Yarrowia lipolytica Strains and Their Biotechnological Applications: How Natural Biodiversity and Metabolic Engineering Could Contribute to Cell Factories Improvement
Abstract
:1. Introduction
2. Everything You Always Wanted to Know about Yarrowia lipolytica (Briefly Resumed)
2.1. Natural Habitats and Safety
2.2. Main Characteristics
2.2.1. Physico-Chemical Conditions for Growth
2.2.2. Ploidy and Morphology
2.2.3. Carbon Sources
2.2.4. Secretion Pathway
2.2.5. Lipid Storage
2.2.6. Genomic Organization
2.3. An Impressive Curriculum Vitae: Short Review of Past, Present and Future Y. lipolytica Uses
2.3.1. Industrial Applications of Wild-Type or Traditionally Improved Strains
2.3.2. Commercial Applications of Genetically Modified Strains
2.3.3. Towards a Bio-Based Economy: Rewiring Strain Metabolism for Alternative Substrates
2.3.4. Potential Applications of Genetically Modified Strains: Bioproducts and Biofuels
2.3.5. Potential Applications of Genetically Modified Strains: Nanoparticles and Biomaterials
2.4. Long-Lost Relatives: Other Yeasts of the Yarrowia Clade
2.4.1. Brief Outline of Phylogeny, Habitat and Characteristics
2.4.2. Potential Applications of Other Yeasts of the Yarrowia Clade
3. Fantastic Yeasts and Where to Find Them: Yarrowia Strains and Yeast Collections
3.1. Oldies but Goodies: Elder Y. lipolytica Strains and Their Derivatives
3.1.1. From Paris Sewer to Worldwide Renown: The Success Story of W29
3.1.2. W29 and ATCC 18942 Progeny: E129 and E150 Strains
3.1.3. W29 Derivatives: The Po1 Series of Strains
3.1.4. Other Derivatives of W29: Obese Strains
3.1.5. Other Derivatives of W29: High-Throughput Expression Platforms for Protein Engineering
3.1.6. Other Derivatives of W29: Glyco-Engineered Strains for Producing Therapeutic Proteins
3.1.7. The Outsider H222
3.2. Gold Diggers: How to Find Nuggets in Old Mines
3.3. Finders Keepers: For a Good Isolate, Help Yourself
3.3.1. A-101 and Derivatives
3.3.2. ACA-DC 50109, Its Derivatives and Other Greek Y. lipolytica Isolates
3.3.3. New Kids on the Block: Y. lipolytica Strains Isolated or Noticed More Recently
4. A Brave New World of Engineered Strains: Tools and Strategies for Building Y. lipolytica Cell Factories
4.1. To Be or Not to Be Integrated: Types of Vectors and Assembly Methods
4.1.1. Episomal Vectors
4.1.2. Integrative Vectors and Cassettes
4.1.3. Multiple Transcription Unit Vectors, In Vitro DNA Assembly Methods and Y. lipolytica Toolboxes
4.1.4. In Vivo DNA Assembly Methods by Homologous Recombination and Artificial Chromosomes
4.2. Functional Elements for the Design of Expression Cassettes
4.2.1. Natural Y. lipolytica Promoters and Promoter Engineering
4.2.2. Natural and Synthetic Terminators
4.2.3. Targeting the Secretion Pathway: Secretion Signals and Surface Display Systems
4.2.4. Targeting Organelles for Subcellular Compartment Engineering
4.2.5. Selection Marker Genes and Marker Rescue Systems
4.3. Gene Editing and Whole Genome Analysis Technologies: CRISPR and Other Tools
4.3.1. CRISPR Tools and Y. lipolytica CRISPR-Cas9 Toolboxes for Gene Editing
4.3.2. TALEN Tools for Gene Editing in Y. lipolytica
4.3.3. Other CRISPR Tools for Base Editing and For Gene Repression or Activation
4.3.4. Transposomics and CRISPR-Derived Tools for Whole Genome Analysis in Y. lipolytica
4.4. How Gene Editing Can Leverage Strain Biodiversity and Be a Source of New Engineering Strategies
4.4.1. Increasing the Homologous Recombination Efficiency in Y. lipolytica
4.4.2. Diploid Strain Formation and Sexual Hybridization Following Mating Type Switching
4.5. Towards a Holistic View of Cell Factories and Bioprocesses Development
4.5.1. The Y. lipolytica Pan-Genome
4.5.2. Genome-Scale Omics Tools and Metabolic Models
4.5.3. Adaptative Evolution Strategies and Bioprocess Engineering
5. Conclusions in the Shape of a Question Mark: What Future for GMOs in Our Societies?
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Groenewald, M.; Boekhout, T.; Neuvéglise, C.; Gaillardin, C.; van Dijck, P.W.; Wyss, M. Yarrowia lipolytica: Safety assessment of an oleaginous yeast with a great industrial potential. Crit. Rev. Microbiol. 2014, 40, 187–206. [Google Scholar] [CrossRef]
- Fröhlich-Wyder, M.T.; Arias-Roth, E.; Jakob, E. Cheese yeasts. Yeast 2019, 36, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Butinar, L.; Santos, S.; Spencer-Martins, I.; Oren, A.; Gunde-Cimerman, N. Yeast diversity in hypersaline habitats. FEMS Microbiol. Lett. 2005, 244, 229–234. [Google Scholar] [CrossRef] [Green Version]
- Chi, Z.; Wang, F.; Wang, L.; Li, J.; Wang, X. Selection of Yarrowia lipolytica strains with high protein content from yeasts isolated from different marine environments. J. Ocean. Univ. China 2007, 6, 360–364. [Google Scholar] [CrossRef]
- Sibanda, T.; Selvarajan, R.; Tekere, M.; Nyoni, H.; Meddows-Taylor, S. Potential biotechnological capabilities of cultivable mycobiota from carwash effluents. Microbiologyopen 2017, 6, e00498. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.P.; Plata, C.; Reichelt, M.; Steiger, S.; Heckel, D.G.; Kaltenpoth, M.; Vilcinskas, A.; Vogel, H. Microbiome-assisted carrion preservation aids larval development in a burying beetle. Proc. Natl. Acad. Sci. USA 2018, 115, 11274–11279. [Google Scholar] [CrossRef] [Green Version]
- Mamaev, D.; Zvyagilskaya, R. Yarrowia lipolytica: A multitalented yeast species of ecological significance. FEMS Yeast Res. 2021, 21, foab008. [Google Scholar] [CrossRef] [PubMed]
- Desnos-Ollivier, M.; Letscher-Bru, V.; Neuvéglise, C.; Dromer, F. Yarrowia lipolytica causes sporadic cases and local outbreaks of infections and colonisation. Mycoses 2020, 63, 737–745. [Google Scholar] [CrossRef]
- Sutherland, J.B.; Cornelison, C.; Crow, S.A. CANDIDA | Yarrowia Lipolytica (Candida Lipolytica). In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 374–378. [Google Scholar]
- Barth, G.; Gaillardin, C. Yarrowia Lipolytica. In Genetics, Biochemistry and Molecular Biology of Non Conventional Yeasts in Biotechnology; Wolf, W.K., Ed.; Springer: Berlin, Germany, 1996; Volume 1, pp. 313–388. [Google Scholar]
- Barth, G.; Gaillardin, C. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol. Rev. 1997, 19, 219–237. [Google Scholar] [CrossRef]
- Thevenieau, F.; Nicaud, J.M.; Gaillardin, C. Applications of the Non-Conventional Yeast Yarrowia lipolytica. In Yeast Biotechnology: Diversity and Applications; Satyanarayana, T., Kunze, G., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 589–613. [Google Scholar]
- Pérez-Campo, F.M.; Domínguez, A. Factors affecting the morphogenetic switch in Yarrowia lipolytica. Curr. Microbiol. 2001, 43, 429–433. [Google Scholar] [CrossRef] [Green Version]
- Timoumi, A.; Guillouet, S.E.; Molina-Jouve, C.; Fillaudeau, L.; Gorret, N. Impacts of environmental conditions on product formation and morphology of Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2018, 102, 3831–3848. [Google Scholar] [CrossRef]
- Nicaud, J.M.; Fabre, E.; Gaillardin, C. Expression of invertase activity in Yarrowia lipolytica and its use as a selective marker. Curr. Genet. 1989, 16, 253–260. [Google Scholar] [CrossRef]
- Swennen, D.; Beckerich, J.M. Yarrowia lipolytica vesicle-mediated protein transport pathways. BMC Evol. Biol. 2007, 7, 219. [Google Scholar] [CrossRef] [Green Version]
- Beopoulos, A.; Chardot, T.; Nicaud, J.M. Yarrowia lipolytica: A model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie 2009, 91, 692–696. [Google Scholar] [CrossRef]
- Madzak, C.; Gaillardin, C.; Beckerich, J.M. Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: A review. J. Biotechnol. 2004, 109, 63–81. [Google Scholar] [CrossRef] [PubMed]
- Madzak, C.; Beckerich, J.M. Heterologous Protein Expression and Secretion in Yarrowia Lipolytica. In Yarrowia Lipolytica; Barth, G., Ed.; Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2013; Volume 25, pp. 1–76. [Google Scholar]
- Madzak, C. Yarrowia lipolytica: Recent achievements in heterologous protein expression and pathway engineering. Appl. Microbiol. Biotechnol. 2015, 99, 4559–4577. [Google Scholar] [CrossRef]
- De Pourcq, K.; Vervecken, W.; Dewerte, I.; Valevska, A.; Van Hecke, A.; Callewaert, N. Engineering the yeast Yarrowia lipolytica for the production of therapeutic proteins homogeneously glycosylated with Man8GlcNAc2 and Man5GlcNAc2. Microb Cell Fact. 2012, 11, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Yoo, S.J.; Kang, H.A. Yeast synthetic biology for the production of recombinant therapeutic proteins. FEMS Yeast Res. 2015, 15, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Athenaki, M.; Gardeli, C.; Diamantopoulou, P.; Tchakouteu, S.S.; Sarris, D.; Philippoussis, A.; Papanikolaou, S. Lipids from yeasts and fungi: Physiology, production and analytical considerations. J. Appl. Microbiol. 2018, 124, 336–367. [Google Scholar] [CrossRef] [Green Version]
- Andre, A.; Chatzifragkou, A.; Diamantopoulou, P.; Sarris, D.; Philippoussis, A.; Galiotou-Panayotou, M.; Komaitis, M.; Papanikolaou, S. Biotechnological conversions of bio-diesel-derived crude glycerol by Yarrowia lipolytica strains. Eng. Life Sci. 2009, 9, 468–478. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Chatzifragkou, A.; Fakas, S.; Galiotou-Panayotou, M.; Komaitis, M.; Nicaud, J.M.; Aggelis, G. Biosynthesis of lipids and organic acids by Yarrowia lipolytica strains cultivated on glucose. Eur. J. Lipid Sci. Technol. 2009, 111, 1221–1232. [Google Scholar] [CrossRef]
- Makri, A.; Fakas, S.; Aggelis, G. Metabolic activities of biotechnological interest in Yarrowia lipolytica grown on glycerol in repeated batch cultures. Bioresour. Technol. 2010, 101, 2351–2358. [Google Scholar] [CrossRef]
- Bellou, S.; Triantaphyllidou, I.E.; Aggeli, D.; Elazzazy, A.M.; Baeshen, M.N.; Aggelis, G. Microbial oils as food additives: Recent approaches for improving microbial oil production and its polyunsaturated fatty acid content. Curr. Opin. Biotechnol. 2016, 37, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Fickers, P.; Benetti, P.H.; Waché, Y.; Marty, A.; Mauersberger, S.; Smit, M.S.; Nicaud, J.M. Hydrophobic substrate utilisation by the yeast Yarrowia lipolytica, and its potential applications. FEMS Yeast Res. 2005, 5, 527–543. [Google Scholar] [CrossRef] [Green Version]
