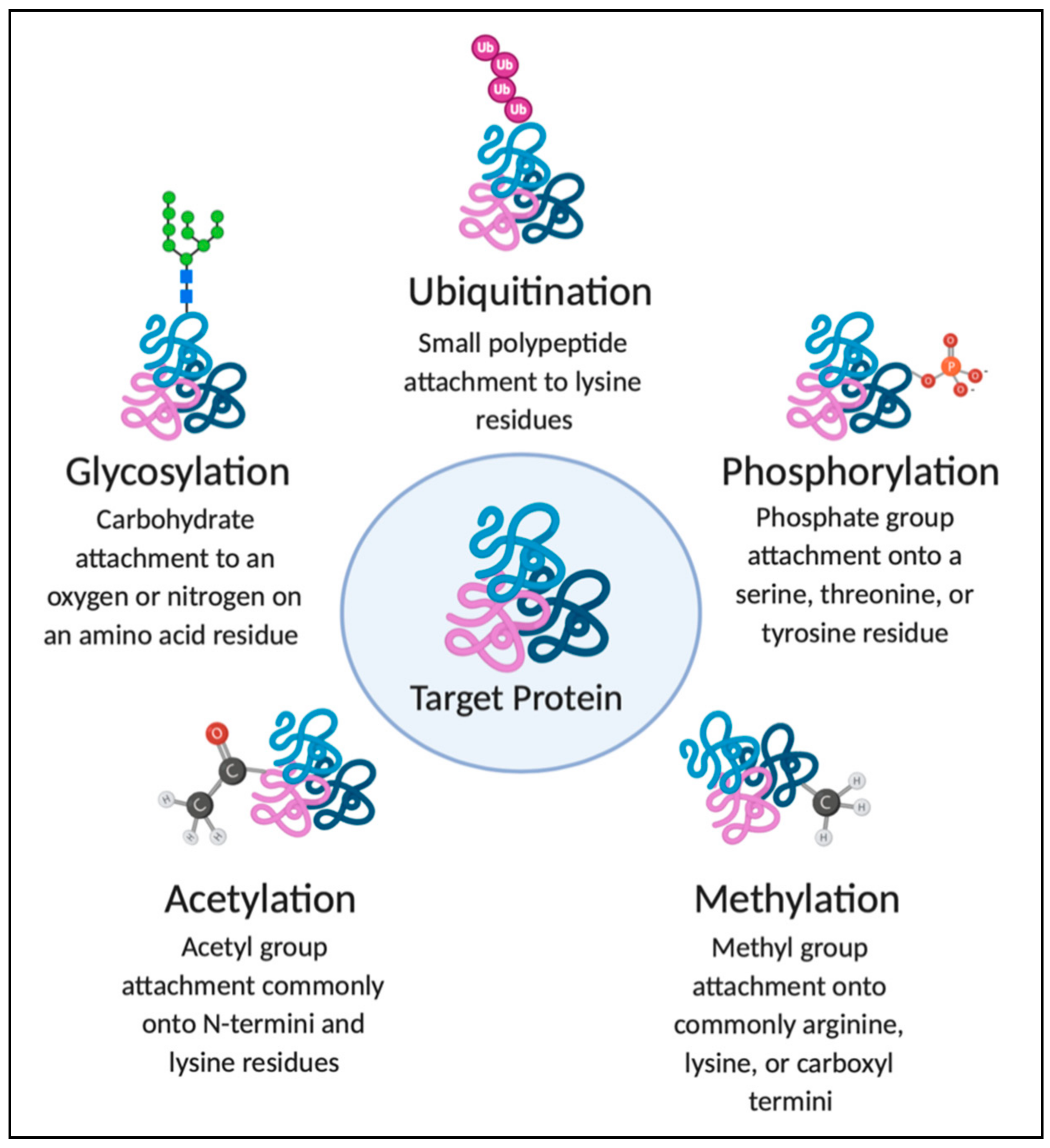
1. Introduction
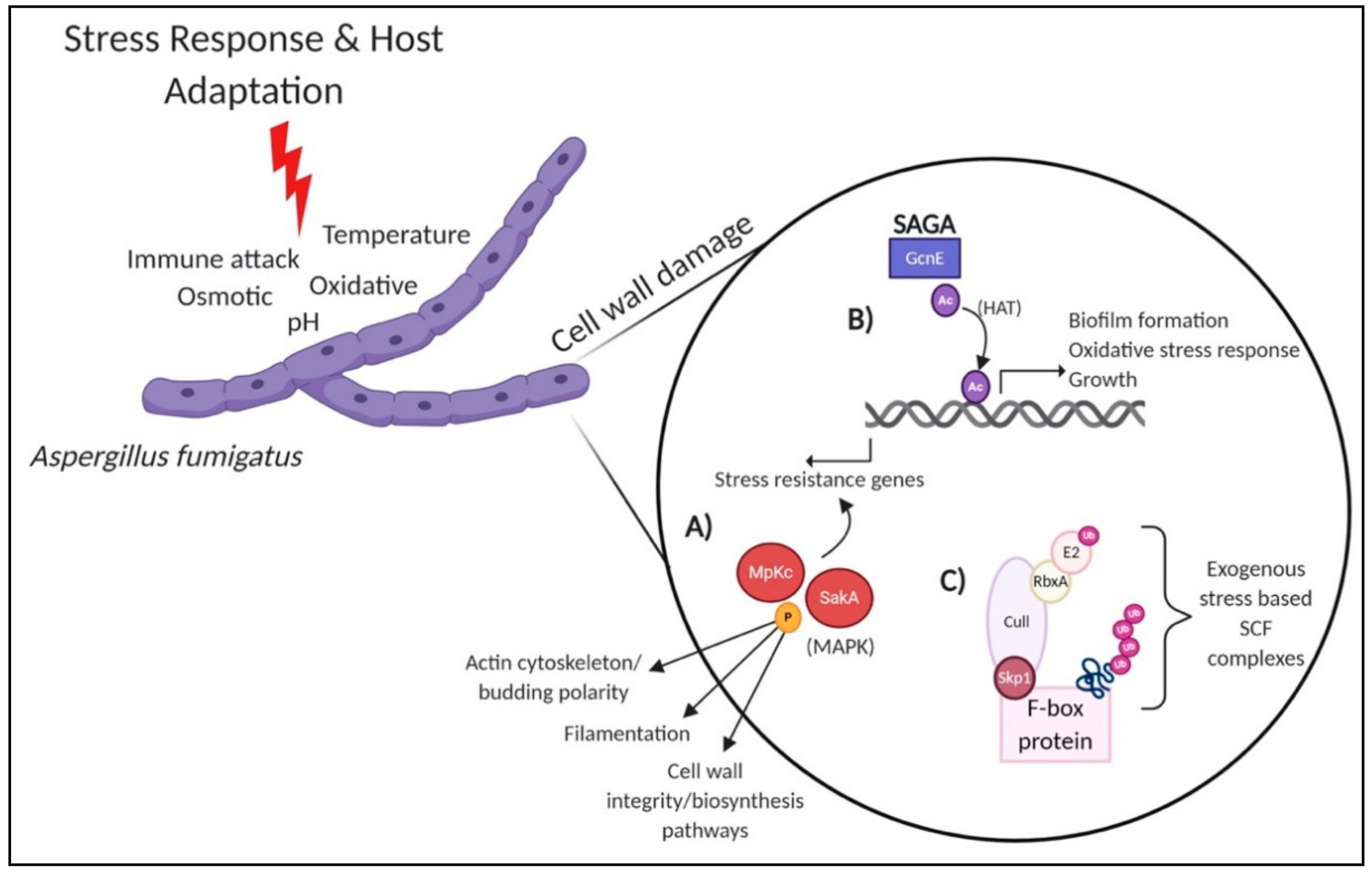
2. PTMs in Stress Response and Host Adaptation
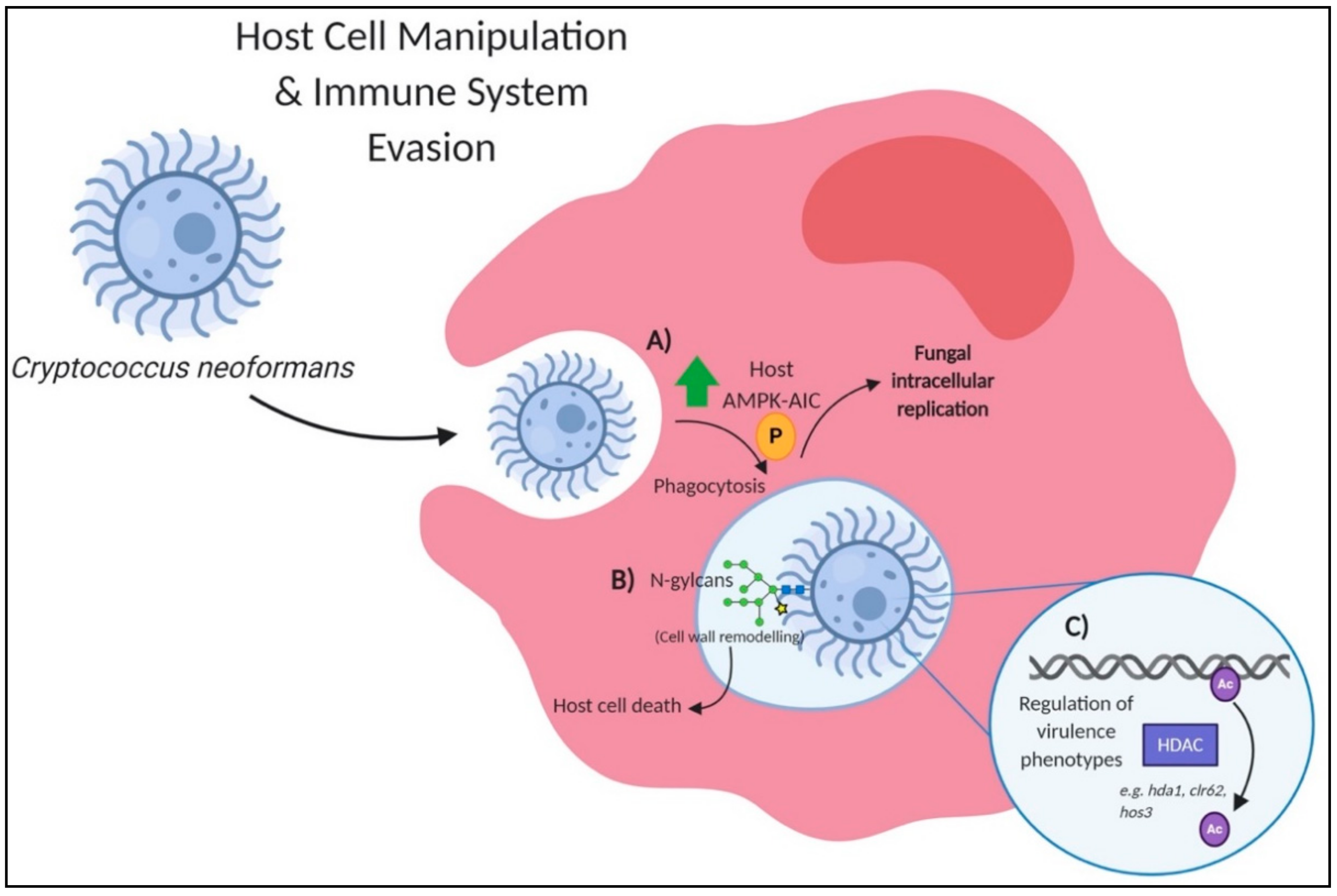
3. PTMs for Host Cell Manipulation and Immune System Evasion
4. PTMs Influence Antifungal Resistance
5. Perspective and Future Outlook
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Leach, M.D.; Brown, A.J.P. Posttranslational modifications of proteins in the pathobiology of medically relevant fungi. Eukaryot. Cell 2011, 11, 98–108. [Google Scholar] [CrossRef]
- Salomon, D.; Orth, K. What pathogens have taught us about posttranslational modifications. Cell Host Microbe 2013, 14, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Ball, B.; Bermas, A.; Carruthers-Lay, D.; Geddes-McAlister, J. Mass spectrometry-based proteomics of fungal pathogenesis, host-fungal interactions, and antifungal development. J. Fungi 2019, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, A.; MacNeil, A.J.; Yeung, J.; Muselius, B.; Gadjeva, M.; Geddes-McAlister, J.; Geddes-McAlister, J. Decoding communication patterns of the innate immune system by quantitative proteomics. J. Leukoc. Biol. 2019, 106, 1221–1232. [Google Scholar] [CrossRef]
- Ball, B.; Langille, M.; Geddes-McAlister, J. Fun(gi)omics: Advanced and diverse technologies to explore emerging fungal pathogens and define mechanisms of antifungal resistance. mBio 2020, 11, 01020-20. [Google Scholar] [CrossRef]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and multi-national prevalence of fungal diseases—Estimate precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.S.; Mannino, D.M.; Eaton, S.; Moss, M. The epidemiology of sepsis in the United States from 1979 through 2000. N. Engl. J. Med. 2003, 348, 1546–1554. [Google Scholar] [CrossRef] [PubMed]
- Kronstad, J.W.; Attarian, R.; Cadieux, B.; Choi, J.; D’Souza, C.A.; Griffiths, E.J.; Geddes, J.M.H.; Hu, G.; Jung, W.H.; Kretschmer, M.; et al. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat. Rev. Genet. 2011, 9, 193–203. [Google Scholar] [CrossRef]
- Perfect, J.R. The antifungal pipeline: A reality check. Nat. Rev. Drug Discov. 2017, 16, 603–616. [Google Scholar] [CrossRef]
- Geddes-McAlister, J.; Shapiro, R.S. New pathogens, new tricks: Emerging, drug-resistant fungal pathogens and future prospects for antifungal therapeutics. Ann. N. Y. Acad. Sci. 2019, 1435, 57–78. [Google Scholar] [CrossRef]
- Wiederhold, N.P. Antifungal resistance: Current trends and future strategies to combat. Infect. Drug Resist. 2017, 10, 249–259. [Google Scholar] [CrossRef]
- Bermas, A.; Geddes-McAlister, J. Combatting the evolution of anti-fungal resistance in Cryptococcus neoformans. Mol. Microbiol. 2020, 114, 1–14. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J. Rare and emerging opportunistic fungal pathogens: Concern for resistance beyond Candida albicans and Aspergillus fumigatus. J. Clin. Microbiol. 2004, 42, 4419–4431. [Google Scholar] [CrossRef] [PubMed]
