Repositioning Lopinavir, an HIV Protease Inhibitor, as a Promising Antifungal Drug: Lessons Learned from Candida albicans—In Silico, In Vitro and In Vivo Approaches
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Fungal Strain and Growth Conditions
2.3. Effects of Lopinavir on C. albicans Sap2: In Silico Approach
2.4. Effects of Lopinavir on Saps: In Vitro Assay
2.5. Effects of Lopinavir on Growth
2.6. Lopinavir Treatment: Looking for Possible Molecular Targets in C. albicans Cells Other Than Saps and Role in Crucial Physiopathology Events
2.7. Effects of Lopinavir on Morphometrical Parameters and Morphology
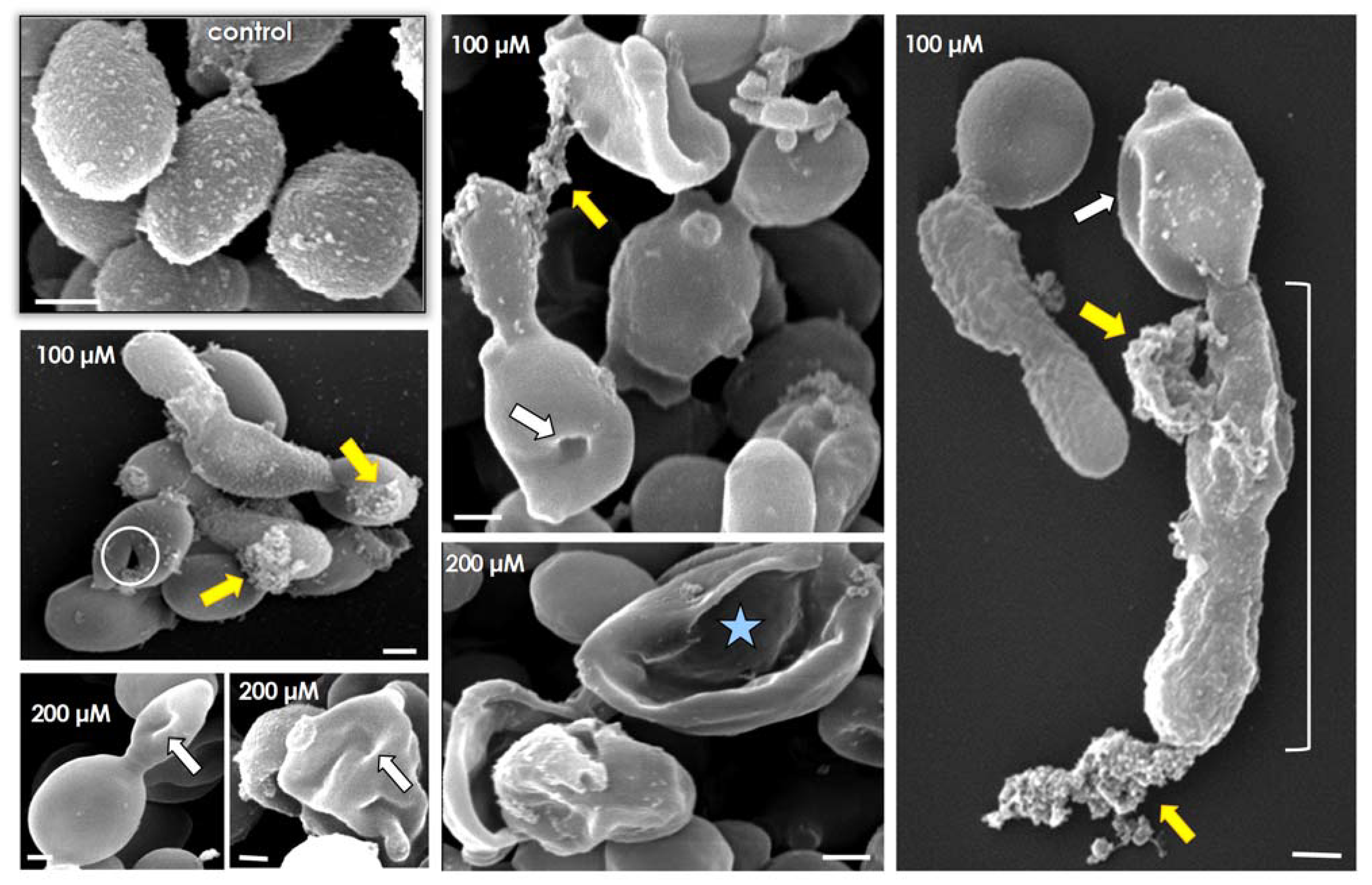
2.8. Effects of Lopinavir on Ultrastructural Architecture
2.9. Effects of Lopinavir on Morphogenesis
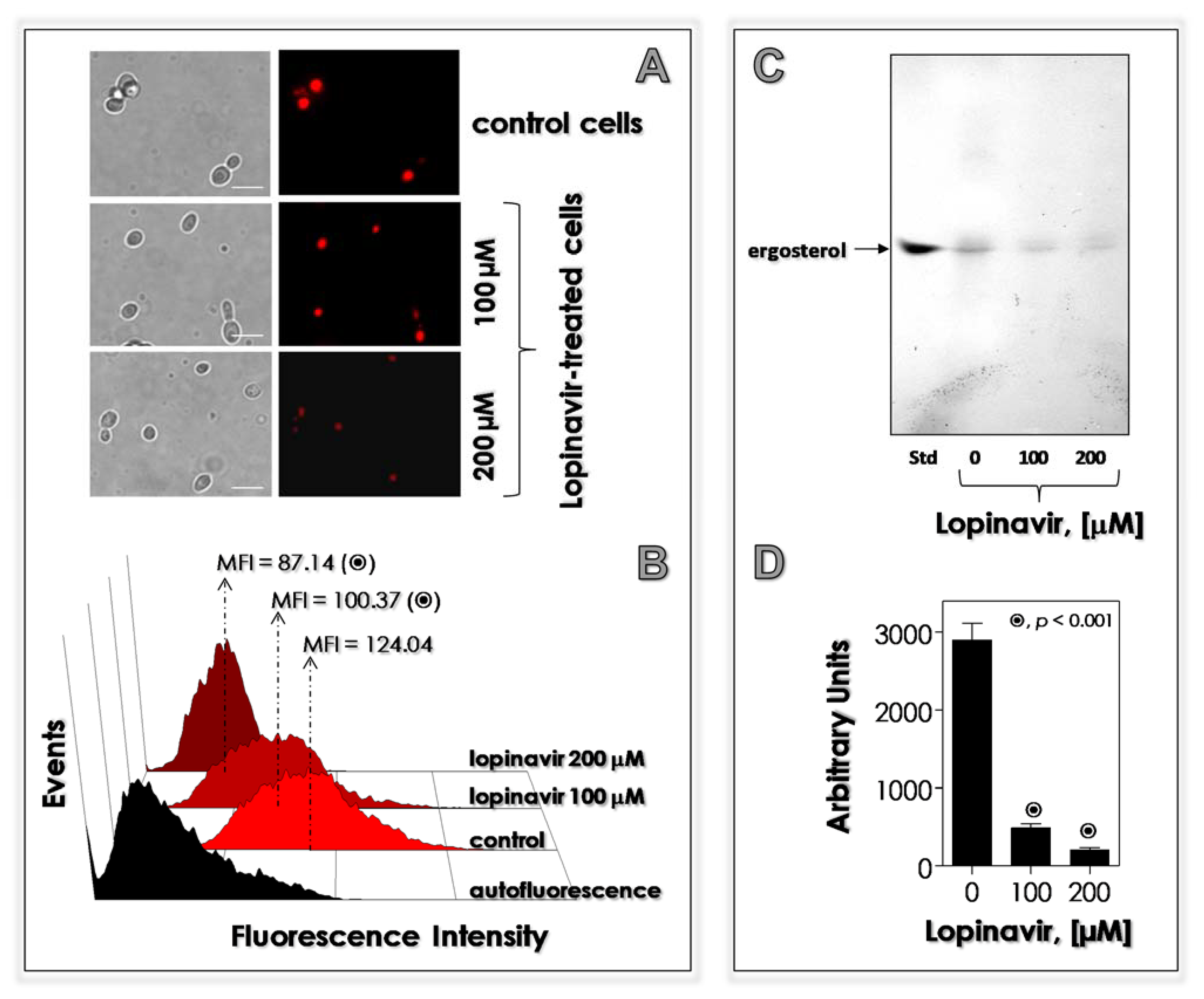
2.10. Effects of Lopinavir on Neutral Lipids
2.10.1. Sterol Content
2.10.2. Nile Red Staining
2.11. Effects of Lopinavir on Cell Wall-Located Surface Molecules
2.12. Effects of Lopinavir on Secreted Hydrolytic Enzymes
2.13. Effects of Lopinavir on Biofilm
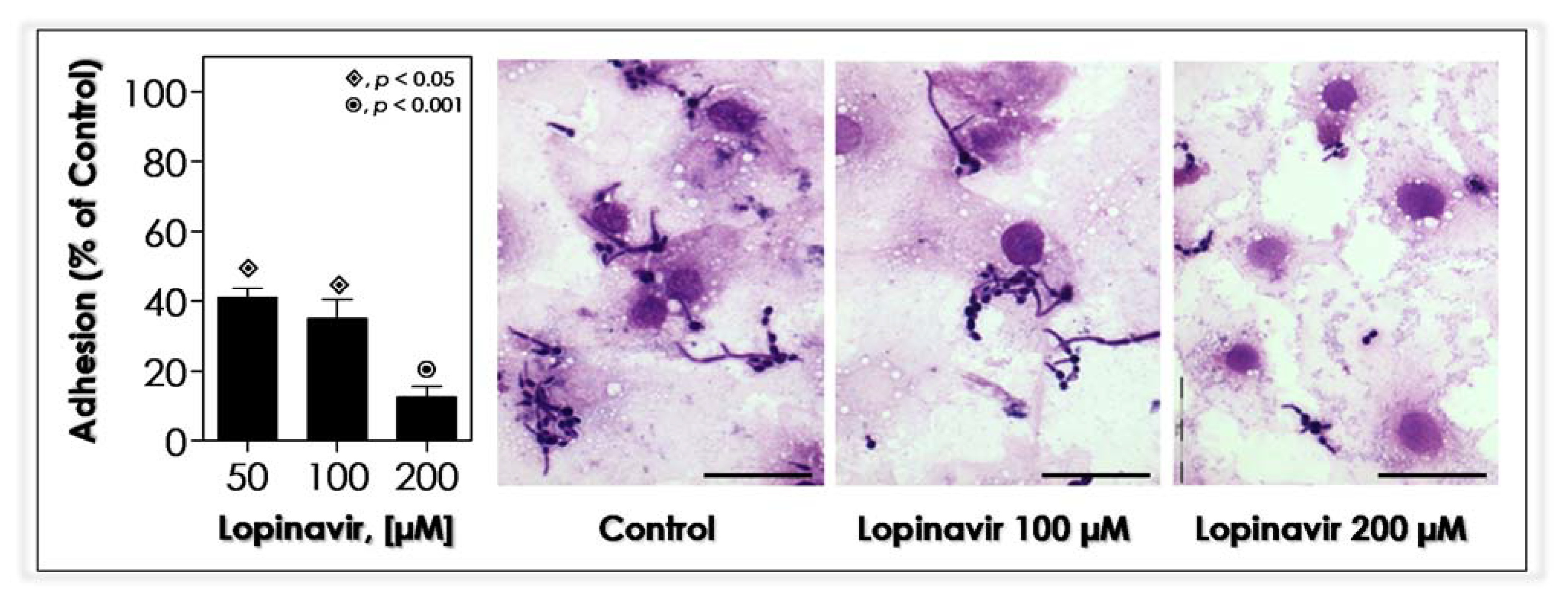
2.14. Effects of Lopinavir on In Vitro Interaction with Epithelial Cells
2.15. BALB/c Mice for In Vivo Infections
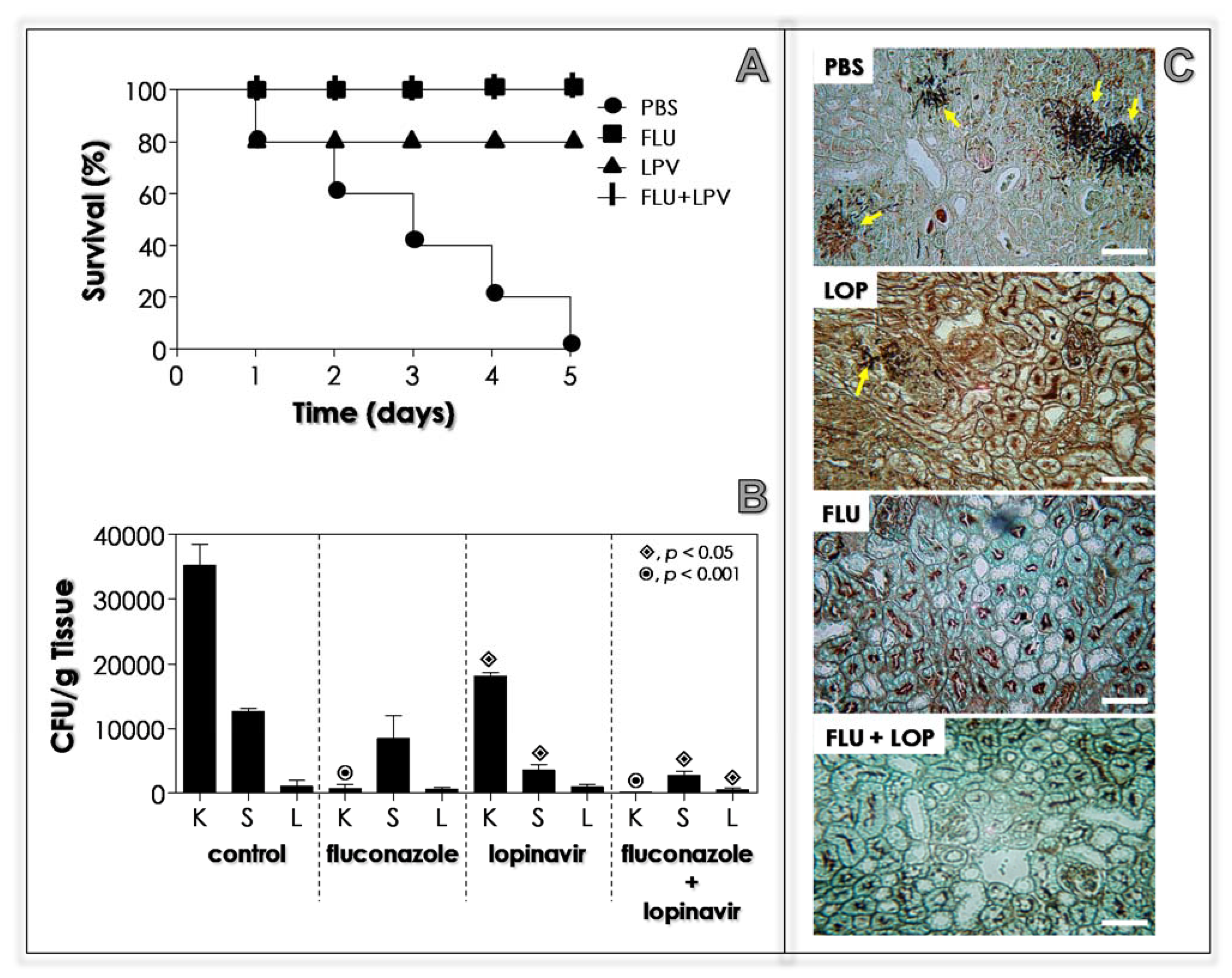
2.16. Effects of Lopinavir on In Vivo Infection of Immunocompetent Mice
2.17. Effects of Lopinavir on In Vivo Infection of Immunosuppressed Mice
2.18. Effects of Lopinavir on the Expression of Sap1-3 Antigens in Post-Mice Passage
2.19. Statistics
3. Results and Discussion
3.1. Lopinavir Binds to Sap2
3.2. Lopinavir Inhibits Sap Activity
3.3. Lopinavir Interferes with Growth Behavior, Morphometrical Parameters and Morphology
3.4. Lopinavir Alters Ultrastructural Architecture
3.5. Lopinavir Arrests the Yeasts-into-Hyphae Transformation
3.6. Lopinavir Disturbs the Synthesis of Neutral Lipids
3.7. Lopinavir Modulates the Surface-Located Molecules
3.8. Lopinavir Diminishes the Secretion of Hydrolytic Enzymes
3.9. Lopinavir Influences the Biofilm on Abiotic Surface
3.10. Lopinavir Blocks the In Vitro Adhesion to Epithelial Cells
3.11. Lopinavir Contains the In Vivo Infection in Immunocompetent Mice
3.12. Lopinavir Contains the In Vivo Infection in Immunosuppressed Mice
3.13. Lopinavir Reduces the Sap Production by Yeasts Recovered from Kidney of Infected Mice
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ohmit, S.E.; Sobel, J.D.; Schuman, P.; Duerr, A.; Mayer, K.; Rompalo, A.; Klein, R.S.; HIV Epidemiology Research Study (HERS) Group. Longitudinal Study of Mucosal Candida Species Colonization and Candidiasis among Human Immunodeficiency Virus (HIV)–Seropositive and At-Risk HIV-Seronegative Women. J. Infect. Dis. 2003, 188, 118–127. [Google Scholar] [CrossRef]
- Palella, F.J., Jr.; Delaney, K.M.; Moorman, A.C.; Loveless, M.O.; Fuhrer, J.; Satten, G.A.; Aschman, D.J.; Holmberg, S.D. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 1998, 338, 853–860. [Google Scholar] [CrossRef]
- Portela, M.B.; Souza, I.P.R.; Costa, E.M.M.D.B.; Hagler, A.N.; Soares, R.M.A.; Santos, A.L.S. Differential Recovery of Candida Species from Subgingival Sites in Human Immunodeficiency Virus-Positive and Healthy Children from Rio de Janeiro, Brazil. J. Clin. Microbiol. 2004, 42, 5925–5927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barchiesi, F.; Maracci, M.; Radi, B.; Arzeni, D.; Baldassarri, I.; Giacometti, A.; Scalise, G. Point prevalence, microbiology and fluconazole susceptibility patterns of yeast isolates colonizing the oral cavities of HIV-infected patients in the era of highly active antiretroviral therapy. J. Antimicrob. Chemother. 2002, 50, 999–1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottfredsson, M.; Cox, G.M.; Indridason, Ó.S.; De Almeida, G.M.D.; Heald, A.E.; Perfect, J.R. Association of Plasma Levels of Human Immunodeficiency Virus Type 1 RNA and Oropharyngeal Candida Colonization. J. Infect. Dis. 1999, 180, 534–537. [Google Scholar] [CrossRef] [Green Version]