- Lasserre, J.P.; Nicaud, J.M.; Pagot, Y.; Joubert-Caron, R.; Caron, M.; Hardouin, J. First complexomic study of alkane-binding protein complexes in the yeast Yarrowia lipolytica. Talanta 2010, 80, 1576–1585. [Google Scholar] [CrossRef]
- Liu, H.; Song, Y.; Fan, X.; Wang, C.; Lu, X.; Tian, Y. Yarrowia lipolytica as an Oleaginous Platform for the Production of Value-Added Fatty Acid-Based Bioproducts. Front. Microbiol. 2021, 11, 608662. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Muniglia, L.; Chevalot, I.; Aggelis, G.; Marc, I. Accumulation of a cocoa-butter-like lipid by Yarrowia lipolytica cultivated on agro-industrial residues. Curr Microbiol 2003, 46, 124–130. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Aggelis, G. Yarrowia lipolytica: A model microorganism used for the production of tailor-made lipids. Eur. J. Lipid Sci. Technol. 2010, 112, 639–654. [Google Scholar] [CrossRef]
- Casarégola, S.; Feynerol, C.; Diez, M.; Fournier, P.; Gaillardin, C. Genomic organization of the yeast Yarrowia lipolytica. Chromosoma 1997, 106, 380–390. [Google Scholar] [CrossRef]
- Dujon, B.; Sherman, D.; Fischer, G.; Durrens, P.; Casaregola, S.; Lafontaine, I.; de Montigny, J.; Marck, C.; Neuvéglise, C.; Talla, E.; et al. Genome evolution in yeasts. Nature 2004, 430, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Neuvéglise, C.; Marck, C.; Gaillardin, C. The intronome of budding yeasts. C. R. Biol. 2011, 334, 662–670. [Google Scholar] [CrossRef]
- Magnan, C.; Yu, J.; Chang, I.; Jahn, E.; Kanomata, Y.; Wu, J.; Zeller, M.; Oakes, M.; Baldi, P.; Sandmeyer, S. Sequence Assembly of Yarrowia lipolytica Strain W29/CLIB89 Shows Transposable Element Diversity. PLoS ONE 2016, 11, e0162363. [Google Scholar] [CrossRef]
- Devillers, H.; Neuvéglise, C. Genome Sequence of the Oleaginous Yeast Yarrowia lipolytica H222. Microbiol. Resour. Announc. 2019, 8, e01547-18. [Google Scholar] [CrossRef] [Green Version]
- Casaregola, S.; Neuvéglise, C.; Lépingle, A.; Bon, E.; Feynerol, C.; Artiguenave, F.; Wincker, P.; Gaillardin, C. Genomic exploration of the hemiascomycetous yeasts: 17. Yarrowia lipolytica. FEBS Lett. 2000, 487, 95–100. [Google Scholar] [CrossRef] [Green Version]
- Schmid-Berger, N.; Schmid, B.; Barth, G. Ylt1, a highly repetitive retrotransposon in the genome of the dimorphic fungus Yarrowia lipolytica. J. Bacteriol. 1994, 176, 2477–2482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casaregola, S.; Neuvéglise, C.; Bon, E.; Gaillardin, C. Ylli, a non-LTR retrotransposon L1 family in the dimorphic yeast Yarrowia lipolytica. Mol. Biol. Evol. 2002, 19, 664–677. [Google Scholar] [CrossRef] [Green Version]
- Kovalchuk, A.; Senam, S.; Mauersberger, S.; Barth, G. Tyl6, a novel Ty3/gypsy-like retrotransposon in the genome of the dimorphic fungus Yarrowia lipolytica. Yeast 2005, 22, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Nicaud, J.M.; Madzak, C.; van den Broek, P.; Gysler, C.; Duboc, P.; Niederberger, P.; Gaillardin, C. Protein expression and secretion in the yeast Yarrowia lipolytica. FEMS Yeast Res. 2002, 2, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Madzak, C. New tools for heterologous protein production in the yeast yarrowia lipolytica. In Recent Research Developments in Microbiology; Pandalai, S.G., Ed.; Research Signpost: Trivandrum, India, 2003; Volume 7, pp. 453–479. [Google Scholar]
- Bankar, A.V.; Kumar, A.R.; Zinjarde, S.S. Environmental and industrial applications of Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2009, 84, 847–865. [Google Scholar] [CrossRef] [PubMed]
- Sibirny, A.; Madzak, C.; Fickers, P. Genetic engineering of non-conventional yeast for the production of valuable compounds. In Microbial Biotechnology: Progress and Trends; Harzevili, F.D., Chen, H., Eds.; CRC Press: Boca Raton, FL, USA, 2014; pp. 63–111. [Google Scholar]
- Papanikolaou, S.; Diamantopoulou, P.; Blanchard, F.; Lambrinea, E.; Chevalot, I.; Stoforos, N.G.; Rondags, E. Physiological Characterization of a Novel Wild-Type Yarrowia lipolytica Strain Grown on Glycerol: Effects of Cultivation Conditions and Mode on Polyols and Citric Acid Production. Appl. Sci. 2020, 10, 7373. [Google Scholar] [CrossRef]
- Rymowicz, W.; Rywińska, A.; Żarowska, B.; Juszczyk, P. Citric acid production from raw glycerol by acetate mutants of Yarrowia lipolytica. Chem. Pap. 2006, 60, 391–394. [Google Scholar] [CrossRef]
- Arslan, N.P.; Aydogan, M.N.; Taskin, M. Citric acid production from partly deproteinized whey under non-sterile culture conditions using immobilized cells of lactose-positive and cold-adapted Yarrowia lipolytica B9. J. Biotechnol. 2016, 231, 32–39. [Google Scholar] [CrossRef]
- Kamzolova, S.V.; Morgunov, I.G. Microbial production of (2R,3S)-isocitric acid: State of the arts and prospects. Appl. Microbiol. Biotechnol. 2019, 103, 9321–9333. [Google Scholar] [CrossRef] [PubMed]
- Zinjarde, S.S. Food-related applications of Yarrowia lipolytica. Food Chem. 2014, 152, 1–10. [Google Scholar] [CrossRef]
- Żogała, B.; Robak, M.; Rymowicz, W.; Wzientek, K.; Rusin, M.; Maruszczak, J. Geoelectrical observation of Yarrowia lipolytica bioremediation of petrol contaminated soil. Pol. J. Environ. Stud. 2005, 14, 665–669. [Google Scholar]
- Robak, M.; Boruczkowski, T.; Drozdz, W.; Lazar, Z.; Baranowska, M.; Przado, D. Application of the yeasts Yarrowia lipolytica for in-situ bioremediation of soil contaminated with creosote oil—A case study. Ochrona Srodowiska 2011, 33, 27–33. [Google Scholar]
- Fickers, P.; Nicaud, J.M.; Destain, J.; Thonart, P. Overproduction of lipase by Yarrowia lipolytica mutants. Appl. Microbiol. Biotechnol. 2003, 63, 136–142. [Google Scholar] [CrossRef]
- Madzak, C. Yarrowia lipolytica engineering as a source of microbial cell factories. In Microbial Cell Factories Engineering for Production of Biomolecules; Singh, V., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 345–380. [Google Scholar]
- Madzak, C. Engineering Yarrowia lipolytica for Use in Biotechnological Applications: A Review of Major Achievements and Recent Innovations. Mol. Biotechnol. 2018, 60, 621–635. [Google Scholar] [CrossRef]
- Abdel-Mawgoud, A.M.; Markham, K.A.; Palmer, C.M.; Liu, N.; Stephanopoulos, G.; Alper, H.S. Metabolic engineering in the host Yarrowia lipolytica. Metab. Eng. 2018, 50, 192–208. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Sharpe, P.L.; Hong, S.P.; Yadav, N.S.; Xie, D.; Short, D.R.; Damude, H.G.; Rupert, R.A.; Seip, J.E.; Wang, J.; et al. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat. Biotechnol. 2013, 31, 734–740. [Google Scholar] [CrossRef]
- Xie, D.; Jackson, E.N.; Zhu, Q. Sustainable source of omega-3 eicosapentaenoic acid from metabolically engineered Yarrowia lipolytica: From fundamental research to commercial production. Appl. Microbiol. Biotechnol. 2015, 99, 1599–1610. [Google Scholar] [CrossRef] [Green Version]
- Hollands, K.; Baron, C.M.; Gibson, K.J.; Kelly, K.J.; Krasley, E.A.; Laffend, L.A.; Lauchli, R.M.; Maggio-Hall, L.A.; Nelson, M.J.; Prasad, J.C.; et al. Engineering two species of yeast as cell factories for 2’-fucosyllactose. Metab. Eng. 2019, 52, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Pignède, G.; Wang, H.J.; Fudalej, F.; Seman, M.; Gaillardin, C.; Nicaud, J.M. Autocloning and amplification of LIP2 in Yarrowia lipolytica. Appl. Environ. Microbiol. 2000, 66, 3283–3289. [Google Scholar] [CrossRef] [Green Version]
- Tiels, P.; Baranova, E.; Piens, K.; De Visscher, C.; Pynaert, G.; Nerinckx, W.; Stout, I.; Fudalej, F.; Hulpiau, P.; Tännler, S.; et al. A bacterial glycosidase enables mannose-6-phosphate modification and improved cellular uptake of yeast-produced recombinant human lysosomal enzymes. Nat. Biotechnol. 2012, 30, 1225–1231. [Google Scholar] [CrossRef] [PubMed]
- Ledesma-Amaro, R.; Nicaud, J.M. Metabolic Engineering for Expanding the Substrate Range of Yarrowia lipolytica. Trends Biotechnol. 2016, 34, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Spagnuolo, M.; Shabbir Hussain, M.; Gambill, L.; Blenner, M. Alternative Substrate Metabolism in Yarrowia lipolytica. Front. Microbiol. 2018, 9, 1077. [Google Scholar] [CrossRef]
- Miller, K.K.; Alper, H.S. Yarrowia lipolytica: More than an oleaginous workhorse. Appl. Microbiol. Biotechnol. 2019, 103, 9251–9262. [Google Scholar] [CrossRef] [PubMed]
- Soong, Y.V.; Liu, N.; Yoon, S.; Lawton, C.; Xie, D. Cellular and metabolic engineering of oleaginous yeast Yarrowia lipolytica for bioconversion of hydrophobic substrates into high-value products. Eng. Life Sci. 2019, 19, 423–443. [Google Scholar] [CrossRef] [Green Version]
- Gottardi, D.; Siroli, L.; Vannini, L.; Patrignani, F.; Lanciotti, R. Recovery and valorization of agri-food wastes and by-products using the non-conventional yeast Yarrowia lipolytica. Trends Food Sci. Technol. 2021, 115, 74–86. [Google Scholar] [CrossRef]
- Carsanba, E.; Papanikolaou, S.; Fickers, P.; Erten, H. Screening various Yarrowia lipolytica strains for citric acid production. Yeast 2019, 36, 319–327. [Google Scholar] [CrossRef]
- Lopes, M.; Miranda, S.M.; Costa, A.R.; Pereira, A.S.; Belo, I. Yarrowia lipolytica as a biorefinery platform for effluents and solid wastes valorization-challenges and opportunities. Crit. Rev. Biotechnol. 2021. [online ahead of print]. [Google Scholar] [CrossRef]
- Mano, J.; Liu, N.; Hammond, J.H.; Currie, D.H.; Stephanopoulos, G. Engineering Yarrowia lipolytica for the utilization of acid whey. Metab. Eng. 2020, 57, 43–50. [Google Scholar] [CrossRef]
- Hapeta, P.; Rakicka, M.; Dulermo, R.; Gamboa-Meléndez, H.; Cruz-Le Coq, A.M.; Nicaud, J.M.; Lazar, Z. Transforming sugars into fat-lipid biosynthesis using different sugars in Yarrowia lipolytica. Yeast 2017, 34, 293–304. [Google Scholar] [CrossRef] [Green Version]
- Niehus, X.; Crutz-Le Coq, A.M.; Sandoval, G.; Nicaud, J.M.; Ledesma-Amaro, R. Engineering Yarrowia lipolytica to enhance lipid production from lignocellulosic materials. Biotechnol. Biofuels 2018, 11, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, S.; Hipp, J.; Trinh, C.T. Activating and Elucidating Metabolism of Complex Sugars in Yarrowia lipolytica. Appl. Environ. Microbiol. 2015, 82, 1334–1345. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Alper, H.S. Enabling xylose utilization in Yarrowia lipolytica for lipid production. Biotechnol. J. 2016, 11, 1230–1240. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Alper, H.S. Producing Biochemicals in Yarrowia lipolytica from Xylose through a Strain Mating Approach. Biotechnol. J. 2020, 15, e1900304. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Su, S.; Madzak, C.; Zhou, J.; Chen, H.; Chen, G. Applying pathway engineering to enhance production of alpha-ketoglutarate in Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2016, 100, 9875–9884. [Google Scholar] [CrossRef]