- Janbon, G.; Quintin, J.; Lanternier, F.; d’Enfert, C. Studying fungal pathogens of humans and fungal infections: Fungal diversity and diversity of approaches. Genes Immun. 2019, 21, 237–245. [Google Scholar]
- Calderone, R.; Fonzi, W.A. Virulence factors of Candida albicans. Trends Microbiol. 2001, 9, 327–335. [Google Scholar] [CrossRef]
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive candidiasis. Nat. Rev. Dis. Prim. 2018, 4, 18026. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.C.-A.; Meyer, W.; Sorrell, T.C. Cryptococcus gattii Infections. Clin. Microbiol. Rev. 2014, 27, 980–1024. [Google Scholar] [CrossRef]
- Alspaugh, J.A. Virulence mechanisms and Cryptococcus neoformans pathogenesis. Fungal Genet. Biol. 2015, 78, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Coelho, C.; Alanio, A. Mechanisms of Cryptococcus neoformans-Mediated Host Damage. Front. Immunol. 2018, 9, 855. [Google Scholar] [CrossRef]
- Hohl, T.M.; Feldmesser, M. Aspergillus fumigatus: Principles of pathogenesis and host defense. Eukaryot. Cell 2007, 6, 1953–1963. [Google Scholar] [CrossRef] [PubMed]
- Dagenais, T.R.T.; Keller, N.P. Pathogenesis of Aspergillus fumigatus in Invasive Aspergillosis. Clin. Microbiol. Rev. 2009, 22, 447–465. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, B.B.; Mylonakis, E. Our paths might cross: The role of the fungal cell wall integrity pathway in stress response and cross talk with other stress response pathways. Eukaryot. Cell 2009, 8, 1616–1625. [Google Scholar] [CrossRef]
- Hopke, A.; Brown, A.J.; Hall, R.; Wheeler, R.T. Dynamic fungal cell wall architecture in stress adaptation and immune evasion. Trends Microbiol. 2018, 26, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Sanz, A.B.; García, R.; Rodríguez-Peña, J.M.; Arroyo, J. The CWI pathway: Regulation of the transcriptional adaptive response to cell wall stress in yeast. J. Fungi 2018, 4, 1. [Google Scholar] [CrossRef]
- Bahn, Y.-S. Master and commander in fungal pathogens: The two-component system and the HOG signaling pathway. Eukaryot. Cell 2008, 7, 2017–2036. [Google Scholar] [CrossRef]
- Zhao, X.; Mehrabi, R.; Xu, J.-R. Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot. Cell 2007, 6, 1701–1714. [Google Scholar] [CrossRef] [PubMed]
- Román, E.; Arana, D.M.; Nombela, C.; Alonso-Monge, R.; Pla, J. MAP kinase pathways as regulators of fungal virulence. Trends Microbiol. 2007, 15, 181–190. [Google Scholar] [CrossRef]
- Juvvadi, P.R.; Lee, S.C.; Heitman, J.; Steinbach, W.J. Calcineurin in fungal virulence and drug resistance: Prospects for harnessing targeted inhibition of calcineurin for an antifungal therapeutic approach. Virulence 2017, 8, 186–197. [Google Scholar] [CrossRef]