- Gajdács, M.; Dóczi, I.; Ábrók, M.; Lázár, A.; Burián, K. Epidemiology of candiduria and Candida urinary tract infections in inpatients and outpatients: Results from a 10-year retrospective survey. Cent. Eur. J. Urol. 2019, 72, 209–214. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Sae-Tia, S.; Fries, B.C. Candidiasis and Mechanisms of Antifungal Resistance. Antibiotics 2020, 9, 312. [Google Scholar] [CrossRef]
- Selmecki, A.; Forche, A.; Berman, J. Genomic Plasticity of the Human Fungal Pathogen Candida albicans. Eukaryot. Cell 2010, 9, 991–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patton, L.L.; van der Horst, C. Oral infections and other manifestations of HIV disease. Infect. Dis. Clin. N. Am. 1999, 13, 879–900. [Google Scholar] [CrossRef]
- Dios, P.D.; Ocampo, A.; Miralles, C.; Otero, I.; Iglesias, I.; Rayo, N. Frequency of oropharyngeal candidiasis in HIV-infected patients on protease inhibitor therapy. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 1999, 87, 437–441. [Google Scholar] [CrossRef]
- Dios, P.D.; Hermida, A.O.; Llerena, N.R.; Martin, I.I.; Varela, I.O.; Vazquez, C.M. Effect of Plasma Human Immunodeficiency Virus Type 1 RNA Levels on Neutrophil Oxidative Burst. J. Infect. Dis. 2000, 181, 1214–1215. [Google Scholar] [CrossRef] [Green Version]
- Patton, L.L.; McKaig, R.; Strauss, R.; Rogers, D.; Eron, J.J., Jr. Changing prevalence of oral manifestations of human immunodeficiency virus in the era of protease inhibitor therapy. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2000, 89, 299–304. [Google Scholar] [CrossRef]
- Ho, D.D.; Neumann, A.U.; Perelson, A.S.; Chen, W.; Leonard, J.M.; Markowitz, M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nat. Cell Biol. 1995, 373, 123–126. [Google Scholar] [CrossRef]
- Wendland, T.; Furrer, H.; Vernazza, P.L.; Frutig, K.; Christen, A.; Matter, L.; Malinverni, R.; Pichler, W.J. HAART in HIV-infected patients: Restoration of antigen-specific CD4 T-cell responses in vitro is correlated with CD4 memory T-cell re-constitution, whereas improvement in delayed type hypersensitivity is related to a decrease in viraemia. AIDS 1999, 13, 1857–1862. [Google Scholar] [CrossRef] [Green Version]
- Mastroianni, C.M.; Lichtner, M.; Mengoni, F.; D’Agostino, C.; Forcina, G.; D’Ettorre, G.; Santopadre, P.; Vullo, V. Improvement in neutrophil and monocyte function during highly active antiretroviral treatment of HIV-1-infected patients. AIDS 1999, 13, 883–890. [Google Scholar] [CrossRef]
- Pericolini, E.; Cenci, E.; Gabrielli, E.; Perito, S.; Mosci, P.; Bistoni, F.; Vecchiarelli, A. Indinavir influences biological function of dendritic cells and stimulates antifungal immunity. J. Leukoc. Biol. 2008, 83, 1286–1294. [Google Scholar] [CrossRef]
- Powderly, W.G.; Landay, A.; Lederman, M.M. Recovery of the immune system with antiretroviral therapy: The end of opportunism? JAMA 1998, 280, 72–77. [Google Scholar] [CrossRef]
- Sepkowitz, K.A. Effect of HAART on natural history of AIDS-related opportunistic disorders. Lancet 1998, 351, 228–230. [Google Scholar] [CrossRef]
- Zingman, B.S. Resolution of Refractory AIDS-Related Mucosal Candidiasis after Initiation of Didanosine plus Saquinavir. N. Engl. J. Med. 1996, 334, 1674–1675. [Google Scholar] [CrossRef]
- Cauda, R.; Tacconelli, E.; Tumbarello, M.; Morace, G.; De Bernardis, F.; Torosantucci, A.; Cassone, A. Role of Protease Inhibitors in Preventing Recurrent Oral Candidosis in Patients With HIV Infection: A Prospective Case-Control Study. JAIDS J. Acquir. Immune Defic. Syndr. 1999, 21, 20–25. [Google Scholar] [CrossRef]
- Braga-Silva, L.A.; Santos, A.L.S. Aspartic protease inhibitors as potential anti-Candida albicans drugs: Impacts on fungal biology, virulence and pathogenesis. Curr. Med. Chem. 2011, 18, 2401–2419. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.L.S. Aspartic proteases of human pathogenic fungi are prospective targets for the generation of novel and effective antifungal inhibitors. Curr. Enzym. Inhib. 2011, 7, 96–118. [Google Scholar] [CrossRef]