- Braga, A.; Belo, I. Biotechnological production of γ-decalactone, a peach like aroma, by Yarrowia lipolytica. World J. Microbiol. Biotechnol. 2016, 32, 169. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Han, B.; Gui, X.; Wang, G.; Xu, L.; Yan, Y.; Madzak, C.; Pan, D.; Wang, Y.; Zha, G.; et al. Engineering Yarrowia lipolytica to Simultaneously Produce Lipase and Single Cell Protein from Agro-industrial Wastes for Feed. Sci. Rep. 2018, 8, 758. [Google Scholar] [CrossRef] [Green Version]
- Shi, N.; Mao, W.; He, X.; Chi, Z.; Chi, Z.; Liu, G. Co-expression of Exo-inulinase and Endo-inulinase Genes in the Oleaginous Yeast Yarrowia lipolytica for Efficient Single Cell Oil Production from Inulin. Appl. Biochem. Biotechnol. 2018, 185, 334–346. [Google Scholar] [CrossRef] [PubMed]
- Worland, A.M.; Czajka, J.J.; Li, Y.; Wang, Y.; Tang, Y.J.; Su, W.W. Biosynthesis of terpene compounds using the non-model yeast Yarrowia lipolytica: Grand challenges and a few perspectives. Curr. Opin. Biotechnol. 2020, 64, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, A.; Feng, X.; Rasool, A.; Sun, W.; Li, C. Production of plant natural products through engineered Yarrowia lipolytica. Biotechnol. Adv. 2020, 43, 107555. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.J.; Wang, Y.Z.; Wang, L.R.; Shi, T.Q.; Sun, X.M.; Huang, H. Advanced Strategies for the Synthesis of Terpenoids in Yarrowia lipolytica. J. Agric. Food Chem. 2021, 69, 2367–2381. [Google Scholar] [CrossRef]
- Tong, Y.; Zhou, J.; Zhang, L.; Xu, P. A Golden-Gate Based Cloning Toolkit to Build Violacein Pathway Libraries in Yarrowia lipolytica. ACS Synth. Biol. 2021, 10, 115–124. [Google Scholar] [CrossRef]
- Fickers, P.; Cheng, H.; Sze Ki Lin, C. Sugar Alcohols and Organic Acids Synthesis in Yarrowia lipolytica: Where Are We? Microorganisms 2020, 8, 574. [Google Scholar] [CrossRef]
- Abbasi, A.R.; Liu, J.; Wang, Z.; Zhao, A.; Ying, H.; Qu, L.; Alam, M.A.; Xiong, W.; Xu, J.; Lv, Y. Recent Advances in Producing Sugar Alcohols and Functional Sugars by Engineering Yarrowia lipolytica. Front. Bioeng. Biotechnol. 2021, 9, 648382. [Google Scholar] [CrossRef]
- Bilal, M.; Xu, S.; Iqbal, H.M.N.; Cheng, H. Yarrowia lipolytica as an emerging biotechnological chassis for functional sugars biosynthesis. Crit. Rev. Food Sci. Nutr. 2021, 61, 535–552. [Google Scholar] [CrossRef]
- Gao, Q.; Yang, J.L.; Zhao, X.R.; Liu, S.C.; Liu, Z.J.; Wei, L.J.; Hua, Q. Yarrowia lipolytica as a Metabolic Engineering Platform for the Production of Very-Long-Chain Wax Esters. J. Agric. Food Chem. 2020, 68, 10730–10740. [Google Scholar] [CrossRef]
- Gao, C.; Qi, Q.; Madzak, C.; Lin, C.S. Exploring medium-chain-length polyhydroxyalkanoates production in the engineered yeast Yarrowia lipolytica. J. Ind. Microbiol. Biotechnol. 2015, 42, 1255–1262. [Google Scholar] [CrossRef]
- Lajus, S.; Dusséaux, S.; Verbeke, J.; Rigouin, C.; Guo, Z.; Fatarova, M.; Bellvert, F.; Borsenberger, V.; Bressy, M.; Nicaud, J.M.; et al. Engineering the Yeast Yarrowia lipolytica for Production of Polylactic Acid Homopolymer. Front. Bioeng. Biotechnol. 2020, 8, 954. [Google Scholar] [CrossRef]
- Wang, L.; Zong, Z.; Liu, Y.; Zheng, M.; Li, D.; Wang, C.; Zheng, F.; Madzak, C.; Liu, Z. Metabolic engineering of Yarrowia lipolytica for the biosynthesis of crotonic acid. Bioresour. Technol. 2019, 287, 121484. [Google Scholar] [CrossRef] [PubMed]
- Ledesma-Amaro, R.; Nicaud, J.M. Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids. Prog. Lipid Res. 2016, 61, 40–50. [Google Scholar] [CrossRef]
- Abghari, A.; Madzak, C.; Chen, S. Combinatorial Engineering of Yarrowia lipolytica as a Promising Cell Biorefinery Platform for the de novo Production of Multi-Purpose Long Chain Dicarboxylic Acids. Fermentation 2017, 3, 40. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Yan, Y.; Madzak, C.; Han, B. Harnessing biodiesel-producing microbes: From genetic engineering of lipase to metabolic engineering of fatty acid biosynthetic pathway. Crit. Rev. Biotechnol. 2017, 37, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Spagnuolo, M.; Yaguchi, A.; Blenner, M. Oleaginous yeast for biofuel and oleochemical production. Curr. Opin. Biotechnol. 2019, 57, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Gu, Y.; Marsafari, M.; Xu, P. Synthetic biology, systems biology, and metabolic engineering of Yarrowia lipolytica toward a sustainable biorefinery platform. J. Ind. Microbiol. Biotechnol. 2020, 47, 845–862. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Moradi, H.; Shi, S.; Darvishi, F. Yeasts as microbial cell factories for sustainable production of biofuels. Renew. Sustain. Energy Rev. 2021, 143, 110907. [Google Scholar] [CrossRef]
- Blazeck, J.; Liu, L.; Knight, R.; Alper, H.S. Heterologous production of pentane in the oleaginous yeast Yarrowia lipolytica. J. Biotechnol. 2013, 165, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Bruder, S.; Moldenhauer, E.J.; Lemke, R.D.; Ledesma-Amaro, R.; Kabisch, J. Drop-in biofuel production using fatty acid photodecarboxylase from Chlorella variabilis in the oleaginous yeast Yarrowia lipolytica. Biotechnol. Biofuels 2019, 12, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Ma, Y.; Liu, N.; Eser, B.E.; Guo, Z.; Jensen, P.R.; Stephanopoulos, G. Synthesis of high-titer alka(e)nes in Yarrowia lipolytica is enabled by a discovered mechanism. Nat. Commun. 2020, 11, 6198. [Google Scholar] [CrossRef]
- Xu, P.; Qiao, K.; Ahn, W.S.; Stephanopoulos, G. Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proc. Natl. Acad. Sci. USA 2016, 113, 10848–10853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Qiao, Y.; Li, F.; Xu, Y.; Yan, Y.; Madzak, C.; Yan, J. Subcellular engineering of lipase dependent pathways directed towards lipid related organelles for highly effectively compartmentalized biosynthesis of triacylglycerol derived products in Yarrowia lipolytica. Metab. Eng. 2019, 55, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, F.; Qiao, Y.; Zhou, Q.; Hu, Z.; He, Y.; Yan, Y.; Xu, L.; Madzak, C.; Yan, J. Design of a New Multienzyme Complex Synthesis System Based on Yarrowia lipolytica Simultaneously Secreted and Surface Displayed Fusion Proteins for Sustainable Production of Fatty Acid-Derived Hydrocarbons. ACS Sustain. Chem. Eng. 2018, 6, 17035–17043. [Google Scholar] [CrossRef]
- Pimprikar, P.S.; Joshi, S.S.; Kumar, A.R.; Zinjarde, S.S.; Kulkarni, S.K. Influence of biomass and gold salt concentration on nanoparticle synthesis by the tropical marine yeast Yarrowia lipolytica NCIM 3589. Colloids Surf. B Biointerfaces 2009, 74, 309–316. [Google Scholar] [CrossRef]
- Ben Tahar, I.; Fickers, P.; Dziedzic, A.; Płoch, D.; Skóra, B.; Kus-Liśkiewicz, M. Green pyomelanin-mediated synthesis of gold nanoparticles: Modelling and design, physico-chemical and biological characteristics. Microb. Cell. Fact. 2019, 18, 210. [Google Scholar] [CrossRef]
- Han, Z.; Madzak, C.; Su, W.W. Tunable nano-oleosomes derived from engineered Yarrowia lipolytica. Biotechnol. Bioeng. 2013, 110, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, Z.; Liu, G.; Cheng, X.; Chi, Z.; Madzak, C.; Liu, C.; Chi, Z. Genetical Surface Display of Silicatein on Yarrowia lipolytica Confers Living and Renewable Biosilica-Yeast Hybrid Materials. ACS Omega 2020, 5, 7555–7566. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.; Xi, W.; Cheng, Z.; Yin, L.; Li, R.; Wu, G.; Liu, W.; Xu, J.; Xiang, S.; Zheng, Y.; et al. A Living Eukaryotic Autocementation Kit from Surface Display of Silica Binding Peptides on Yarrowia lipolytica. ACS Synth. Biol. 2016, 5, 1466–1474. [Google Scholar] [CrossRef]
- Péter, G.; Dlauchy, D.; Vasdinyei, R.; Tornai-Lehoczki, J.; Deák, T. Candida galli sp. nov., a new yeast from poultry. Antonie Van Leeuwenhoek 2004, 86, 105–110. [Google Scholar] [CrossRef]
- Knutsen, A.K.; Robert, V.; Poot, G.A.; Epping, W.; Figge, M.; Holst-Jensen, A.; Skaar, I.; Smith, M.T. Polyphasic re-examination of Yarrowia lipolytica strains and the description of three novel Candida species: Candida oslonensis sp. nov., Candida alimentaria sp. nov. and Candida hollandica sp. nov. Int. J. Syst. Evol. Microbiol. 2007, 57 Pt 10, 2426–2435. [Google Scholar] [CrossRef]
- Limtong, S.; Youngmanitchai, W.; Kawasaki, H.; Seki, T. Candida phangngensis sp. nov., an anamorphic yeast species in the Yarrowia clade, isolated from water in mangrove forests in Phang-Nga Province, Thailand. Int. J. Syst. Evol. Microbiol. 2008, 58 Pt 2, 515–519. [Google Scholar] [CrossRef]
- Chang, C.F.; Chen, C.C.; Lee, C.F.; Liu, S.M. Identifying and characterizing Yarrowia keelungensis sp. nov., an oil-degrading yeast isolated from the sea surface microlayer. Antonie Van Leeuwenhoek 2013, 104, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Groenewald, M.; Smith, M.T. The teleomorph state of Candida deformans Langeron & Guerra and description of Yarrowia yakushimensis comb. nov. Antonie Van Leeuwenhoek 2013, 103, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Nagy, E.; Niss, M.; Dlauchy, D.; Arneborg, N.; Nielsen, D.S.; Péter, G. Yarrowia divulgata f.a., sp. nov., a yeast species from animal-related and marine sources. Int. J. Syst. Evol. Microbiol. 2013, 63 Pt 12, 4818–4823. [Google Scholar] [CrossRef] [Green Version]
- Nagy, E.; Dlauchy, D.; Medeiros, A.O.; Péter, G.; Rosa, C.A. Yarrowia porcina sp. nov. and Yarrowia bubula f.a. sp. nov., two yeast species from meat and river sediment. Antonie Van Leeuwenhoek 2014, 105, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Gaillardin, C.; Neuvéglise, C.; Kerscher, S.; Nicaud, J.M. Mitochondrial genomes of yeasts of the Yarrowia clade. FEMS Yeast Res. 2012, 12, 317–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bing, J.; You, Z.; Zheng, Q.; Tang, J.; Ran, Y.; Huang, G. Biological and genomic analyses of a clinical isolate of Yarrowia galli from China. Curr. Genet. 2020, 66, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.F.; Li, X.H.; Hui, F.L. Yarrowia brassicae f.a., sp. nov., a new yeast species from traditional Chinese sauerkraut. Int. J. Syst. Evol. Microbiol. 2018, 68, 2024–2027. [Google Scholar] [CrossRef] [PubMed]