- Mattos, E.C.; Silva, L.P.; Valero, C.; De Castro, P.A.; Dos Reis, T.F.; Ribeiro, L.F.C.; Marten, M.R.; Silva-Rocha, R.; Westmann, C.; Silva, C.H.T.d.P.d.; et al. The Aspergillus fumigatus phosphoproteome reveals roles of high-osmolarity glycerol mitogen-activated protein kinases in promoting cell wall damage and caspofungin tolerance. mBio 2020, 11, e02962-19. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-J.; Hou, Y.-H.; Chen, Y.-L. The histone acetyltransferase GcnE regulates conidiation and biofilm formation in Aspergillus fumigatus. Med. Mycol. 2019, 58, 248–259. [Google Scholar] [CrossRef]
- Frawley, D.; Bayram, Ö. Identification of SkpA-CulA-F-box E3 ligase complexes in pathogenic Aspergilli. Fungal Genet. Biol. 2020, 140, 103396. [Google Scholar] [CrossRef]
- Shivarathri, R.; Tscherner, M.; Zwolanek, F.; Singh, N.K.; Chauhan, N.; Kuchler, K. The fungal histone acetyl transferase Gcn5 controls virulence of the human pathogen Candida albicans through multiple pathways. Sci. Rep. 2019, 9, 1–18. [Google Scholar] [CrossRef]
- Lan, H.; Sun, R.; Fan, K.; Yang, K.; Zhang, F.; Nie, X.Y.; Wang, X.; Zhuang, Z.; Wang, S. The Aspergillus flavus histone Acetyltransferase AflGcnE regulates morphogenesis, aflatoxin biosynthesis, and pathogenicity. Front. Microbiol. 2016, 7, 1324. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.; Fan, X.; Chen, J. Function and subcellular localization of Gcn5, a histone acetyltransferase in Candida albicans. Fungal Genet. Biol. 2015, 81, 132–141. [Google Scholar] [CrossRef]
- O’Meara, T.R.; Hay, C.; Price, M.S.; Giles, S.; Alspaugh, J.A. Cryptococcus neoformans Histone Acetyltransferase Gcn5 Regulates Fungal Adaptation to the Host. Eukaryot. Cell 2010, 9, 1193–1202. [Google Scholar] [CrossRef]
- Roig, P.; Gozalbo, D. Depletion of polyubiquitin encoded by the UBI4 gene confers pleiotropic phenotype to Candida albicans cells. Fungal Genet. Biol. 2003, 39, 70–81. [Google Scholar] [CrossRef]
- Leach, M.D.; Stead, D.A.; Argo, E.; Maccallum, D.M.; Brown, A.J.P. Molecular and proteomic analyses highlight the importance of ubiquitination for the stress resistance, metabolic adaptation, morphogenetic regulation and virulence of Candida albicans. Mol. Microbiol. 2011, 79, 1574–1593. [Google Scholar] [CrossRef]
- Geddes, J.M.H.; Caza, M.; Croll, D.; Stoynov, N.; Foster, L.J.; Kronstad, J.W. Analysis of the protein kinase a-regulated proteome of Cryptococcus neoformans identifies a role for the ubiquitin-proteasome pathway in capsule formation. mBio 2016, 7, e01862-15. [Google Scholar] [CrossRef]