- Saag, M.S.; Gandhi, R.T.; Hoy, J.F.; Landovitz, R.J.; Thompson, M.A.; Sax, P.E.; Smith, D.M.; Benson, C.A.; Buchbinder, S.P.; Del Rio, C.; et al. Antiretroviral drugs for treatment and prevention of HIV infection in adults: 2020 recommendations of the International Antiviral Society-USA Panel. JAMA 2020, 324, 1651–1669. [Google Scholar] [CrossRef]
- Otto, S.B.J.; George, P.E.; Mercedes, R.; Nabukeera-Barungi, N. Cryptococcal meningitis and immune reconstitution inflammatory syndrome in a pediatric patient with HIV after switching to second line antiretroviral therapy: A case report. BMC Infect. Dis. 2020, 20, 68. [Google Scholar] [CrossRef] [PubMed]
- Braga-Silva, L.A.; Mesquita, D.G.; Ribeiro, M.D.; Carvalho, S.M.; Fracalanzza, S.E.; Santos, A.L.S. Trailing end-point pheno-type antibiotic-sensitive strains of Candida albicans produce different amounts of aspartyl peptidases. Braz. J. Med. Biol. Res. 2009, 42, 765–770. [Google Scholar] [CrossRef] [Green Version]
- White, T.; Agabian, N. Candida albicans secreted aspartyl proteinases: Isoenzyme pattern is determined by cell type, and levels are determined by environmental factors. J. Bacteriol. 1995, 177, 5215–5221. [Google Scholar] [CrossRef] [Green Version]
- Santos, A.L.S.; Carvalho, I.M.; Silva, B.A.; Portela, M.B.; Alviano, C.S.; Soares, R.M.A. Secretion of serine peptidase by a clinical strain of Candida albicans: Influence of growth conditions and cleavage of human serum proteins and extracellular matrix components. FEMS Immunol. Med. Microbiol. 2006, 46, 209–220. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cutfield, S.M.; Dodson, E.J.; Anderson, B.F.; Moody, P.C.; Marshall, C.J.; Sullivan, P.A.; Cutfield, J.F.; Moody, P.; Moody, P.; Marshall, C. The crystal structure of a major secreted aspartic proteinase from Candida albicans in complexes with two inhibitors. Struct. 1995, 3, 1261–1271. [Google Scholar] [CrossRef] [Green Version]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2010, 7, 525–537. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Olson, A.J. Using AutoDock for Ligand-Receptor Docking. Curr. Protoc. Bioinform. 2008, 24, 8–14. [Google Scholar] [CrossRef]
- Burokerkilgore, M.; Wang, K. A Coomassie Brilliant Blue G-250-Based Colorimetric Assay for Measuring Activity of Calpain and Other Proteases. Anal. Biochem. 1993, 208, 387–392. [Google Scholar] [CrossRef]
- Braga-Silva, L.A.; Mogami, S.S.; Valle, R.S.; Silva-Neto, I.D.; Santos, A.L.S. Multiple effects of amprenavir against Candida albicans. FEMS Yeast Res. 2010, 10, 221–224. [Google Scholar] [CrossRef]
- Palmeira, V.F.; Kneipp, L.F.; Rozental, S.; Alviano, C.S.; Santos, A.L.S. Beneficial Effects of HIV Peptidase Inhibitors on Fonsecaea pedrosoi: Promising Compounds to Arrest Key Fungal Biological Processes and Virulence. PLoS ONE 2008, 3, e3382. [Google Scholar] [CrossRef] [Green Version]
- Hazen, K.C.; Cutler, J.E. Autoregulation of germ tube formation by Candida albicans. Infect. Immun. 1979, 24, 661–666. [Google Scholar] [CrossRef] [Green Version]
- Braga-Silva, L.A.; Santos, A.L.S.; Portela, M.B.; Souto-Padron, N.T.; Soares, R.M.A. Effect of suramin on the human pathogen Candida albicans: Implications on the fungal development and virulence. FEMS Immunol. Med. Microbiol. 2007, 51, 399–406. [Google Scholar] [CrossRef] [Green Version]
- Soares, M.C.; Aléssio, M.L.; Léger, C.L.; Bluet-Pajot, M.T.; Clauser, H.; Enjalbert, A.; Kordon, C.; Wandscheer, D.E. Effect of essential fatty acid deficiency on membrane fatty acid content and growth hormone stimulation of rat pituitaries during postnatal development. J. Lipid Res. 1995, 36, 1401–1406. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane Stanley, G.H. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Larsen, T.; Axelsen, J.; Ravn, H.W. Simplified and rapid method for extraction of ergosterol from natural samples and detection with quantitative and semi-quantitative methods using thin-layer chromatography. J. Chromatogr. A 2004, 1026, 301–304. [Google Scholar] [CrossRef]