- Rakicka, M.; Kieroń, A.; Hapeta, P.; Neuvéglise, C.; Lazar, Z. Sweet and sour potential of yeast from the Yarrowia clade. Biomass Bioenergy 2016, 92, 48–54. [Google Scholar] [CrossRef]
- Quarterman, J.; Slininger, P.J.; Kurtzman, C.P.; Thompson, S.R.; Dien, B.S. A survey of yeast from the Yarrowia clade for lipid production in dilute acid pretreated lignocellulosic biomass hydrolysate. Appl. Microbiol. Biotechnol. 2017, 101, 3319–3334. [Google Scholar] [CrossRef]
- Morin, N.; Czerwiec, Q.; Nicaud, J.M.; Neuvéglise, C.; Rossignol, T. Transforming Candida hispaniensis, a promising oleaginous and flavogenic yeast. Yeast 2020, 37, 348–355. [Google Scholar] [CrossRef]
- Červenák, F.; Juríková, K.; Devillers, H.; Kaffe, B.; Khatib, A.; Bonnell, E.; Sopkovičová, M.; Wellinger, R.J.; Nosek, J.; Tzfati, Y.; et al. Identification of telomerase RNAs in species of the Yarrowia clade provides insights into the co-evolution of telomerase, telomeric repeats and telomere-binding proteins. Sci. Rep. 2019, 9, 13365. [Google Scholar] [CrossRef] [Green Version]
- Michely, S.; Gaillardin, C.; Nicaud, J.M.; Neuvéglise, C. Comparative physiology of oleaginous species from the Yarrowia clade. PLoS ONE 2013, 8, e63356. [Google Scholar] [CrossRef] [PubMed]
- Quarterman, J.C.; Slininger, P.J.; Hector, R.E.; Dien, B.S. Engineering Candida phangngensis-an oleaginous yeast from the Yarrowia clade-for enhanced detoxification of lignocellulose-derived inhibitors and lipid overproduction. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef] [PubMed]
- Wojtatowicz, M.; Rymowicz, W.; Kautola, H. Comparison of different strains of the yeast Yarrowia lipolytica for citric acid production from glucose hydrol. Appl. Biochem. Biotechnol. 1991, 31, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Rywińska, A.; Rymowicz, W.; Zarowska, B.; Skrzypiński, A. Comparison of citric acid production from glycerol and glucose by different strains of Yarrowia lipolytica. World J. Microbiol. Biotechnol. 2010, 26, 1217–1224. [Google Scholar] [CrossRef] [PubMed]
- Devillers, H.; Brunel, F.; Połomska, X.; Sarilar, V.; Lazar, Z.; Robak, M.; Neuvéglise, C. Draft genome sequence of Yarrowia lipolytica strain A-101 isolated from polluted soil in Poland. Genome Announc. 2016, 4, e01094-16. [Google Scholar] [CrossRef] [Green Version]
- Lazar, Z.; Rossignol, T.; Verbeke, J.; Crutz-Le Coq, A.-M.; Nicaud, J.-M.; Robak, M. Optimized invertase expression and secretion cassette for improving Yarrowia lipolytica growth on sucrose for industrial applications. J. Ind. Microbiol. Biotechnol. 2013, 40, 1273–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rywińska, A.; Juszczyk, P.; Wojtatowicz, M.; Robak, M.; Lazar, Z.; Tomaszewska, L.; Rymowicz, W. Glycerol as a promising substrate for Yarrowia lipolytica biotechnological applications. Biomass Bioenergy 2013, 48, 148–166. [Google Scholar] [CrossRef]
- Rzechonek, D.A.; Neuvéglise, C.; Devillers, H.; Rymowicz, W.; Mirończuk, A.M. EUF1-a newly identified gene involved in erythritol utilization in Yarrowia lipolytica. Sci. Rep. 2017, 7, 12507. [Google Scholar] [CrossRef] [Green Version]
- Papanikolaou, S.; Galiotou-Panayotou, M.; Chevalot, I.; Komaitis, M.; Marc, I.; Aggelis, G. Influence of glucose and saturated free-fatty acid mixtures on citric acid and lipid production by Yarrowia lipolytica. Curr. Microbiol. 2006, 52, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Bellou, S.; Triantaphyllidou, I.E.; Mizerakis, P.; Aggelis, G. High lipid accumulation in Yarrowia lipolytica cultivated under double limitation of nitrogen and magnesium. J. Biotechnol. 2016, 234, 116–126. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Fakas, S.; Fick, M.; Chevalot, I.; Galiotou-Panayotou, M.; Komaitis, M.; Marc, I.; Aggelis, G. Biotechnological valorisation of raw glycerol discharged after bio-diesel (fatty acid methyl esters) manufacturing process: Production of 1,3-propanediol, citric acid and single cell oil. Biomass Bioenergy 2008, 32, 60–71. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Galiotou-Panayotou, M.; Fakas, S.; Komaitis, M.; Aggelis, G. Citric acid production by Yarrowia lipolytica cultivated on olive-mill wastewater-based media. Bioresour. Technol. 2008, 99, 2419–2428. [Google Scholar] [CrossRef]
- Zhao, C.H.; Cui, W.; Liu, X.Y.; Chi, Z.M.; Madzak, C. Expression of inulinase gene in the oleaginous yeast Yarrowia lipolytica and single cell oil production from inulin-containing materials. Metab. Eng. 2010, 12, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Y.; Zhang, Y.; Chi, Z.; Liu, G.L.; Wang, Z.P.; Chi, Z.M. Role of pyruvate carboxylase in accumulation of intracellular lipid of the oleaginous yeast Yarrowia lipolytica ACA-DC 50109. Appl. Microbiol. Biotechnol. 2015, 99, 1637–1645. [Google Scholar] [CrossRef]
- Daskalaki, A.; Perdikouli, N.; Aggeli, D.; Aggelis, G. Laboratory evolution strategies for improving lipid accumulation in Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2019, 103, 8585–8596. [Google Scholar] [CrossRef] [PubMed]
- Paramithiotis, S.; Mueller, M.R.A.; Ehrmann, M.A.; Tsakalidou, E.; Seiler, H.; Vogel, R.; Kalantzopoulos, G. Polyphasic identification of wild yeast strains isolated from Greek sourdoughs. Syst. Appl. Microbiol. 2000, 23, 156–164. [Google Scholar] [CrossRef]
- Sarantou, S.; Stoforos, N.G.; Kalantzi, O.; Papanikolaou, S. Biotechnological valorization of biodiesel-derived glycerol: Trials with the non-conventional yeasts Yarrowia lipolytica and Rhodosporidium sp. Carbon Resour. Convers. 2021, 4, 61–75. [Google Scholar] [CrossRef]
- von Arx, J.A. On endomyces, Endomycopsis and related yeast-like fungi. Antonie Van Leeuwenhoek 1972, 38, 289–309. [Google Scholar] [CrossRef]
- Dos Santos, E.O.; Michelon, M.; Furlong, E.B.; Burkert, J.F.; Kalil, S.J.; Burkert, C.A. Evaluation of the composition of culture medium for yeast biomass production using raw glycerol from biodiesel synthesis. Braz. J. Microbiol. 2012, 43, 432–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, K.; Machida, H. Studies on the production of lipase by microorganisms. Part IV. Identification of a new strain and determination of its lipase activity. Nippon Nogeikagaku Kaishi 1963, 37, 649–652. [Google Scholar] [CrossRef]
- Briffaud, J.; Engasser, J.M. Citric acid production from glucose. I. Growth and excretion kinetics in a stirred fermentor. Biotechnol. Bioeng. 1979, 21, 2083–2092. [Google Scholar] [CrossRef]
- Briffaud, J.; Engasser, J. Citric acid production from glucose. II. Growth and excretion kinetics in a trickle-flow fermentor. Biotechnol. Bioeng. 1979, 21, 2093–2111. [Google Scholar] [CrossRef]
- Wentworth, S.D.; Cooper, D.G. Self-cycling fermentation of a citric acid producing strain of Candida lipolytica. J. Ferment. Bioeng. 1996, 81, 400–405. [Google Scholar] [CrossRef]
- Barth, G.; Weber, H. Genetic studies in the yeast Saccharomyces lipolytica: Inactivation and mutagenesis. Z. Allg. Mikrobiol. 1983, 23, 147–157. [Google Scholar] [CrossRef]
- Förster, A.; Aurich, A.; Mauersberger, S.; Barth, G. Citric acid production from sucrose using a recombinant strain of the yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2007, 75, 1409–1417. [Google Scholar] [CrossRef]
- Moeller, L.; Zehnsdorf, A.; Aurich, A.; Bley, T.; Strehlitz, B. Substrate utilization by recombinant Yarrowia lipolytica growing on sucrose. Appl. Microbiol. Biotechnol. 2012, 93, 1695–1702. [Google Scholar] [CrossRef]
- Zinjarde, S.S.; Pant, A.A. Hydrocarbon degraders from tropical marine environments. Mar. Pollut. Bull. 2002, 44, 118–121. [Google Scholar] [CrossRef]
- Dusane, D.H.; Nancharaiah, Y.V.; Venugopalan, V.P.; Kumar, A.R.; Zinjarde, S.S. Biofilm formation by a biotechnologically important tropical marine yeast isolate, Yarrowia lipolytica NCIM 3589. Water Sci. Technol. 2008, 58, 2467–2475. [Google Scholar] [CrossRef]
- Zinjarde, S.S.; Pant, A. Emulsifier from a tropical marine yeast, yarrowia lipolytica NCIM 3589. J. Basic Microbiol. 2002, 42, 67–73. [Google Scholar] [CrossRef]
- Liu, X.; Wang, X.; Xu, J.; Xia, J.; Lv, J.; Zhang, T.; Wu, Z.; Deng, Y.; He, J. Citric acid production by Yarrowia lipolytica SWJ-1b using corn steep liquor as a source of organic nitrogen and vitamins. Ind. Crop. Prod. 2015, 78, 154–160. [Google Scholar] [CrossRef]
- Tan, M.J.; Chen, X.; Wang, Y.K.; Liu, G.L.; Chi, Z.M. Enhanced citric acid production by a yeast Yarrowia lipolytica over-expressing a pyruvate carboxylase gene. Bioprocess. Biosyst. Eng. 2016, 39, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Wang, Q.; Zhang, F.; Zhang, S.C.; Chi, Z.M.; Madzak, C. Direct conversion of inulin into single cell protein by the engineered Yarrowia lipolytica carrying inulinase gene. Process Biochem. 2011, 46, 1442–1448. [Google Scholar] [CrossRef]
- Liu, X.; Lv, J.; Xu, J.; Xia, J.; Dai, B.; Xu, X.; Xu, J.l. Erythritol production by Yarrowia lipolytica mutant strain M53 generated through atmospheric and room temperature plasma mutagenesis. Food Sci. Biotechnol. 2017, 26, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Gaillardin, C.M.; Charoy, V.; Heslot, H. A study of copulation, sporulation and meiotic segregation in Candida lipolytica. Arch. Mikrobiol 1973, 92, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Ogrydziak, D.M.; Nicaud, J.M. Characterization of Yarrowia lipolytica XPR2 multi-copy strains over-producing alkaline extracellular protease—A system for rapidly increasing secretory pathway cargo loads. FEMS Yeast Res. 2012, 12, 938–948. [Google Scholar] [CrossRef] [Green Version]
- Pomraning, K.R.; Baker, S.E. Draft genome sequence of the dimorphic yeast Yarrowia lipolytica strain W29. Genome Announc. 2015, 3, e01211-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickerham, L.J.; Kurtzman, C.P.; Herman, A.I. Sexual reproduction in Candida lipolytica. Science 1970, 167, 1141. [Google Scholar] [CrossRef]