- Masso-Silva, J.; Espinosa, V.; Liu, T.-B.; Wang, Y.; Xue, C.; Rivera, A. The F-box protein Fbp1 shapes the immunogenic potential of Cryptococcus neoformans. mBio 2018, 9, e01828-17. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.-B.; Xue, C. Fbp1-mediated ubiquitin-proteasome pathway controls Cryptococcus neoformans virulence by regulating fungal intracellular growth in macrophages. Infect. Immun. 2014, 82, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.M.; Perrin, R.M.; Dagenais, T.R.T.; Keller, N.P. H3K9 methylation regulates growth and development in Aspergillus fumigatus. Eukaryot. Cell 2008, 7, 2052–2060. [Google Scholar] [CrossRef]
- Bok, J.W.; Chiang, Y.-M.; Szewczyk, E.; Reyes-Dominguez, Y.; Davidson, A.D.; Sanchez, J.F.; Lo, H.-C.; Watanabe, K.; Strauss, J.; Oakley, B.R.; et al. Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 2009, 5, 462–464. [Google Scholar] [CrossRef]
- Palmer, J.M.; Bok, J.W.; Lee, S.; Dagenais, T.R.T.; Andes, D.R.; Kontoyiannis, D.P.; Keller, N.P. Loss of CclA, required for histone 3 lysine 4 methylation, decreases growth but increases secondary metabolite production in Aspergillus fumigatus. PeerJ 2013, 1, e4. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ami, R.; Lewis, R.E.; Leventakos, K.; Kontoyiannis, D.P. Aspergillus fumigatus inhibits angiogenesis through the production of gliotoxin and other secondary metabolites. Blood 2009, 114, 5393–5399. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Ding, S.L.; Qin, Q.-M.; Gupta, R.; Gomez, G.; Lin, F.; Feng, X.; Da Costa, L.F.; Chaki, S.P.; Katepalli, M.; et al. Global reprogramming of host kinase signaling in response to fungal infection. Cell Host Microbe 2017, 21, 637–649.e6. [Google Scholar] [CrossRef]
- Reales-Calderón, J.; Sylvester, M.; Strijbis, K.; Jensen, O.N.; Nombela, C.; Molero, G.; Gil, C. Candida albicans induces pro-inflammatory and anti-apoptotic signals in macrophages as revealed by quantitative proteomics and phosphoproteomics. J. Proteom. 2013, 91, 106–135. [Google Scholar] [CrossRef]
- Alers, S.; Loffler, A.S.; Wesselborg, S.; Stork, B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: Cross talk, shortcuts, and feedbacks. Mol. Cell Biol. 2012, 32, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Thak, E.J.; Lee, S.-B.; Xu-Vanpala, S.; Lee, D.-J.; Chung, S.-Y.; Bahn, Y.-S.; Oh, D.-B.; Shinohara, M.L.; Kang, H.A. Core N-Glycan structures are critical for the pathogenicity of Cryptococcus neoformans by modulating host cell death. mBio 2020, 11, e00711-20. [Google Scholar] [CrossRef]
- Brandão, F.; Esher, S.K.; Ost, K.S.; Pianalto, K.; Nichols, C.B.; Fernandes, L.; Bocca, A.L.; Poças-Fonseca, M.J.; Alspaugh, J.A. HDAC genes play distinct and redundant roles in Cryptococcus neoformans virulence. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef]
- Tavares, A.H.; Bürgel, P.H.; Bocca, A.L. Turning up the heat: Inflammasome activation by fungal pathogens. PLoS Pathog. 2015, 11, e1004948. [Google Scholar] [CrossRef] [PubMed]