- Granato, M.Q.; Gonçalves, D.S.; Seabra, S.H.; McCann, M.; Devereux, M.; Santos, A.L.S.; Kneipp, L.F. 1,10-Phenanthroline-5,6-dione-based compounds are effective in disturbing crucial physiological events of Phialophora verrucosa. Front. Microbiol. 2017, 8, 76. [Google Scholar] [CrossRef] [Green Version]
- Palmeira, V.F.; Alviano, D.S.; Braga-Silva, L.A.; Goulart, F.R.V.; Granato, M.Q.; Rozental, S.; Alviano, C.S.; Santos, A.L.S.; Kneipp, L.F. HIV aspartic peptidase inhibitors modulate surface molecules and enzyme activities involved with physiopathological events in Fonsecaea pedrosoi. Front. Microbiol. 2017, 8, 918. [Google Scholar] [CrossRef]
- Rüchel, R.; Tegeler, R.; Trost, M. A comparison of secretory proteinases from different strains of Candida albicans. Med. Mycol. 1982, 20, 233–244. [Google Scholar] [CrossRef]
- Price, M.F.; Wilkinson, I.D.; Gentry, L.O. Plate method for detection of phospholipase activity in Candida albicans. Med. Mycol. 1982, 20, 7–14. [Google Scholar] [CrossRef]
- Aktas, E.; Yigit, N.; Ayyildiz, A. Esterase Activity in Various Candida Species. J. Int. Med. Res. 2002, 30, 322–324. [Google Scholar] [CrossRef]
- Peeters, E.; Nelis, H.J.; Coenye, T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J. Microbiol. Methods 2008, 72, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Gandra, R.M.; Silva, L.N.; Souto, X.M.; Sangenito, L.S.; Cruz, L.P.S.; Braga-Silva, L.A.; Gonçalves, D.S.; Seabra, S.H.; Branquinha, M.H.; Santos, A.L.S. The serine peptidase inhibitor TPCK induces several morphophysiological changes in the opportunistic fungal pathogen Candida parapsilosis sensu stricto. Med. Mycol. 2019, 57, 1024–1037. [Google Scholar] [CrossRef]
- Muñoz, J.E.; Ramirez, L.M.; Dias, L.D.S.; Rivas, L.A.; Ramos, L.S.; Santos, A.L.S.; Taborda, C.P.; Parra-Giraldo, C.M. Pathogenicity levels of Colombian strains of Candida auris and Brazilian strains of Candida haemulonii species complex in both murine and Galleria mellonella experimental models. J. Fungi 2020, 6, 104. [Google Scholar] [CrossRef]
- Rossi, D.C.; Muñoz, J.E.; Carvalho, D.D.; Belmonte, R.; Faintuch, B.; Borelli, P.; Miranda, A.; Taborda, C.P.; Daffre, S. Thrapeutic use of a cationic antimicrobial peptide from the spider Acanthoscurria gomesiana in the control of experimental candidiasis. BMC Microbiol. 2012, 12, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andes, D.; Diekema, D.J.; Pfaller, M.A.; Prince, R.A.; Marchillo, K.; Ashbeck, J.; Hou, J. In vivo pharmacodynamic characterization of anidulafungin in a neutropenic murine candidiasis model. Antimicrob. Agents Chemother. 2008, 52, 539–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naglik, J.R.; Challacombe, S.J.; Hube, B. Candida albicans Secreted Aspartyl Proteinases in Virulence and Pathogenesis. Microbiol. Mol. Biol. Rev. 2003, 67, 400–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monika, S.; Malgorzata, B.; Zbigniew, O. Contribution of Aspartic Proteases in Candida Virulence. Protease Inhibitors against Candida Infections. Curr. Protein Pept. Sci. 2017, 18, 1050–1062. [Google Scholar] [CrossRef] [PubMed]
- Na, B.K.; Song, C.Y. Use of monoclonal antibody in diagnosis of candidiasis caused by Candida albicans: Detection of circulating aspartyl proteinase antigen. Clin. Diagn. Lab. Immunol. 1999, 6, 924–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, K.; Abad-Zapatero, C. Candida proteases and their inhibition: Prospects for antifungal therapy. Curr. Med. Chem. 2001, 8, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Monod, M.; Borg-von, Z.M. Secreted aspartic proteases as virulence factors of Candida species. Biol. Chem. 2002, 383, 1087–1093. [Google Scholar] [CrossRef]
- Cvetkovic, R.S.; Goa, K.L. Lopinavir/ritonavir: A review of its use in the management of HIV infection. Drugs 2003, 63, 769–802. [Google Scholar] [CrossRef]