- Le Dall, M.T.; Nicaud, J.M.; Gaillardin, C. Multiple-copy integration in the yeast Yarrowia lipolytica. Curr. Genet. 1994, 26, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Madzak, C.; Tréton, B.; Blanchin-Roland, S. Strong hybrid promoters and integrative expression/secretion vectors for quasi-constitutive expression of heterologous proteins in the yeast Yarrowia lipolytica. J. Mol. Microbiol. Biotechnol. 2000, 2, 207–216. [Google Scholar] [PubMed]
- Liu, L.; Alper, H.S. Draft Genome Sequence of the Oleaginous Yeast Yarrowia lipolytica PO1f, a Commonly Used Metabolic Engineering Host. Genome Announc. 2014, 2, e00652-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, C.; Ryu, S.; Haridas, S.; Na, H.; Zane, M.; LaButti, K.; Barry, K.; Grigoriev, I.V.; Trinh, C.T. Draft Genome Assemblies of Ionic Liquid-Resistant Yarrowia lipolytica PO1f and Its Superior Evolved Strain, YlCW001. Microbiol. Resour. Announc. 2020, 9, e01356-19. [Google Scholar] [CrossRef] [Green Version]
- Leplat, C.; Nicaud, J.M.; Rossignol, T. High-throughput transformation method for Yarrowia lipolytica mutant library screening. FEMS Yeast Res. 2015, 15, fov052. [Google Scholar] [CrossRef] [Green Version]
- Kamzolova, S.V.; Dedyukhina, E.G.; Samoilenko, V.A.; Lunina, J.N.; Puntus, I.F.; Allayarov, R.L.; Chiglintseva, M.N.; Mironov, A.A.; Igor, G.; Morgunov, I.G. Isocitric acid production from rapeseed oil by Yarrowia lipolytica yeast. Appl. Microbiol. Biotechnol. 2013, 97, 9133–9144. [Google Scholar] [CrossRef] [PubMed]
- Kamzolova, S.V.; Morgunov, I.G. Metabolic peculiarities of the citric acid overproduction from glucose in yeasts Yarrowia lipolytica. Bioresour. Technol. 2017, 243, 433–440. [Google Scholar] [CrossRef]
- Kamzolova, S.V.; Morgunov, I.G. α-Ketoglutaric acid production from rapeseed oil by Yarrowia lipolytica yeast. Appl. Microbiol. Biotechnol. 2013, 97, 5517–5525. [Google Scholar] [CrossRef]
- Zhou, J.W.; Zhou, H.Y.; Du, G.C.; Liu, L.M.; Chen, J. Screening of a thiamine-auxotrophic yeast for alpha-ketoglutaric acid overproduction. Lett. Appl. Microbiol. 2010, 51, 264–271. [Google Scholar] [CrossRef]
- Zeng, W.; Zhang, H.; Xu, S.; Fang, F.; Zhou, J. Biosynthesis of keto acids by fed-batch culture of Yarrowia lipolytica WSH-Z06. Bioresour. Technol. 2017, 243, 1037–1043. [Google Scholar] [CrossRef]
- Zeng, W.; Fang, F.; Liu, S.; Du, G.; Chen, J.; Zhou, J. Comparative genomics analysis of a series of Yarrowia lipolytica WSH-Z06 mutants with varied capacity for α-ketoglutarate production. J. Biotechnol. 2016, 239, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Madzak, C.; Du, G.; Zhou, J.; Chen, J. Enhanced alpha-ketoglutaric acid production in Yarrowia lipolytica WSH-Z06 by regulation of the pyruvate carboxylation pathway. Appl. Microbiol. Biotechnol. 2012, 96, 1527–1537. [Google Scholar] [CrossRef]
- Zhou, J.; Yin, X.; Madzak, C.; Du, G.; Chen, J. Enhanced α-ketoglutarate production in Yarrowia lipolytica WSH-Z06 by alteration of the acetyl-CoA metabolism. J. Biotechnol. 2012, 161, 257–264. [Google Scholar] [CrossRef]
- Walker, C.; Ryu, S.; Na, H.; Zane, M.; LaButti, K.; Lipzen, A.; Haridas, S.; Barry, K.; Grigoriev, I.V.; Quarterman, J.; et al. Draft Genome Assemblies of Five Robust Yarrowia lipolytica Strains Exhibiting High Lipid Production, Pentose Sugar Utilization, and Sugar Alcohol Secretion from Undetoxified Lignocellulosic Biomass Hydrolysates. Microbiol. Resour. Announc. 2018, 7, e01040-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerscher, S.; Durstewitz, G.; Casaregola, S.; Gaillardin, C.; Brandt, U. The complete mitochondrial genome of yarrowia lipolytica. Comp. Funct. Genom. 2001, 2, 80–90. [Google Scholar] [CrossRef] [Green Version]
- Blanchin-Roland, S.; Cordero Otero, R.R.; Gaillardin, C. Two upstream activation sequences control the expression of the XPR2 gene in the yeast Yarrowia lipolytica. Mol. Cell. Biol. 1994, 14, 327–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madzak, C.; Mimmi, M.C.; Caminade, E.; Brault, A.; Baumberger, S.; Briozzo, P.; Mougin, C.; Jolivalt, C. Shifting the optimal pH of activity for a laccase from the fungus Trametes versicolor by structure-based mutagenesis. Protein Eng. Des. Sel. 2006, 9, 77–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galli, C.; Gentili, P.; Jolivalt, C.; Madzak, C.; Vadalà, R. How is the reactivity of laccase affected by single-point mutations? Engineering laccase for improved activity towards sterically demanding substrates. Appl. Microbiol. Biotechnol. 2011, 91, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Bredeweg, E.L.; Pomraning, K.R.; Dai, Z.; Nielsen, J.; Kerkhoven, E.J.; Baker, S.E. A molecular genetic toolbox for Yarrowia lipolytica. Biotechnol. Biofuels 2017, 10, 2. [Google Scholar] [CrossRef] [Green Version]
- Wagner, J.M.; Williams, E.V.; Alper, H.S. Developing a piggyBac Transposon System and Compatible Selection Markers for Insertional Mutagenesis and Genome Engineering in Yarrowia lipolytica. Biotechnol. J. 2018, 13, e1800022. [Google Scholar] [CrossRef]
- Wang, J.; Ledesma-Amaro, R.; Wei, Y.; Ji, B.; Ji, X.J. Metabolic engineering for increased lipid accumulation in Yarrowia lipolytica—A Review. Bioresour. Technol. 2020, 313, 123707. [Google Scholar] [CrossRef] [PubMed]
- Dulermo, T.; Nicaud, J.M. Involvement of the G3P shuttle and β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab. Eng. 2011, 13, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Hill, A.; Liu, L.; Knight, R.; Miller, J.; Pan, A.; Otoupal, P.; Alper, H.S. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 2014, 5, 3131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, K.; Imam Abidi, S.H.; Liu, H.; Zhang, H.; Chakraborty, S.; Watson, N.; Ajikumar, P.K.; Stephanopoulos, G. Engineering lipid overproduction in the oleaginous yeast Yarrowia lipolytica. Metab. Eng. 2015, 29, 56–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mlickova, K.; Luo, Y.; d’Andrea, S.; Pec, P.; Chardot, T.; Nicaud, J.M. Acyl-CoA oxidase, a key step for lipid accumulation in the yeast Yarrowia lipolytica. J. Mol. Catal. B Enzym. 2004, 28, 81–85. [Google Scholar] [CrossRef]
- Lazar, Z.; Dulermo, T.; Neuvéglise, C.; Crutz-Le Coq, A.M.; Nicaud, J.M. Hexokinase—A limiting factor in lipid production from fructose in Yarrowia lipolytica. Metab. Eng. 2014, 26, 89–99. [Google Scholar] [CrossRef]
- Beopoulos, A.; Haddouche, R.; Kabran, P.; Dulermo, T.; Chardot, T.; Nicaud, J.M. Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA:diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl. Microbiol. Biotechnol. 2012, 93, 1523–1537. [Google Scholar] [CrossRef] [Green Version]
- Papanikolaou, S.; Beopoulos, A.; Koletti, A.; Theveniau, F.; Koutinas, A.A.; Nicaud, J.M.; Aggelis, G. Importance of the methyl-citrate cycle on glycerol metabolism in the yeast Yarrowia lipolytica. J. Biotechnol. 2013, 168, 303–314. [Google Scholar] [CrossRef]
- Qiao, K.; Wasylenko, T.M.; Zhou, K.; Xu, P.; Stephanopoulos, G. Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat. Biotechnol. 2017, 35, 173–177. [Google Scholar] [CrossRef]
- Lazar, Z.; Gamboa-Meléndez, H.; Le Coq, A.M.; Neuvéglise, C.; Nicaud, J.M. Awakening the endogenous Leloir pathway for efficient galactose utilization by Yarrowia lipolytica. Biotechnol. Biofuels 2015, 8, 185. [Google Scholar] [CrossRef]
- Liu, L.; Pan, A.; Spofford, C.; Zhou, N.; Alper, H.S. An evolutionary metabolic engineering approach for enhancing lipogenesis in Yarrowia lipolytica. Metab. Eng. 2015, 29, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Bordes, F.; Fudalej, F.; Dossat, V.; Nicaud, J.M.; Marty, A. A new recombinant protein expression system for high-throughput screening in the yeast Yarrowia lipolytica. J. Microbiol. Methods 2007, 70, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Emond, S.; Montanier, C.; Nicaud, J.M.; Marty, A.; Monsan, P.; André, I.; Remaud-Siméon, M. New efficient recombinant expression system to engineer Candida antarctica lipase B. Appl. Environ. Microbiol. 2010, 76, 2684–2687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leplat, C.; Nicaud, J.M.; Rossignol, T. Overexpression screen reveals transcription factors involved in lipid accumulation in Yarrowia lipolytica. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Pourcq, K.; Tiels, P.; Van Hecke, A.; Geysens, S.; Vervecken, W.; Callewaert, N. Engineering Yarrowia lipolytica to produce glycoproteins homogeneously modified with the universal Man3GlcNAc2 N-glycan core. PLoS ONE 2012, 7, e39976. [Google Scholar] [CrossRef] [Green Version]
- Jost, B.; Holz, M.; Aurich, A.; Barth, G.; Bley, T.; Müller, R.A. The influence of oxygen limitation for the production of succinic acid with recombinant strains of Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2015, 99, 1675–1686. [Google Scholar] [CrossRef]
- Yovkova, V.; Otto, C.; Aurich, A.; Mauersberger, S.; Barth, G. Engineering the α-ketoglutarate overproduction from raw glycerol by overexpression of the genes encoding NADP+-dependent isocitrate dehydrogenase and pyruvate carboxylase in Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2014, 98, 2003–2013. [Google Scholar] [CrossRef]
- Lazar, Z.; Neuvéglise, C.; Rossignol, T.; Devillers, H.; Morin, N.; Robak, M.; Nicaud, J.M.; Crutz-Le Coq, A.M. Characterization of hexose transporters in Yarrowia lipolytica reveals new groups of Sugar Porters involved in yeast growth. Fungal Genet. Biol. 2017, 100, 1–12. [Google Scholar] [CrossRef]
- Wang, G.; Guo, L.; Liang, W.; Chi, Z.; Liu, L. Systematic analysis of the lysine acetylome reveals diverse functions of lysine acetylation in the oleaginous yeast Yarrowia lipolytica. AMB Express 2017, 7, 94. [Google Scholar] [CrossRef] [Green Version]
- Diamantopoulou, P.; Filippousi, R.; Antoniou, D.; Varfi, E.; Xenopoulos, E.; Sarris, D.; Papanikolaou, S. Production of added-value microbial metabolites during growth of yeast strains on media composed of biodiesel-derived crude glycerol and glycerol/xylose blends. FEMS Microbiol. Lett. 2020, 367, fnaa063. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Kampisopoulou, E.; Blanchard, F.; Rondags, E.; Gardeli, C.; Koutinas, A.A.; Chevalot, I.; Aggelis, G. Production of secondary metabolites through glycerol fermentation under carbon-excess conditions by the yeasts Yarrowia lipolytica and Rhodosporidium toruloides. Eur. J. Lipid. Sci. Technol. 2017, 119, 1600507. [Google Scholar] [CrossRef]