- Krysan, D.J.; Sutterwala, F.S.; Wellington, M. Catching fire: Candida albicans, macrophages, and pyroptosis. PLoS Pathog. 2014, 10, e1004139. [Google Scholar] [CrossRef]
- Uwamahoro, N.; Verma-Gaur, J.; Shen, H.-H.; Qu, Y.; Lewis, R.; Lu, J.; Bambery, K.; Masters, S.L.; Vince, J.E.; Naderer, T.; et al. The pathogen Candida albicans Hijacks pyroptosis for escape from macrophages. mBio 2014, 5, e00003-14. [Google Scholar] [CrossRef]
- O’Meara, T.R.; Veri, A.O.; Ketela, T.; Jiang, B.; Roemer, T.; Cowen, L.E. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat. Commun. 2015, 6, 6741. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, T.R.; Duah, K.; Guo, C.X.; Maxson, M.E.; Gaudet, R.G.; Koselny, K.; Wellington, M.; Powers, M.E.; MacAlpine, J.; O’Meara, M.J.; et al. High-throughput screening identifies genes required for Candida albicans induction of macrophage pyroptosis. mBio 2018, 9, e01581-18. [Google Scholar] [CrossRef]
- Kronstad, J.W.; Saikia, S.; Nielson, E.D.; Kretschmer, M.; Jung, W.; Hu, G.; Geddes, J.M.H.; Griffiths, E.J.; Choi, J.; Cadieux, B.; et al. Adaptation of Cryptococcus neoformans to mammalian hosts: Integrated regulation of metabolism and virulence. Eukaryot. Cell 2011, 11, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Donlin, M.J.; Upadhya, R.; Gerik, K.J.; Lam, W.; VanArendonk, L.G.; Specht, C.A.; Sharma, N.K.; Lodge, J.K. Cross talk between the cell wall integrity and cyclic AMP/protein kinase A pathways in Cryptococcus neoformans. mBio 2014, 5, e01573-14. [Google Scholar] [CrossRef] [PubMed]
- Bahn, Y.-S.; Jung, K.-W. Stress signaling pathways for the pathogenicity of Cryptococcus. Eukaryot. Cell 2013, 12, 1564–1577. [Google Scholar] [CrossRef] [PubMed]
- DeLeon-Rodriguez, C.M.; Casadevall, A. Cryptococcus neoformans: Tripping on acid in the phagolysosome. Front. Microbiol. 2016, 7, 164. [Google Scholar] [CrossRef]
- Brandão, F.A.S.; Derengowski, L.S.; Albuquerque, P.; Nicola, A.M.; Silva-Pereira, I.; Poças-Fonseca, M.J. Histone deacetylases inhibitors effects on Cryptococcus neoformans major virulence phenotypes. Virulence 2015, 6, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Helenius, A.; Aebi, M. Roles of N-linked Glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 2004, 73, 1019–1049. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rubio, R.; De Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The fungal cell wall: Candida, Cryptococcus, and Aspergillus species. Front. Microbiol. 2020, 10, 2993. [Google Scholar] [CrossRef]
- Bates, S.; Hughes, H.B.; Munro, C.A.; Thomas, W.P.; Maccallum, D.M.; Bertram, G.; Atrih, A.; Ferguson, M.A.; Brown, A.J.; Odds, F.C.; et al. Outer chain N-Glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 2006, 281, 90–98. [Google Scholar] [CrossRef] [PubMed]