- Hermann, J.C.; Ghanem, E.; Li, Y.; Raushel, F.M.; Irwin, J.J.; Shoichet, B.K. Predicting substrates by docking high-energy intermediates to enzyme structures. J. Am. Chem. Soc. 2006, 128, 15882–15891. [Google Scholar] [CrossRef]
- Jorgensen, W.L. Efficient drug lead discovery and optimization. Acc. Chem. Res. 2009, 42, 724–733. [Google Scholar] [CrossRef] [Green Version]
- Calugi, C.; Guarna, A.; Trabocchi, A. Insight into the structural similarity between HIV protease and secreted aspartic protease-2 and binding mode analysis of HIV-Candida albicans inhibitors. J. Enzyme Inhib. Med. Chem. 2013, 28, 936–943. [Google Scholar] [CrossRef] [Green Version]
- Mastrolorenzo, A.; Rusconi, S.; Scozzafava, A.; Barbaro, G.; Supuran, C. Inhibitors of HIV-1 Protease: Current State of the Art 10 Years after their Introduction. From Antiretroviral Drugs to Antifungal, Antibacterial and Antitumor Agents Based on Aspartic Protease Inhibitors. Curr. Med. Chem. 2007, 14, 2734–2748. [Google Scholar] [CrossRef]
- Kumar, S.P.; Kulkarni, V.M. Insights into the selective inhibition of Candida albicans secreted aspartyl protease: A docking analysis study. Bioorganic Med. Chem. 2002, 10, 1153–1170. [Google Scholar] [CrossRef]
- Dostál, J.; Brynda, J.; Hrušková-Heidingsfeldová, O.; Pachl, P.; Pichová, I.; Řezáčová, P. The crystal structure of protease Sapp1p from Candida parapsilosis in complex with the HIV protease inhibitor ritonavir. J. Enzym. Inhib. Med. Chem. 2011, 27, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.; Speth, C.; Lukasser-Vogl, E.; Zangerle, R.; Borg-von Zepelin, M.; Dierich, M.P.; Würzner, R. Human immunode-ficiency virus type 1 protease inhibitor attenuates Candida albicans virulence properties in vitro. Immunopharmacology 1999, 41, 227–234. [Google Scholar] [CrossRef]
- Blasi, E.; Colombari, B.; Orsi, C.F.; Pinti, M.; Troiano, L.; Cossarizza, A.; Esposito, R.; Peppoloni, S.; Mussini, C.; Neglia, R. The human immunodeficiency virus (HIV) protease inhibitor indinavir directly affects the opportunistic fungal pathogen Cryptococcus neoformans. FEMS Immunol. Med. Microbiol. 2004, 42, 187–195. [Google Scholar] [CrossRef]
- Valle, R.S.; Ramos, L.S.; Reis, V.J.; Ziccardi, M.; Dornelas-Ribeiro, M.; Sodré, C.L.; Branquinha, M.H.; Santos, A.L.S. Trichosporon asahii secretes a 30-kDa aspartic peptidase. Microbiol. Res. 2017, 205, 66–72. [Google Scholar] [CrossRef]
- Palmeira, V.F.; Kneipp, L.F.; Alviano, C.S.; Santos, A.L.S. Secretory aspartyl peptidase activity from mycelia of the human fungal pathogen Fonsecaea pedrosoi: Effect of HIV aspartyl proteolytic inhibitors. Res. Microbiol. 2006, 157, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Palmeira, V.F.; Goulart, F.R.V.; Granato, M.Q.; Alviano, D.S.; Alviano, C.S.; Kneipp, L.F.; Santos, A.L.S. Fonsecaea pedrosoi Sclerotic Cells: Secretion of Aspartic-Type Peptidase and Susceptibility to Peptidase Inhibitors. Front. Microbiol. 2018, 9, 1383. [Google Scholar] [CrossRef] [PubMed]
- Granato, M.Q.; Sousa, I.S.; Rosa, T.L.S.A.; Gonçalves, D.S.; Seabra, S.H.; Alviano, D.S.; Pessolani, M.C.V.; Santos, A.L.S.; Kneipp, L.F. Aspartic peptidase of Phialophora verrucosa as target of HIV peptidase inhibitors: Blockage of its enzymatic activity and interference with fungal growth and macrophage interaction. J. Enzym. Inhib. Med. Chem. 2020, 35, 629–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gehrt, A.; Peter, J.; Pizzo, P.A.; Walsh, T.J. Effect of increasing inoculum sizes of pathogenic filamentous fungi on MICs of antifungal agents by broth microdilution method. J. Clin. Microbiol. 1995, 33, 1302–1307. [Google Scholar] [CrossRef] [Green Version]
- Lass-Flörl, C.; Speth, C.; Kofler, G.; Dierch, M.P.; Gunsilius, E.; Würzner, R. Effect of increasing inoculum sizes of Aspergillus hyphae on MICs and MFCs of antifungal agents by broth microdilution method. Int. J. Antimicrob. Agents 2003, 21, 229–233. [Google Scholar] [CrossRef]
- Lerner, C.G.; Goldman, R.C. Stimuli that induce production of Candida albicans extracellular aspartyl proteinase. J. Gen. Microbiol. 1993, 139, 1643–1651. [Google Scholar] [CrossRef] [Green Version]