- Guo, H.; Wan, H.; Chen, H.; Fang, F.; Liu, S.; Zhou, J. Proteomic analysis of the response of α-ketoglutarate-producer Yarrowia lipolytica WSH-Z06 to environmental pH stimuli. Appl. Microbiol. Biotechnol. 2016, 100, 8829–8841. [Google Scholar] [CrossRef]
- Taskin, M.; Saghafian, A.; Aydogan, M.N.; Arslan, N.P. Microbial lipid production by cold-adapted oleaginous yeast Yarrowia lipolytica B9 in non-sterile whey medium. Biofuels. Bioprod. Biorefin. 2015, 9, 595–605. [Google Scholar] [CrossRef]
- Kamzolova, S.V.; Morgunov, I.G. Optimization of medium composition and fermentation conditions for α-ketoglutaric acid production from biodiesel waste by Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2020, 104, 7979–7989. [Google Scholar] [CrossRef]
- Kuttiraja, M.; Krishna, S.; Dhouha, A.; Tyagi, R.D. A substrate-based approach for the selection of oil-bearing heterotrophs from nitrogen-deficient soil for lipid production. Appl. Biochem. Biotechnol. 2015, 175, 1926–1937. [Google Scholar] [CrossRef] [PubMed]
- Mathiazhakan, K.; Ayed, D.; Tyagi, R.D. Kinetics of lipid production at lab scale fermenters by a new isolate of Yarrowia lipolytica SKY7. Bioresour. Technol. 2016, 221, 234–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larroude, M.; Rossignol, T.; Nicaud, J.M.; Ledesma-Amaro, R. Synthetic biology tools for engineering Yarrowia lipolytica. Biotechnol. Adv. 2018, 36, 2150–2164. [Google Scholar] [CrossRef]
- Darvishi, F.; Ariana, M.; Marella, E.R.; Borodina, I. Advances in synthetic biology of oleaginous yeast Yarrowia lipolytica for producing non-native chemicals. Appl. Microbiol. Biotechnol. 2018, 102, 5925–5938. [Google Scholar] [CrossRef] [PubMed]
- Markham, K.A.; Alper, H.S. Synthetic Biology Expands the Industrial Potential of Yarrowia lipolytica. Trends Biotechnol. 2018, 36, 1085–1095. [Google Scholar] [CrossRef]
- Shi, T.Q.; Huang, H.; Kerkhoven, E.J.; Ji, X.J. Advancing metabolic engineering of Yarrowia lipolytica using the CRISPR/Cas system. Appl. Microbiol. Biotechnol. 2018, 102, 9541–9548. [Google Scholar] [CrossRef] [Green Version]
- Ganesan, V.; Spagnuolo, M.; Agrawal, A.; Smith, S.; Gao, D.; Blenner, M. Advances and opportunities in gene editing and gene regulation technology for Yarrowia lipolytica. Microb. Cell. Fact. 2019, 18, 208. [Google Scholar] [CrossRef]
- Vandermies, M.; Fickers, P. Bioreactor-Scale Strategies for the Production of Recombinant Protein in the Yeast Yarrowia lipolytica. Microorganisms 2019, 7, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.C.; Beckerich, J.M.; Gaillardin, C. One-step transformation of the dimorphic yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 1997, 48, 232–235. [Google Scholar] [CrossRef]
- Wang, J.H.; Hung, W.; Tsai, S.H. High efficiency transformation by electroporation of Yarrowia lipolytica. J. Microbiol. 2011, 49, 469–472. [Google Scholar] [CrossRef]
- Markham, K.A.; Vazquez, S.; Alper, H.S. High-efficiency transformation of Yarrowia lipolytica using electroporation. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef]
- Fournier, P.; Abbas, A.; Chasles, M.; Kudla, B.; Ogrydziak, D.M.; Yaver, D.; Xuan, J.W.; Peito, A.; Ribet, A.M.; Feynerol, C. Colocalization of centromeric and replicative functions on autonomously replicating sequences isolated from the yeast Yarrowia lipolytica. Proc. Natl. Acad. Sci. USA 1993, 90, 4912–4916. [Google Scholar] [CrossRef] [Green Version]
- Vernis, L.; Abbas, A.; Chasles, M.; Gaillardin, C.M.; Brun, C.; Huberman, J.A.; Fournier, P. An origin of replication and a centromere are both needed to establish a replicative plasmid in the yeast Yarrowia lipolytica. Mol. Cell. Biol. 1997, 17, 1995–2004. [Google Scholar] [CrossRef] [Green Version]
- Fickers, P.; Le Dall, M.T.; Gaillardin, C.; Thonart, P.; Nicaud, J.M. New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J. Microbiol. Methods 2003, 55, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.; Engel, J.; Jin, E.; Holdridge, B.; Xu, P. YaliBricks, a versatile genetic toolkit for streamlined and rapid pathway engineering in Yarrowia lipolytica. Metab. Eng. Commun. 2017, 5, 68–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, L.; Holdridge, B.; Engel, J.; Xu, P. Genetic Tools for Streamlined and Accelerated Pathway Engineering in Yarrowia lipolytica. Methods Mol. Biol. 2019, 1927, 155–177. [Google Scholar] [CrossRef]
- Liu, L.; Otoupal, P.; Pan, A.; Alper, H.S. Increasing expression level and copy number of a Yarrowia lipolytica plasmid through regulated centromere function. FEMS Yeast Res. 2014, 14, 1124–1127. [Google Scholar] [CrossRef] [Green Version]
- Larroude, M.; Park, Y.K.; Soudier, P.; Kubiak, M.; Nicaud, J.M.; Rossignol, T. A modular Golden Gate toolkit for Yarrowia lipolytica synthetic biology. Microb. Biotechnol. 2019, 12, 1249–1259. [Google Scholar] [CrossRef] [Green Version]
- Juretzek, T.; Le Dall, M.; Mauersberger, S.; Gaillardin, C.; Barth, G.; Nicaud, J. Vectors for gene expression and amplification in the yeast Yarrowia lipolytica. Yeast 2001, 18, 97–113. [Google Scholar] [CrossRef]
- Yue, L.; Chi, Z.; Wang, L.; Liu, J.; Madzak, C.; Li, J.; Wang, X. Construction of a new plasmid for surface display on cells of Yarrowia lipolytica. J. Microbiol. Methods 2008, 72, 116–123. [Google Scholar] [CrossRef]
- Blazeck, J.; Liu, L.; Redden, H.; Alper, H. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl. Environ. Microbiol. 2011, 77, 7905–7914. [Google Scholar] [CrossRef] [Green Version]
- Celińska, E.; Ledesma-Amaro, R.; Larroude, M.; Rossignol, T.; Pauthenier, C.; Nicaud, J.M. Golden Gate Assembly system dedicated to complex pathway manipulation in Yarrowia lipolytica. Microb. Biotechnol. 2017, 10, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Chuang, L.T.; Chen, D.C.; Nicaud, J.M.; Madzak, C.; Chen, Y.H.; Huang, Y.S. Co-expression of heterologous desaturase genes in Yarrowia lipolytica. New Biotechnol. 2010, 27, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Celińska, E.; Grajek, W. A novel multigene expression construct for modification of glycerol metabolism in Yarrowia lipolytica. Microb. Cell Fact. 2013, 12, 102. [Google Scholar] [CrossRef] [Green Version]
- Celińska, E.; Borkowska, M.; Korpys-Woźniak, P.; Kubiak, M.; Nicaud, J.M.; Kubiak, P.; Gorczyca, M.; Bialas, W. Optimization of Yarrowia lipolytica-based consolidated biocatalyst through synthetic biology approach: Transcription units and signal peptides shuffling. Appl. Microbiol. Biotechnol. 2020, 104, 5845–5859. [Google Scholar] [CrossRef] [PubMed]
- Holkenbrink, C.; Dam, M.I.; Kildegaard, K.R.; Beder, J.; Dahlin, J.; Belda, D.D.; Borodina, I. EasyCloneYALI: CRISPR/Cas9-Based Synthetic Toolbox for Engineering of the Yeast Yarrowia lipolytica. Biotechnol. J. 2018, 13, e1700543. [Google Scholar] [CrossRef] [Green Version]
- Larroude, M.; Celinska, E.; Back, A.; Thomas, S.; Nicaud, J.M.; Ledesma-Amaro, R. A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of β-carotene. Biotechnol. Bioeng. 2018, 115, 464–472. [Google Scholar] [CrossRef] [Green Version]
- Gao, S.; Han, L.; Zhu, L.; Ge, M.; Yang, S.; Jiang, Y.; Chen, D. One-step integration of multiple genes into the oleaginous yeast Yarrowia lipolytica. Biotechnol. Lett. 2014, 36, 2523–2528. [Google Scholar] [CrossRef]
- Gao, S.; Tong, Y.; Zhu, L.; Ge, M.; Jiang, Y.; Chen, D.; Yang, S. Production of β-carotene by expressing a heterologous multifunctional carotene synthase in Yarrowia lipolytica. Biotechnol. Lett. 2017, 39, 921–927. [Google Scholar] [CrossRef]
- Liu, H.H.; Madzak, C.; Sun, M.L.; Ren, L.; Song, P.; Huang, H.; Ji, X.J. Engineering Yarrowia lipolytica for arachidonic acid production through rapid assembly of metabolic pathway. Biochem. Eng. J. 2017, 119, 52–58. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.P.; Borsenberger, V.; Croux, C.; Duquesne, S.; Truan, G.; Marty, A.; Bordes, F. An artificial chromosome ylAC enables efficient assembly of multiple genes in Yarrowia lipolytica for biomanufacturing. Commun. Biol. 2020, 3, 199. [Google Scholar] [CrossRef]
- Ogrydziak, D.; Demain, A.; Tannenbaum, S. Regulation of extracellular protease production in Candida lipolytica. Biochim. Biophys. Acta 1977, 497, 525–538. [Google Scholar] [CrossRef]
- Müller, S.; Sandal, T.; Kamp-Hansen, P.; Dalbøge, H. Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Klyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica. Yeast 1998, 14, 1267–1283. [Google Scholar] [CrossRef]
- Juretzek, T.; Wang, H.J.; Nicaud, J.M.; Mauersberger, S.; Barth, G. Comparison of promoters suitable for regulated overexpression of β-galactosidase in the alkane-utilizing yeast Yarrowia lipolytica. Biotechnol. Bioprocess. Eng. 2000, 5, 320–326. [Google Scholar] [CrossRef]
- Trassaert, M.; Vandermies, M.; Carly, F.; Denies, O.; Thomas, S.; Fickers, P.; Nicaud, J.M. New inducible promoter for gene expression and synthetic biology in Yarrowia lipolytica. Microb. Cell. Fact. 2017, 16, 141. [Google Scholar] [CrossRef]
- Tai, M.; Stephanopoulos, G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 2013, 15, 1–9. [Google Scholar] [CrossRef]
- Kamineni, A.; Chen, S.; Chifamba, G.; Tsakraklides, V. Promoters for lipogenesis-specific downregulation in Yarrowia lipolytica. FEMS Yeast Res. 2020, 20, foaa035. [Google Scholar] [CrossRef] [PubMed]
- Madzak, C.; Blanchin-Roland, S.; Cordero Otero, R.R.; Gaillardin, C. Functional analysis of upstream regulating regions from the Yarrowia lipolytica XPR2 promoter. Microbiology 1999, 145, 75–87. [Google Scholar] [CrossRef] [Green Version]
- Kopecný, D.; Pethe, C.; Sebela, M.; Houba-Hérin, N.; Madzak, C.; Majira, A.; Laloue, M. High-level expression and characterization of Zea mays cytokinin oxidase/dehydrogenase in Yarrowia lipolytica. Biochimie 2005, 87, 1011–1022. [Google Scholar] [CrossRef]
- Blazeck, J.; Reed, B.; Garg, R.; Gerstner, R.; Pan, A.; Agarwala, V.; Alper, H.S. Generalizing a hybrid synthetic promoter approach in Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2013, 97, 3037–3052. [Google Scholar] [CrossRef] [PubMed]