- West, L.; Lowman, D.W.; Mora-Montes, H.M.; Grubb, S.; Murdoch, C.; Thornhill, M.H.; Gow, N.A.; Williams, D.; Haynes, K. Differential virulence of Candida glabrata glycosylation mutants. J. Biol. Chem. 2013, 288, 22006–22018. [Google Scholar] [CrossRef] [PubMed]
- Garfoot, A.L.; Goughenour, K.D.; Wüthrich, M.; Rajaram, M.V.S.; Schlesinger, L.S.; Klein, B.S.; Rappleye, C.A. O-Mannosylation of proteins enables Histoplasma yeast survival at mammalian body temperatures. mBio 2018, 9, e02121-17. [Google Scholar] [CrossRef] [PubMed]
- Kotz, A.; Wagener, J.; Engel, J.; Routier, F.H.; Echtenacher, B.; Jacobsen, I.D.; Heesemann, J.; Ebel, F. Approaching the secrets of N-Glycosylation in Aspergillus fumigatus: Characterization of the AfOch1 protein. PLoS ONE 2010, 5, e15729. [Google Scholar] [CrossRef]
- McKenzie, C.G.J.; Koser, U.; Lewis, L.E.; Bain, J.M.; Mora-Montes, H.M.; Barker, R.N.; Gow, N.A.R.; Erwig, L.-P. Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect. Immun. 2010, 78, 1650–1658. [Google Scholar] [CrossRef]
- Wagener, J.; Weindl, G.; De Groot, P.W.J.; De Boer, A.D.; Kaesler, S.; Thavaraj, S.; Bader, O.; Mailänder-Sanchez, D.; Borelli, C.; Weig, M.; et al. Glycosylation of Candida albicans Cell wall proteins is critical for induction of innate immune responses and apoptosis of epithelial cells. PLoS ONE 2012, 7, e50518. [Google Scholar] [CrossRef]
- Taipale, M.; Jarosz, D.F.; Lindquist, S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 515–528. [Google Scholar] [CrossRef]
- Cowen, L.E.; Singh, S.D.; Hler, J.R.K.; Collins, C.; Zaas, A.K.; Schell, W.A.; Aziz, H.; Mylonakis, E.; Perfect, J.R.; Whitesell, L.; et al. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc. Natl. Acad. Sci. USA 2009, 106, 2818–2823. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.D.; Robbins, N.; Zaas, A.K.; Schell, W.A.; Perfect, J.R.; Cowen, L.E. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via Calcineurin. PLoS Pathog. 2009, 5, e1000532. [Google Scholar] [CrossRef]
- Robbins, N.; Uppuluri, P.; Nett, J.; Rajendran, R.; Ramage, G.; Lopez-Ribot, J.L.; Andes, D.; Cowen, L.E. Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 2011, 7, e1002257. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, T.R.; Robbins, N.; Cowen, L.E. The Hsp90 chaperone network modulates Candida virulence traits. Trends Microbiol. 2017, 25, 809–819. [Google Scholar] [CrossRef]
- Robbins, N.; Leach, M.D.; Cowen, L.E. Lysine Deacetylases Hda1 and Rpd3 regulate Hsp90 function thereby governing fungal drug resistance. Cell Rep. 2012, 2, 878–888. [Google Scholar] [CrossRef]
- Li, X.; Robbins, N.; O’Meara, T.R.; Cowen, L.E. Extensive functional redundancy in the regulation of Candida albicans drug resistance and morphogenesis by lysine deacetylases Hos2, Hda1, Rpd3 and Rpd31. Mol. Microbiol. 2017, 103, 635–656. [Google Scholar] [CrossRef]