- Hube, B.; Monod, M.; Schofield, D.A.; Brown, A.J.; Gow, N.A. Expression of seven members of the gene family encoding se-cretory aspartyl proteinases in Candida albicans. Mol. Microbiol. 1994, 14, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Mata-Essayag, S.; Magaldi, S.; Hartung de Capriles, C.; Deibis, L.; Verde, G.; Perez, C. "In vitro" antifungal activity of protease inhibitors. Mycopathologia 2001, 152, 135–142. [Google Scholar] [CrossRef]
- Cordeiro, R.D.A.; Serpa, R.; Mendes, P.B.L.; Evangelista, A.J.D.J.; Andrade, A.R.C.; Franco, J.D.S.; Pereira, V.D.S.; De Alencar, L.P.; Oliveira, J.; De Camargo, Z.P.; et al. The HIV aspartyl protease inhibitor ritonavir impairs planktonic growth, biofilm formation and proteolytic activity in Trichosporon spp. Biofouling 2017, 33, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Brilhante, R.S.; Caetano, É.P.; Riello, G.B.; Guedes, G.M.; Castelo-Branco, D.D.S.; Fechine, M.A.; Oliveira, J.S.; Camargo, Z.P.; Mesquita, J.R.; Monteiro, A.J.; et al. Antiretroviral drugs saquinavir and ritonavir reduce inhibitory concentration values of itraconazole against Histoplasma capsulatum strains in vitro. Braz. J. Infect. Dis. 2016, 20, 155–159. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Alspaugh, J.A.; Liu, H.; Harris, S. Fungal Morphogenesis. Cold Spring Harb. Perspect. Med. 2014, 5, a019679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riquelme, M.; Aguirre, J.; Bartnicki-García, S.; Braus, G.H.; Feldbrügge, M.; Fleig, U.; Hansberg, W.; Herrera-Estrella, A.; Kämper, J.; Kück, U.; et al. Fungal Morphogenesis, from the Polarized Growth of Hyphae to Complex Reproduction and Infection Structures. Microbiol. Mol. Biol. Rev. 2018, 82, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basso, V.; D’Enfert, C.; Znaidi, S.; Bachellier-Bassi, S. From Genes to Networks: The Regulatory Circuitry Controlling Candida albicans Morphogenesis. Curr. Topics Microbiol. Immunol. 2018, 422, 61–99. [Google Scholar] [CrossRef]
- Kadosh, D. Regulatory mechanisms controlling morphology and pathogenesis in Candida albicans. Curr. Opin. Microbiol. 2019, 52, 27–34. [Google Scholar] [CrossRef]
- Cenci, E.; Francisci, D.; Belfiori, B.; Pierucci, S.; Baldelli, F.; Bistoni, F.; Vecchiarelli, A. Tipranavir exhibits different effects on opportunistic pathogenic fungi. J. Infect. 2008, 56, 58–64. [Google Scholar] [CrossRef]
- Margolis, A.M.; Heverling, H.; Pham, P.A.; Stolbach, A. A review of the toxicity of HIV medications. J. Med. Toxicol. 2014, 10, 26–39. [Google Scholar] [CrossRef] [Green Version]
- Guaraldi, G.; Stentarelli, C.; Zona, S.; Santoro, A. HIV-Associated Lipodystrophy: Impact of Antiretroviral Therapy. Drugs 2013, 73, 1431–1450. [Google Scholar] [CrossRef] [PubMed]
- Graef, M. Lipid droplet-mediated lipid and protein homeostasis in budding yeast. FEBS Lett. 2018, 592, 1291–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The fungal cell wall: Candida, Cryptococcus, and Aspergillus species. Front. Microbiol. 2019, 10, 2993. [Google Scholar] [CrossRef]
- Rollenhagen, C.; Mamtani, S.; Ma, D.; Dixit, R.; Eszterhas, S.; Lee, S.A. The Role of Secretory Pathways in Candida albicans Pathogenesis. J. Fungi 2020, 6, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rapala-Kozik, M.; Bochenska, O.; Zajac, D.; Karkowska-Kuleta, J.; Gogol, M.; Zawrotniak, M.; Kozik, A. Extracellular proteinases of Candida species pathogenic yeasts. Mol. Oral Microbiol. 2018, 33, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Cassone, A.; De Bernardis, F.; Torosantucci, A.; Tacconelli, E.; Tumbarello, M.; Cauda, R. In vitro and In vivo Anticandidal Activity of Human Immunodeficiency Virus Protease Inhibitors. J. Infect. Dis. 1999, 180, 448–453. [Google Scholar] [CrossRef]
- Monari, C.; Pericolini, E.; Bistoni, G.; Cenci, E.; Bistoni, F.; Vecchiarelli, A. Influence of Indinavir on Virulence and Growth of Cryptococcus neoformans. J. Infect. Dis. 2005, 191, 307–311. [Google Scholar] [CrossRef]