- Shabbir Hussain, M.; Wheeldon, I.; Blenner, M.A. A Strong Hybrid Fatty Acid Inducible Transcriptional Sensor Built From Yarrowia lipolytica Upstream Activating and Regulatory Sequences. Biotechnol. J. 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.K.; Korpys, P.; Kubiak, M.; Celińska, E.; Soudier, P.; Trébulle, P.; Larroude, M.; Rossignol, T.; Nicaud, J.M. Engineering the architecture of erythritol-inducible promoters for regulated and enhanced gene expression in Yarrowia lipolytica. FEMS Yeast Res. 2019, 19. [Google Scholar] [CrossRef]
- Dulermo, R.; Brunel, F.; Dulermo, T.; Ledesma-Amaro, R.; Vion, J.; Trassaert, M.; Thomas, S.; Nicaud, J.M.; Leplat, C. Using a vector pool containing variable-strength promoters to optimize protein production in Yarrowia lipolytica. Microb. Cell. Fact. 2017, 16, 31. [Google Scholar] [CrossRef]
- Liu, R.; Liu, L.; Li, X.; Liu, D.; Yuan, Y. Engineering yeast artificial core promoter with designated base motifs. Microb. Cell Fact. 2020, 19, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curran, K.A.; Morse, N.J.; Markham, K.A.; Wagman, A.M.; Gupta, A.; Alper, H.S. Short Synthetic Terminators for Improved Heterologous Gene Expression in Yeast. ACS Synth. Biol. 2015, 4, 824–832. [Google Scholar] [CrossRef]
- Swennen, D.; Paul, M.F.; Vernis, L.; Beckerich, J.M.; Fournier, A.; Gaillardin, C. Secretion of active anti-Ras single-chain Fv antibody by the yeasts Yarrowia lipolytica and Kluyveromyces lactis. Microbiology 2002, 148, 41–50. [Google Scholar] [CrossRef] [Green Version]
- Gasmi, N.; Fudalej, F.; Kallel, H.; Nicaud, J.M. A molecular approach to optimize hIFN α2b expression and secretion in Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2011, 89, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Celińska, E.; Borkowska, M.; Białas, W.; Korpys, P.; Nicaud, J.M. Robust signal peptides for protein secretion in Yarrowia lipolytica: Identification and characterization of novel secretory tags. Appl. Microbiol. Biotechnol. 2018, 102, 5221–5233. [Google Scholar] [CrossRef] [Green Version]
- Yuzbasheva, E.Y.; Yuzbashev, T.V.; Laptev, I.A.; Konstantinova, T.K.; Sineoky, S.P. Efficient cell surface display of Lip2 lipase using C-domains of glycosylphosphatidylinositol-anchored cell wall proteins of Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2011, 91, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.Y.; Van, T.L.; Cheon, S.A.; Choo, J.; Kim, J.Y.; Kang, H.A. Cell-surface expression of Aspergillus saitoi-derived functional α-1,2-mannosidase on Yarrowia lipolytica for glycan remodeling. J. Microbiol. 2013, 51, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.M.; Wang, C.M.; Men, X.; Yue, T.Q.; Madzak, C.; Xiang, X.H.; Xiang, H.Y.; Zhang, H.B. Construction of arming Yarrowia lipolytica surface-displaying soybean seed coat peroxidase for use as whole-cell biocatalyst. Enzyme. Microb. Technol. 2020, 135, 109498. [Google Scholar] [CrossRef]
- Yang, X.S.; Jiang, Z.B.; Song, H.T.; Jiang, S.J.; Madzak, C.; Ma, L.X. Cell-surface display of the active mannanase in Yarrowia lipolytica with a novel surface-display system. Biotechnol. Appl. Biochem. 2009, 54, 171–176. [Google Scholar] [CrossRef]
- Duquesne, S.; Bozonnet, S.; Borde, F.; Dumon, C.; Nicaud, J.M.; Marty, A. Construction of a highly active xylanase displaying oleaginous yeast: Comparison of anchoring systems. PLoS ONE 2014, 9, e95128. [Google Scholar] [CrossRef] [Green Version]
- Yuzbasheva, E.Y.; Yuzbashev, T.V.; Perkovskaya, N.I.; Mostova, E.B.; Vybornaya, T.V.; Sukhozhenko, A.V.; Toropygin, I.Y.; Sineoky, S.P. Cell surface display of Yarrowia lipolytica lipase Lip2p using the cell wall protein YlPir1p, its characterization, and application as a whole-cell biocatalyst. Appl. Biochem. Biotechnol. 2015, 175, 3888–3900. [Google Scholar] [CrossRef]
- Haddouche, R.; Delessert, S.; Sabirova, J.; Neuvéglise, C.; Poirier, Y.; Nicaud, J.M. Roles of multiple acyl-CoA oxidases in the routing of carbon flow towards β-oxidation and polyhydroxyalkanoate biosynthesis in Yarrowia lipolytica. FEMS Yeast Res. 2010, 10, 917–927. [Google Scholar] [CrossRef] [Green Version]
- Kerkhoven, E.J.; Kim, Y.M.; Wei, S.; Nicora, C.D.; Fillmore, T.L.; Purvine, S.O.; Webb-Robertson, B.J.; Smith, R.D.; Baker, S.E.; Metz, T.O.; et al. Leucine Biosynthesis Is Involved in Regulating High Lipid Accumulation in Yarrowia lipolytica. mBio 2017, 8, e00857-17. [Google Scholar] [CrossRef] [Green Version]
- Boeke, J.D.; Trueheart, J.; Natsoulis, G.; Fink, G.R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987, 154, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Cheon, S.A.; Han, E.J.; Kang, H.A.; Ogrydziak, D.M.; Kim, J.Y. Isolation and characterization of the TRP1 gene from the yeast Yarrowia lipolytica and multiple gene disruption using a TRP blaster. Yeast 2003, 20, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Larroude, M.; Trabelsi, H.; Nicaud, J.M.; Rossignol, T. A set of Yarrowia lipolytica CRISPR/Cas9 vectors for exploiting wild-type strain diversity. Biotechnol. Lett. 2020, 42, 773–785. [Google Scholar] [CrossRef] [Green Version]
- Jang, I.S.; Yu, B.J.; Jang, J.Y.; Jegal, J.; Lee, J.Y. Improving the efficiency of homologous recombination by chemical and biological approaches in Yarrowia lipolytica. PLoS ONE 2018, 13, e0194954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaillardin, C.; Ribet, A.M. LEU2 directed expression of beta-galactosidase activity and phleomycin resistance in Yarrowia lipolytica. Curr. Genet. 1987, 11, 369–375. [Google Scholar] [CrossRef]
- Cordero Otero, R.; Gaillardin, C. Efficient selection of hygromycin-B-resistant Yarrowia lipolytica transformants. Appl Microbiol Biotechnol 1996, 46, 143–148. [Google Scholar] [CrossRef]
- Vorachek-Warren, M.K.; McCusker, J.H. DsdA (D-serine deaminase): A new heterologous MX cassette for gene disruption and selection in Saccharomyces cerevisiae. Yeast 2004, 21, 163–171. [Google Scholar] [CrossRef] [Green Version]
- Egermeier, M.; Sauer, M.; Marx, H. Golden Gate-based metabolic engineering strategy for wild-type strains of Yarrowia lipolytica. FEMS Microbiol. Lett. 2019, 366, fnz022. [Google Scholar] [CrossRef]
- Patra, P.; Das, M.; Kundu, P.; Ghosh, A. Recent advances in systems and synthetic biology approaches for developing novel cell-factories in non-conventional yeasts. Biotechnol. Adv. 2021, 47, 107695. [Google Scholar] [CrossRef]
- Raschmanová, H.; Weninger, A.; Glieder, A.; Kovar, K.; Vogl, T. Implementing CRISPR-Cas technologies in conventional and non-conventional yeasts: Current state and future prospects. Biotechnol. Adv. 2018, 36, 641–665. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Schwartz, C.M.; Hussain, M.S.; Blenner, M.; Wheeldon, I. Synthetic RNA Polymerase III Promoters Facilitate High-Efficiency CRISPR-Cas9-Mediated Genome Editing in Yarrowia lipolytica. ACS Synth. Biol. 2016, 5, 356–359. [Google Scholar] [CrossRef]
- Gao, S.; Tong, Y.; Wen, Z.; Zhu, L.; Ge, M.; Chen, D.; Jiang, Y.; Yang, S. Multiplex gene editing of the Yarrowia lipolytica genome using the CRISPR-Cas9 system. J. Ind. Microbiol. Biotechnol. 2016, 43, 1085–1093. [Google Scholar] [CrossRef]
- Schwartz, C.; Shabbir-Hussain, M.; Frogue, K.; Blenner, M.; Wheeldon, I. Standardized Markerless Gene Integration for Pathway Engineering in Yarrowia lipolytica. ACS Synth. Biol. 2017, 6, 402–409. [Google Scholar] [CrossRef]
- Borsenberger, V.; Onésime, D.; Lestrade, D.; Rigouin, C.; Neuvéglise, C.; Daboussi, F.; Bordes, F. Multiple Parameters Drive the Efficiency of CRISPR/Cas9-Induced Gene Modifications in Yarrowia lipolytica. J. Mol. Biol. 2018, 430, 4293–4306. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mawgoud, A.M.; Stephanopoulos, G. Improving CRISPR/Cas9-mediated genome editing efficiency in Yarrowia lipolytica using direct tRNA-sgRNA fusions. Metab. Eng. 2020, 62, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Edwards, H.; Xu, P. CRISPR-Cas12a/Cpf1-assisted precise, efficient and multiplexed genome-editing in Yarrowia lipolytica. Metab. Eng. Commun. 2019, 10, e00112. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, C.; Cheng, J.F.; Evans, R.; Schwartz, C.A.; Wagner, J.M.; Anglin, S.; Beitz, A.; Pan, W.; Lonardi, S.; Blenner, M.; et al. Validating genome-wide CRISPR-Cas9 function improves screening in the oleaginous yeast Yarrowia lipolytica. Metab. Eng. 2019, 55, 102–110. [Google Scholar] [CrossRef] [Green Version]
- Scholze, H.; Boch, J. TAL effectors are remote controls for gene activation. Curr. Opin. Microbiol. 2011, 14, 47–53. [Google Scholar] [CrossRef]
- Rigouin, C.; Gueroult, M.; Croux, C.; Dubois, G.; Borsenberger, V.; Barbe, S.; Marty, A.; Daboussi, F.; André, I.; Bordes, F. Production of Medium Chain Fatty Acids by Yarrowia lipolytica: Combining Molecular Design and TALEN to Engineer the Fatty Acid Synthase. ACS Synth. Biol. 2017, 6, 1870–1879. [Google Scholar] [CrossRef]
- Rigouin, C.; Croux, C.; Dubois, G.; Daboussi, F.; Bordes, F. Genome Editing in Y. lipolytica Using TALENs. Methods Mol. Biol. 2021, 2307, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.J.; Park, B.G.; Kim, B.G.; Hahn, J.S. Multiplex Gene Disruption by Targeted Base Editing of Yarrowia lipolytica Genome Using Cytidine Deaminase Combined with the CRISPR/Cas9 System. Biotechnol. J. 2020, 15, e1900238. [Google Scholar] [CrossRef]
- Schwartz, C.; Frogue, K.; Ramesh, A.; Misa, J.; Wheeldon, I. CRISPRi repression of nonhomologous end-joining for enhanced genome engineering via homologous recombination in Yarrowia lipolytica. Biotechnol. Bioeng. 2017, 114, 2896–2906. [Google Scholar] [CrossRef]
- Zhang, J.L.; Peng, Y.Z.; Liu, D.; Liu, H.; Cao, Y.X.; Li, B.Z.; Li, C.; Yuan, Y.J. Gene repression via multiplex gRNA strategy in Y. lipolytica. Microb. Cell. Fact. 2018, 17, 62. [Google Scholar] [CrossRef]
- Schwartz, C.; Curtis, N.; Löbs, A.K.; Wheeldon, I. Multiplexed CRISPR Activation of Cryptic Sugar Metabolism Enables Yarrowia Lipolytica Growth on Cellobiose. Biotechnol. J. 2018, 13, e1700584. [Google Scholar] [CrossRef]
- Misa, J.; Schwartz, C. CRISPR Interference and Activation to Modulate Transcription in Yarrowia lipolytica. Methods Mol. Biol. 2021, 2307, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, C.; Wheeldon, I. CRISPR-Cas9-Mediated Genome Editing and Transcriptional Control in Yarrowia lipolytica. Methods Mol. Biol. 2018, 1772, 327–345. [Google Scholar] [CrossRef]