- Diezmann, S.; Michaut, M.; Shapiro, R.S.; Bader, G.D.; Cowen, L.E. Mapping the Hsp90 genetic interaction network in Candida albicans reveals environmental contingency and rewired circuitry. PLoS Genet. 2012, 8, e1002562. [Google Scholar] [CrossRef]
- Li, Y.; Li, H.; Sui, M.; Li, M.; Wang, J.; Meng, Y.; Sun, T.; Liang, Q.; Suo, C.; Gao, X.; et al. Fungal acetylome comparative analysis identifies an essential role of acetylation in human fungal pathogen virulence. Commun. Biol. 2019, 2, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Rocha, M.C.; Minari, K.; Fabri, J.H.T.M.; Kerkaert, J.D.; Gava, L.M.; Da Cunha, A.F.; Cramer, R.A.; Borges, J.C.; Malavazi, I. Aspergillus fumigatus Hsp90 interacts with the main components of the cell wall integrity pathway and cooperates in heat shock and cell wall stress adaptation. Cell. Microbiol. 2021, 23, e13273. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005, 69, 262–291. [Google Scholar] [CrossRef]
- Caplan, T.; Lorente-Macías, Á.; Stogios, P.J.; Evdokimova, E.; Hyde, S.; Wellington, M.A.; Liston, S.; Iyer, K.R.; Puumala, E.; Shekhar-Guturja, T.; et al. Overcoming fungal echinocandin resistance through inhibition of the non-essential stress kinase Yck2. Cell Chem. Biol. 2020, 27, 269–282.e5. [Google Scholar] [CrossRef] [PubMed]
- Kuchler, K.; Jenull, S.; Shivarathri, R.; Chauhan, N. Fungal KATs/KDACs: A New Highway to Better Antifungal Drugs? PLoS Pathog. 2016, 12, e1005938. [Google Scholar] [CrossRef]
- Parente-Rocha, J.A.; Bailão, A.M.; Amaral, A.C.; Taborda, C.P.; Paccez, J.D.; Borges, C.L.; Pereira, M. Antifungal resistance, metabolic routes as drug targets, and new antifungal agents: An overview about endemic dimorphic fungi. Mediat. Inflamm. 2017, 2017, 9870679. [Google Scholar] [CrossRef]
- McCarthy, M.W.; Kontoyiannis, D.P.; Cornely, O.A.; Perfect, J.R.; Walsh, T.J. Novel agents and drug targets to meet the challenges of resistant fungi. J. Infect. Dis. 2017, 216, S474–S483. [Google Scholar] [CrossRef]
- Kaltdorf, M.; Srivastava, M.; Gupta, S.K.; Liang, C.; Binder, J.; Dietl, A.-M.; Meir, Z.; Haas, H.; Osherov, N.; Krappmann, S.; et al. Systematic identification of anti-fungal drug targets by a metabolic network approach. Front. Mol. Biosci. 2016, 3, 22. [Google Scholar] [CrossRef]
- Su, H.; Han, L.; Huang, X. Potential targets for the development of new antifungal drugs. J. Antibiot. 2018, 71, 978–991. [Google Scholar] [CrossRef]
- Ramakrishnan, J.; Rathore, S.S.; Raman, T. Review on fungal enzyme inhibitors—Potential drug targets to manage human fungal infections. RSC Adv. 2016, 6, 42387–42401. [Google Scholar] [CrossRef]
- Bencúrová, E.; Gupta, S.K.; Sarukhanyan, E.; Dandekar, T. Identification of antifungal targets based on computer modeling. J. Fungi 2018, 4, 81. [Google Scholar] [CrossRef] [PubMed]