- Sidrim, J.J.; Perdigão-Neto, L.V.; Cordeiro, R.A.; Brilhante, R.S.; Leite, J.J.; Teixeira, C.E.; Monteiro, A.J.; Freitas, R.M.; Ribeiro, J.F.; Mesquita, J.R.; et al. Viral protease inhibitors affect the production of virulence factors in Cryptococcus neoformans. Can. J. Microbiol. 2012, 58, 932–936. [Google Scholar] [CrossRef]
- Staniszewska, M.; Monika, S. Virulence Factors in Candida species. Curr. Protein Pept. Sci. 2020, 21, 313–323. [Google Scholar] [CrossRef]
- De Mello, T.P.; Ramos, L.D.S.; Braga-Silva, L.A.; Branquinha, M.H.; Santos, A.L.S. Fungal Biofilm—A Real Obstacle against an Efficient Therapy: Lessons from Candida. Curr. Top. Med. Chem. 2017, 17, 1987–2004. [Google Scholar] [CrossRef]
- Tsang, C.S.P.; Hong, I.; Tsang, P.C.S. HIV protease inhibitors differentially inhibit adhesion of Candida albicans to acrylic surfaces. Mycoses 2009, 53, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Bektić, J.; Lell, C.P.; Fuchs, A.; Stoiber, H.; Speth, C.; Lass-Flörl, C.; Borg-von Zepelin, M.; Dierich, M.P.; Würzner, R. HIV protease inhibitors attenuate adherence of Candida albicans to epithelial cells in vitro. FEMS Immunol. Med. Microbiol. 2001, 31, 65–71. [Google Scholar] [CrossRef]
- Zepelin, M.B.-V.; Meyer, I.; Thomssen, R.; Würzner, R.; Sanglard, D.; Telenti, A.; Monod, M. HIV-Protease Inhibitors Reduce Cell Adherence of Candida albicans Strains by Inhibition of Yeast Secreted Aspartic Proteases. J. Investig. Dermatol. 1999, 113, 747–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korting, H.C.; Schaller, M.; Eder, G.; Hamm, G.; Böhmer, U.; Hube, B. Effects of the Human Immunodeficiency Virus (HIV) Proteinase Inhibitors Saquinavir and Indinavir on In vitro Activities of Secreted Aspartyl Proteinases of Candida albicans Isolates from HIV-Infected Patients. Antimicrob. Agents Chemother. 1999, 43, 2038–2042. [Google Scholar] [CrossRef] [Green Version]
- Lipke, P.N. What We Do Not Know about Fungal Cell Adhesion Molecules. J. Fungi 2018, 4, 59. [Google Scholar] [CrossRef] [Green Version]
- Segal, E.; Frenkel, M. Experimental In vivo Models of Candidiasis. J. Fungi 2018, 4, 21. [Google Scholar] [CrossRef] [Green Version]
- Mencacci, A.; Spaccapelo, R.; Del Sero, G.; Enssle, K.H.; Cassone, A.; Bistoni, F.; Romani, L. CD4+ T-helper-cell responses in mice with low-level Candida albicans infection. Infect. Immun. 1996, 64, 4907–4914. [Google Scholar] [CrossRef] [Green Version]
- Pericolini, E.; Cenci, E.; Monari, C.; Perito, S.; Mosci, P.; Bistoni, G.; Vecchiarelli, A. Indinavir-treated Cryptococcus neoformans promotes an efficient antifungal immune response in immunosuppressed hosts. Med. Mycol. 2006, 44, 119–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Normile, T.G.; Bryan, A.M.; Del Poeta, M. Animal models of Cryptococcus neoformans in identifying immune parameters associated with primary infection and reactivation of latent infection. Front. Immunol. 2020, 11, 581750. [Google Scholar] [CrossRef] [PubMed]
- Cassone, A.; Tacconelli, E.; De Bernardis, F.; Tumbarello, M.; Torosantucci, A.; Chiani, P.; Cauda, R. Antiretroviral Therapy with Protease Inhibitors Has an Early, Immune Reconstitution–Independent Beneficial Effect on Candida Virulence and Oral Candidiasis in Human Immunodeficiency Virus–Infected Subjects. J. Infect. Dis. 2002, 185, 188–195. [Google Scholar] [CrossRef] [Green Version]
- De Bernardis, F.; Tacconelli, E.; Mondello, F.; Cataldo, A.; Arancia, S.; Cauda, R.; Cassone, A. Anti-retroviral therapy with protease inhibitors decreases virulence enzyme expression in vivo by Candida albicans without selection of avirulent fungus strains or decreasing their anti-mycotic susceptibility. FEMS Immunol. Med. Microbiol. 2004, 41, 27–34. [Google Scholar] [CrossRef] [Green Version]
- Cassone, A.; Cauda, R. HIV proteinase inhibitors: Do they really work against Candida in a clinical setting? Trends Microbiol. 2002, 10, 177–178. [Google Scholar] [CrossRef]
- Eldesouky, H.E.; Salama, E.A.; Lanman, N.A.; Hazbun, T.R.; Seleem, M.N. Potent Synergistic Interactions between Lopinavir and Azole Antifungal Drugs against Emerging Multidrug-Resistant Candida auris. Antimicrob. Agents Chemother. 2020, 65, e00684-20. [Google Scholar] [CrossRef]
- Bustamante, C.; Ochoa, R.; Asela, C.; Muskus, C. Repurposing of known drugs for leishmaniasis treatment using bioinfor-matic predictions, in vitro validations and pharmacokinetic simulations. J. Comput. Aided Mol. Des. 2019, 33, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Nosengo, N. Can you teach old drugs new tricks? Nat. Cell Biol. 2016, 534, 314–316. [Google Scholar] [CrossRef] [PubMed]
- Gajdács, M.; Spengler, G. The role of drug repurposing in the development of novel antimicrobial drugs: Non-antibiotic phamacological agents as quorum sensing-inhibitors. Antibiotics 2019, 8, 270. [Google Scholar] [CrossRef] [Green Version]
- Stragliotto, G.; Pantalone, M.R.; Rahbar, A.; Söderberg-Nauclér, C. Valganciclovir as Add-On to Standard Therapy in Secondary Glioblastoma. Microorganisms 2020, 8, 1471. [Google Scholar] [CrossRef] [PubMed]
- Quezada, H.; Martínez-Vázquez, M.; López-Jácome, E.; González-Pedrajo, B.; Andrade, A.; Fernández-Presas, A.M.; Tovar-García, A.; García-Contreras, R. Repurposed anti-cancer drugs: The future for anti-infective therapy? Expert Rev. Anti-Infect. Ther. 2020, 18, 609–612. [Google Scholar] [CrossRef] [Green Version]
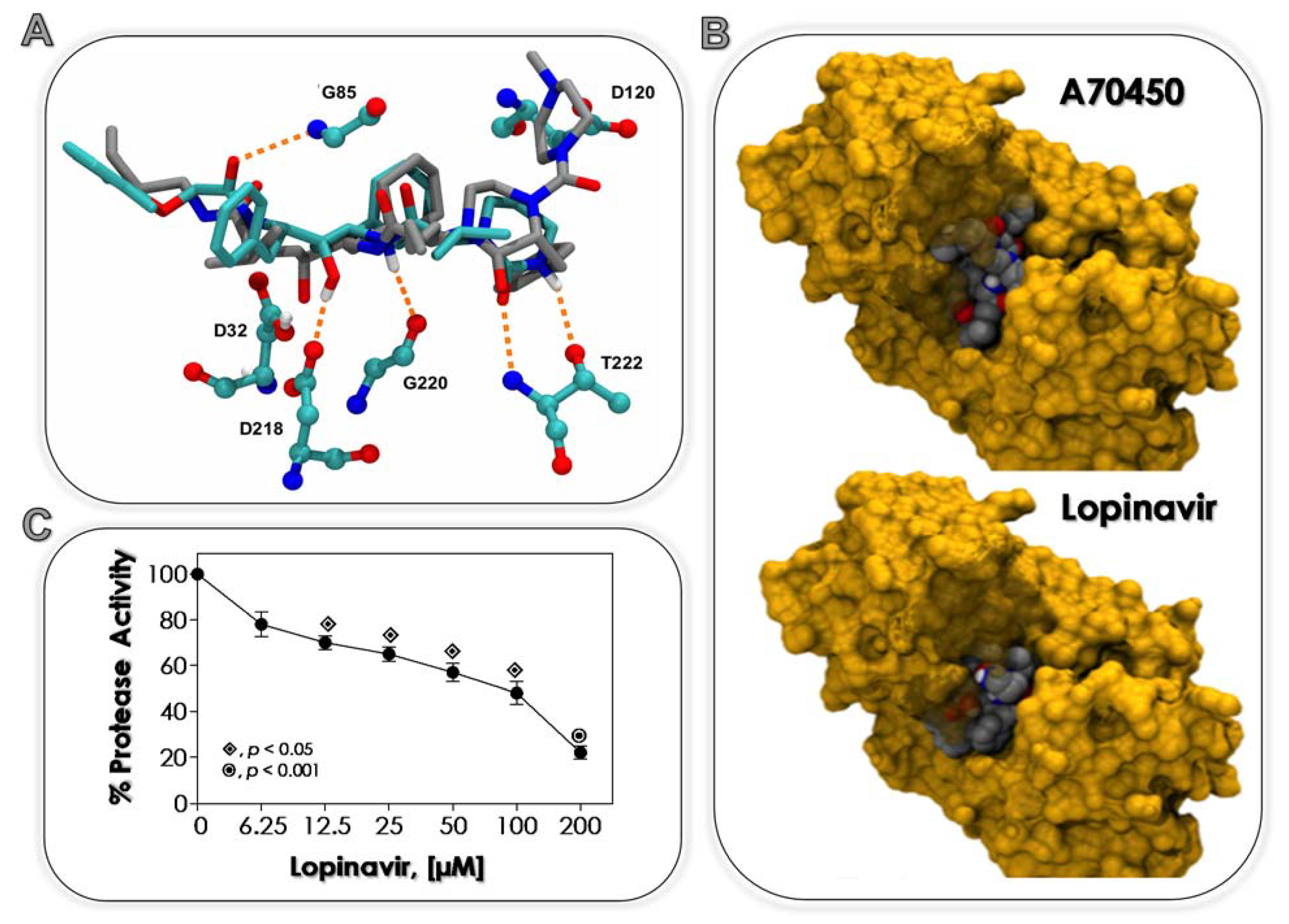
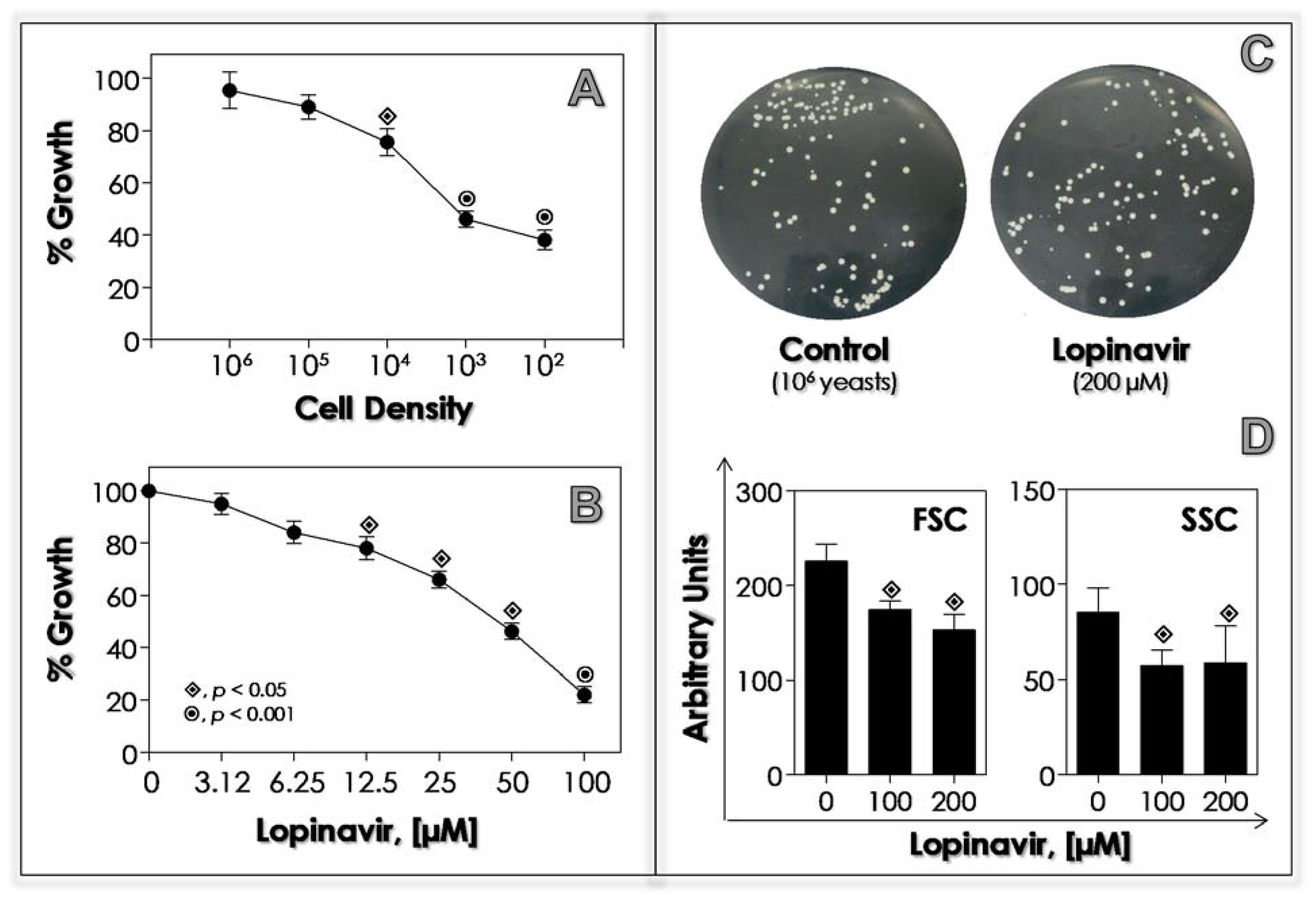
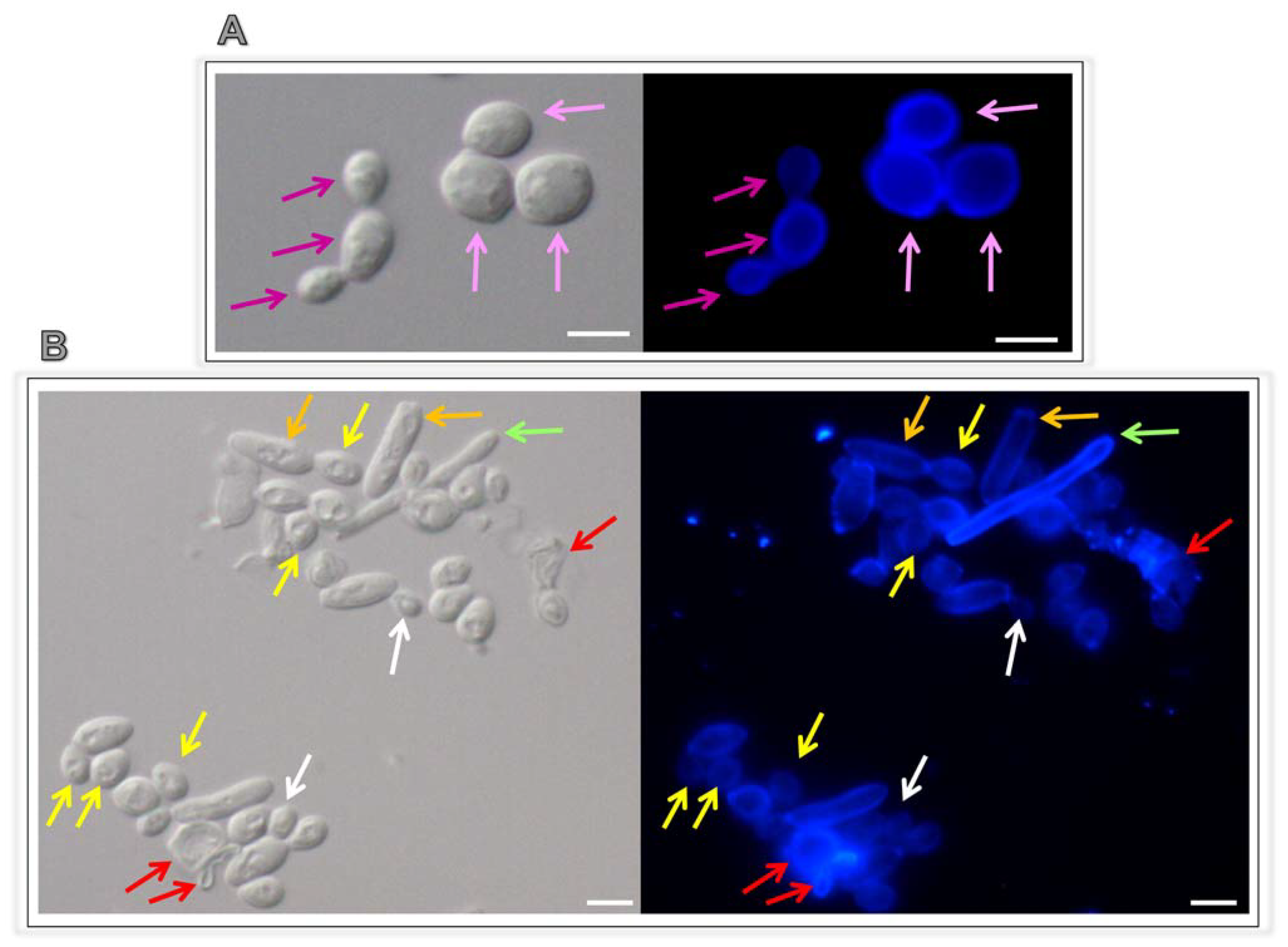
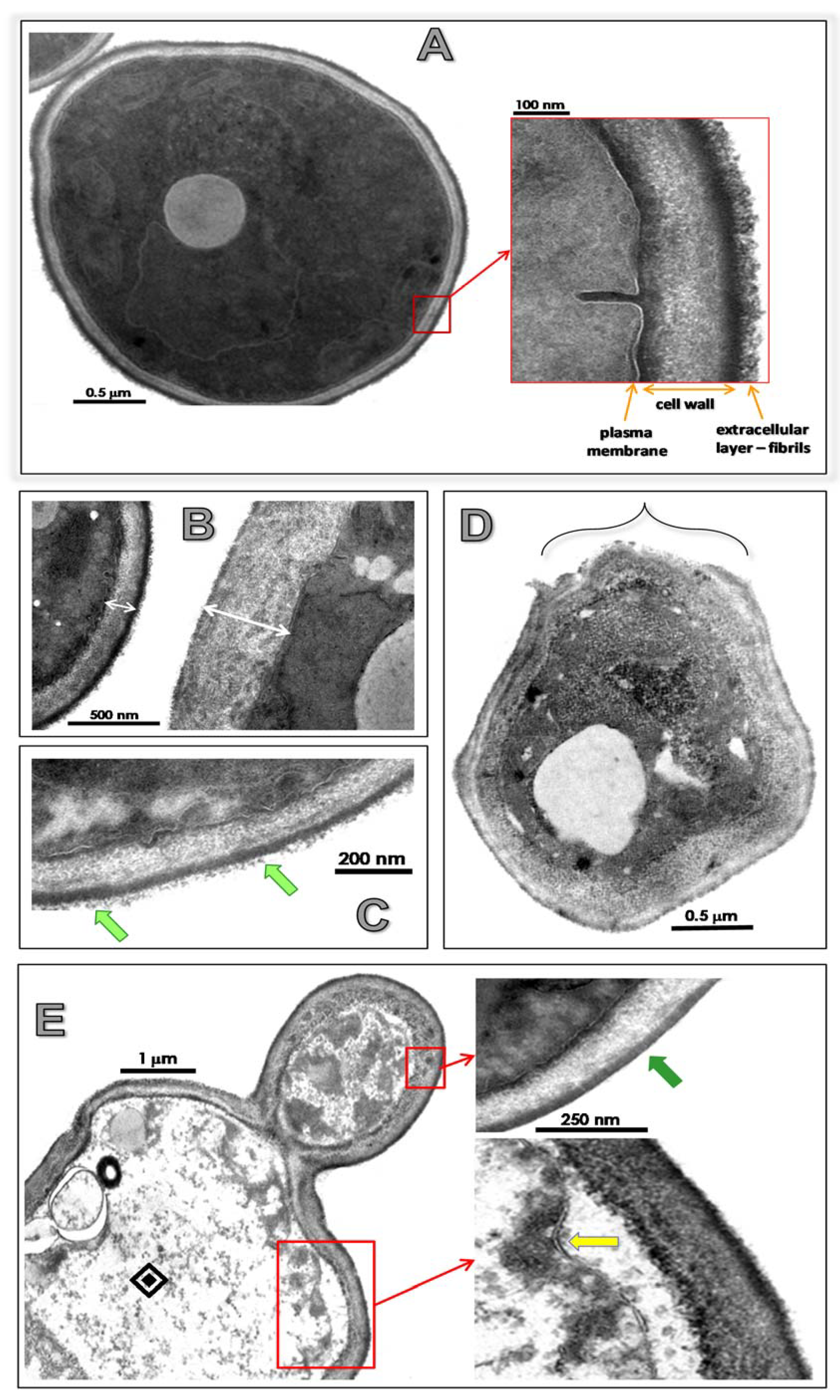




| Ligands | Estimated Binding Free Energy (kcal/mol) | Cluster Ranking |
|---|---|---|
| A70450 (standard inhibitor, protonated form) | −9.5 | 1 |
| Lopinavir (unprotonated) | −9.0 | 1 |
| Lopinavir (protonated) | −8.6 | 3 |
| [Lopinavir] | Sialic Acid-Rich Molecules (LFA-Labeled Cells) a | Mannose-Rich Molecules (ConA-Labeled Cells) a | N-Acetylglucosamine-Rich Molecules (WGA-Labeled Cells) a | Surface-Located Aspartic Proteases (Sap1-3-Labeled Cells) a | ||||
|---|---|---|---|---|---|---|---|---|
| %FC b | MFI b | %FC | MFI | %FC | MFI | %FC | MFI | |
| None | 41.96 ± 3.2 | 542.68 | 99.22 ± 0.4 | 1858.30 | 48.35 ± 1.3 | 114.62 | 88.77 ± 0.3 | 753.58 |
| 100 μM | 30.75 ± 1.3 * | 300.57 ** | 94.86 ± 3.3 | 1502.74 * | 59.72 ± 0.9 * | 161.03 ** | 90.01 ± 1.7 | 620.88 * |
| 200 μM | 25.30 ± 0.5 ** | 236.97 ** | 92.62 ± 2.9 | 598.31 ** | 74.04 ± 2.0 ** | 198.52 ** | 89.39 ± 1.1 | 366.67 ** |
| Lopinavir | Esterase Activity (Pz Value) a | Aspartic Protease Activity (Pz Value) |
|---|---|---|
| None | 0.576 ± 0.011 | 0.444 ± 0.052 |
| 100 μM | 0.629 ± 0.038 * | 0.595 ± 0.012 ** |
| 200 μM | 0.672 ± 0.021 ** | 0.775 ± 0.037 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, A.L.S.; Braga-Silva, L.A.; Gonçalves, D.S.; Ramos, L.S.; Oliveira, S.S.C.; Souza, L.O.P.; Oliveira, V.S.; Lins, R.D.; Pinto, M.R.; Muñoz, J.E.; et al. Repositioning Lopinavir, an HIV Protease Inhibitor, as a Promising Antifungal Drug: Lessons Learned from Candida albicans—In Silico, In Vitro and In Vivo Approaches. J. Fungi 2021, 7, 424. https://doi.org/10.3390/jof7060424
Santos ALS, Braga-Silva LA, Gonçalves DS, Ramos LS, Oliveira SSC, Souza LOP, Oliveira VS, Lins RD, Pinto MR, Muñoz JE, et al. Repositioning Lopinavir, an HIV Protease Inhibitor, as a Promising Antifungal Drug: Lessons Learned from Candida albicans—In Silico, In Vitro and In Vivo Approaches. Journal of Fungi. 2021; 7(6):424. https://doi.org/10.3390/jof7060424
Chicago/Turabian StyleSantos, André L. S., Lys A. Braga-Silva, Diego S. Gonçalves, Lívia S. Ramos, Simone S. C. Oliveira, Lucieri O. P. Souza, Vanessa S. Oliveira, Roberto D. Lins, Marcia R. Pinto, Julian E. Muñoz, and et al. 2021. "Repositioning Lopinavir, an HIV Protease Inhibitor, as a Promising Antifungal Drug: Lessons Learned from Candida albicans—In Silico, In Vitro and In Vivo Approaches" Journal of Fungi 7, no. 6: 424. https://doi.org/10.3390/jof7060424