- Ball, B.; Sukumaran, A.; Geddes-McAlister, J. Label-free quantitative proteomics workflow for discovery-driven host-pathogen interactions. J. Vis. Exp. 2020, 164. [Google Scholar] [CrossRef]
- Sukumaran, A.; Woroszchuk, E.; Ross, T.; Geddes-McAlister, J. Proteomics of host-bacterial interactions: New insights from dual perspectives. Can. J. Microbiol. 2020, 7. [Google Scholar] [CrossRef]
- Lee, K.-T.; So, Y.-S.; Yang, D.-H.; Jung, K.-W.; Choi, J.; Lee, D.-G.; Kwon, H.; Jang, J.; Wang, L.L.; Cha, S.; et al. Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans. Nat. Commun. 2016, 7, 12766. [Google Scholar] [CrossRef] [PubMed]
- Arras, S.D.M.; Chua, S.M.H.; Wizrah, M.S.I.; Faint, J.A.; Yap, A.S.; Fraser, J.A. Targeted genome editing via CRISPR in the pathogen Cryptococcus neoformans. PLoS ONE 2016, 11, e0164322. [Google Scholar] [CrossRef] [PubMed]
- Wensing, L.; Sharma, J.; Uthayakumar, D.; Proteau, Y.; Chavez, A.; Shapiro, R.S. A CRISPR interference platform for efficient genetic repression in Candida albicans. mSphere 2019, 4, e00002-19. [Google Scholar] [CrossRef] [PubMed]
- Kugadas, A.; Geddes-McAlister, J.; Guy, E.; DiGiandomenico, A.; Sykes, D.B.; Mansour, M.K.; Mirchev, R.; Gadjeva, M. Frontline science: Employing enzymatic treatment options for management of ocular biofilm-based infections. J. Leukoc. Biol. 2019, 105, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Hogrebe, A.; Von Stechow, L.; Bekker-Jensen, D.B.; Weinert, B.T.; Kelstrup, C.D.; Olsen, J.V. Benchmarking common quantification strategies for large-scale phosphoproteomics. Nat. Commun. 2018, 9, 1045. [Google Scholar] [CrossRef] [PubMed]
- Munk, S.; Refsgaard, J.C.; Olsen, J.V.; Jensen, L.J. From Phosphosites to Kinases. In Advanced Structural Safety Studies; Springer Nature: Singapore, 2016; Volume 1355, pp. 307–321. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Retanal, C.; Ball, B.; Geddes-McAlister, J. Post-Translational Modifications Drive Success and Failure of Fungal–Host Interactions. J. Fungi 2021, 7, 124. https://doi.org/10.3390/jof7020124
Retanal C, Ball B, Geddes-McAlister J. Post-Translational Modifications Drive Success and Failure of Fungal–Host Interactions. Journal of Fungi. 2021; 7(2):124. https://doi.org/10.3390/jof7020124
Chicago/Turabian StyleRetanal, Charmaine, Brianna Ball, and Jennifer Geddes-McAlister. 2021. "Post-Translational Modifications Drive Success and Failure of Fungal–Host Interactions" Journal of Fungi 7, no. 2: 124. https://doi.org/10.3390/jof7020124
APA StyleRetanal, C., Ball, B., & Geddes-McAlister, J. (2021). Post-Translational Modifications Drive Success and Failure of Fungal–Host Interactions. Journal of Fungi, 7(2), 124. https://doi.org/10.3390/jof7020124