- Patterson, K.; Yu, J.; Landberg, J.; Chang, I.; Shavarebi, F.; Bilanchone, V.; Sandmeyer, S. Functional genomics for the oleaginous yeast Yarrowia lipolytica. Metab. Eng. 2018, 48, 184–196. [Google Scholar] [CrossRef] [Green Version]
- Loira, N.; Dulermo, T.; Nicaud, J.M.; Sherman, D.J. A genome-scale metabolic model of the lipid-accumulating yeast Yarrowia lipolytica. BMC Syst. Biol. 2012, 6, 35. [Google Scholar] [CrossRef] [Green Version]
- Kerkhoven, E.J.; Pomraning, K.R.; Baker, S.E.; Nielsen, J. Regulation of amino-acid metabolism controls flux to lipid accumulation in Yarrowia lipolytica. NPJ Syst. Biol. Appl. 2016, 2, 16005. [Google Scholar] [CrossRef] [Green Version]
- Verbeke, J.; Beopoulos, A.; Nicaud, J.M. Efficient homologous recombination with short length flanking fragments in Ku70 deficient Yarrowia lipolytica strains. Biotechnol. Lett. 2013, 35, 571–576. [Google Scholar] [CrossRef]
- Kretzschmar, A.; Otto, C.; Holz, M.; Werner, S.; Hübner, L.; Barth, G. Increased homologous integration frequency in Yarrowia lipolytica strains defective in non-homologous end-joining. Curr. Genet. 2013, 59, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Mai, J.; Ding, Y.; Wei, Y.; Ledesma-Amaro, R.; Ji, X.J. Improving the homologous recombination efficiency of Yarrowia lipolytica by grafting heterologous component from Saccharomyces cerevisiae. Metab. Eng. Commun. 2020, 11, e00152. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Quijano, R.; Gaillardin, C.; Ruiz-Herrera, J. Functional analysis of the MATB mating-type idiomorph of the dimorphic fungus Yarrowia lipolytica. Curr. Microbiol. 2008, 57, 115–120. [Google Scholar] [CrossRef]
- Han, C.; Kwon, H.; Park, G.; Jang, M.; Lee, H.J.; Seo, S.; Kwon, M.; Jeon, W.; Lee, H.; Lee, H.; et al. Enhanced mating-type switching and sexual hybridization in heterothallic yeast Yarrowia lipolytica. FEMS Yeast Res. 2020, 20, foaa011. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, C.G.P.; Fitzpatrick, D.A. Pangloss: A Tool for Pan-Genome Analysis of Microbial Eukaryotes. Genes 2019, 10, 521. [Google Scholar] [CrossRef] [Green Version]
- Morin, N.; Cescut, J.; Beopoulos, A.; Lelandais, G.; Le Berre, V.; Uribelarrea, J.L.; Molina-Jouve, C.; Nicaud, J.M. Transcriptomic analyses during the transition from biomass production to lipid accumulation in the oleaginous yeast Yarrowia lipolytica. PLoS ONE 2011, 6, e27966. [Google Scholar] [CrossRef] [Green Version]
- Sabra, W.; Bommareddy, R.R.; Maheshwari, G.; Papanikolaou, S.; Zeng, A.P. Substrates and oxygen dependent citric acid production by Yarrowia lipolytica: Insights through transcriptome and fluxome analyses. Microb. Cell Fact. 2017, 16, 78. [Google Scholar] [CrossRef]
- Lazar, Z.; Liu, N.; Stephanopoulos, G. Holistic Approaches in Lipid Production by Yarrowia lipolytica. Trends Biotechnol. 2018, 36, 1157–1170. [Google Scholar] [CrossRef]
- Worland, A.M.; Czajka, J.J.; Xing, Y.; Harper, W.F., Jr.; Moore, A.; Xiao, Z.; Han, Z.; Wang, Y.; Su, W.W.; Tang, Y.J. Analysis of Yarrowia lipolytica growth, catabolism, and terpenoid biosynthesis during utilization of lipid-derived feedstock. Metab. Eng. Commun. 2020, 11, e00130. [Google Scholar] [CrossRef] [PubMed]
- Korpys-Woźniak, P.; Celińska, E. Global transcriptome profiling reveals genes responding to overproduction of a small secretory, a high cysteine- and a high glycosylation-bearing protein in Yarrowia lipolytica. Biotechnol. Rep. 2021, 31, e00646. [Google Scholar] [CrossRef]
- Pan, P.; Hua, Q. Reconstruction and in silico analysis of metabolic network for an oleaginous yeast, Yarrowia lipolytica. PLoS ONE 2012, 7, e51535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pomraning, K.R.; Wei, S.; Karagiosis, S.A.; Kim, Y.M.; Dohnalkova, A.C.; Arey, B.W.; Bredeweg, E.L.; Orr, G.; Metz, T.O.; Baker, S.E. Comprehensive Metabolomic, Lipidomic and Microscopic Profiling of Yarrowia lipolytica during Lipid Accumulation Identifies Targets for Increased Lipogenesis. PLoS ONE 2015, 10, e0123188. [Google Scholar] [CrossRef]
- Trébulle, P.; Nicaud, J.M.; Leplat, C.; Elati, M. Inference and interrogation of a coregulatory network in the context of lipid accumulation in Yarrowia lipolytica. NPJ Syst. Biol. Appl. 2017, 3, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Park, B.G.; Kim, E.J.; Kim, J.; Kim, B.G. In silico identification of metabolic engineering strategies for improved lipid production in Yarrowia lipolytica by genome-scale metabolic modeling. Biotechnol. Biofuels 2019, 12, 187. [Google Scholar] [CrossRef] [PubMed]
- Poorinmohammad, N.; Kerkhoven, E.J. Systems-level approaches for understanding and engineering of the oleaginous cell factory Yarrowia lipolytica. Biotechnol. Bioeng. 2021. [Google Scholar] [CrossRef]
- Wagner, J.M.; Liu, L.; Yuan, S.F.; Venkataraman, M.V.; Abate, A.R.; Alper, H.S. A comparative analysis of single cell and droplet-based FACS for improving production phenotypes: Riboflavin overproduction in Yarrowia lipolytica. Metab. Eng. 2018, 47, 346–356. [Google Scholar] [CrossRef]
- Yang, X.; Wang, H.; Li, C.; Lin, C.S.K. Restoring of Glucose Metabolism of Engineered Yarrowia lipolytica for Succinic Acid Production via a Simple and Efficient Adaptive Evolution Strategy. J. Agric. Food Chem. 2017, 65, 4133–4139. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, K.; Miao, L.; Rong, L.; Zhao, Y.; Li, S.; Ma, L.; Li, J.; Zhang, C.; Xiao, D.; et al. Simultaneous Improvement of Limonene Production and Tolerance in Yarrowia lipolytica through Tolerance Engineering and Evolutionary Engineering. ACS Synth. Biol. 2021, 10, 884–896. [Google Scholar] [CrossRef]
- Qiu, X.; Gu, Y.; Du, G.; Zhang, J.; Xu, P.; Li, J. Conferring thermotolerant phenotype to wild type Yarrowia lipolytica improves cell growth and erythritol production. Biotechnol. Bioeng. 2021. [Google Scholar] [CrossRef]
- Zhou, L.; Xu, Z.; Wen, Z.; Lu, M.; Wang, Z.; Zhang, Y.; Zhou, H.; Jin, M. Combined adaptive evolution and transcriptomic profiles reveal aromatic aldehydes tolerance mechanisms in Yarrowia lipolytica. Bioresour. Technol. 2021, 329, 124910. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, L.; Lu, M.; Zhang, Y.; Perveen, S.; Zhou, H.; Wen, Z.; Xu, Z.; Jin, M. Adaptive laboratory evolution of Yarrowia lipolytica improves ferulic acid tolerance. Appl. Microbiol. Biotechnol. 2021, 105, 1745–1758. [Google Scholar] [CrossRef] [PubMed]
- Xie, D. Integrating Cellular and Bioprocess Engineering in the Non-Conventional Yeast Yarrowia lipolytica for Biodiesel Production: A Review. Front. Bioeng. Biotechnol. 2017, 5, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, A.; Geier, M.; Bühler, B.; Schmid, A.; Mauersberger, S.; Glieder, A. Steroid biotransformations in biphasic systems with Yarrowia lipolytica expressing human liver cytochrome P450 genes. Microb. Cell Fact. 2012, 11, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavšček, M.; Bhutada, G.; Madl, T.; Natter, K. Optimization of lipid production with a genome-scale model of Yarrowia lipolytica. BMC Syst. Biol. 2015, 9, 72. [Google Scholar] [CrossRef] [Green Version]
- Stylianou, E.; Pateraki, C.; Ladakis, D.; Damala, C.; Vlysidis, A.; Latorre-Sánchez, M.; Coll, C.; Lin, C.S.K.; Koutinas, A. Bioprocess development using organic biowaste and sustainability assessment of succinic acid production with engineered Yarrowia lipolytica strain. Biochem. Eng. J. 2021, 108099. [Google Scholar] [CrossRef]
- Gurme, S.T.; Surwase, S.N.; Patil, S.A.; Jadhav, J.P. Evaluation of Various Factors Affecting Bioconversion of l-Tyrosine to l-DOPA by Yeast Yarrowia lipolytica-NCIM 3450 Using Response Surface Methodology. Nat. Prod. Bioprospect. 2014, 4, 141–147. [Google Scholar] [CrossRef] [Green Version]
- Rywińska, A.; Marcinkiewicz, M.; Cibis, E.; Rymowicz, W. Optimization of medium composition for erythritol production from glycerol by Yarrowia lipolytica using response surface methodology. Prep. Biochem. Biotechnol. 2015, 45, 515–529. [Google Scholar] [CrossRef]
- Yellapu, S.K.; Bezawada, J.; Kaur, R.; Kuttiraja, M.; Tyagi, R.D. Detergent assisted lipid extraction from wet yeast biomass for biodiesel: A response surface methodology approach. Bioresour. Technol. 2016, 218, 667–673. [Google Scholar] [CrossRef]
- Darvishi, F.; Moradi, M.; Madzak, C.; Jolivalt, C. Production of Laccase by Recombinant Yarrowia lipolytica from Molasses: Bioprocess Development Using Statistical Modeling and Increase Productivity in Shake-Flask and Bioreactor Cultures. Appl. Biochem. Biotechnol. 2017, 181, 1228–1239. [Google Scholar] [CrossRef]
- Celińska, E.; Nicaud, J.M.; Białas, W. Hydrolytic secretome engineering in Yarrowia lipolytica for consolidated bioprocessing on polysaccharide resources: Review on starch, cellulose, xylan, and inulin. Appl. Microbiol. Biotechnol. 2021, 105, 975–989. [Google Scholar] [CrossRef]
- Berg, P.; Baltimore, D.; Brenner, S.; Roblin, R.O.; Singer, M.F. Summary statement of the Asilomar conference on recombinant DNA molecules. Proc. Natl. Acad. Sci. USA 1975, 72, 1981–1984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaskell, G.; Bauer, M.W.; Durant, J.; Allum, N.C. Worlds apart? The reception of genetically modified foods in Europe and the U.S. Science 1999, 285, 384–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- A CRISPR definition of genetic modification. Nat. Plants 2018, 4, 233. [CrossRef] [PubMed]
- Ruffell, D. The EU Court of Justice extends the GMO Directive to gene-edited organisms. FEBS Lett. 2018, 592, 3653–3657. [Google Scholar] [CrossRef] [Green Version]
- Tagliabue, G. The nonsensical GMO pseudo-category and a precautionary rabbit hole. Nat. Biotechnol. 2015, 33, 907–908. [Google Scholar] [CrossRef] [PubMed]
- Tagliabue, G. The meaningless pseudo-category of “GMOs”: The trouble with the “new techniques” for genetically modifying crops demonstrates the illogical process-based definition of GMOs in EU regulation. EMBO Rep. 2016, 17, 10–13. [Google Scholar] [CrossRef] [Green Version]
- Qiu, X.; Xu, P.; Zhao, X.; Du, G.; Zhang, J.; Li, J. Combining genetically-encoded biosensors with high throughput strain screening to maximize erythritol production in Yarrowia lipolytica. Metab. Eng. 2020, 60, 66–76. [Google Scholar] [CrossRef]
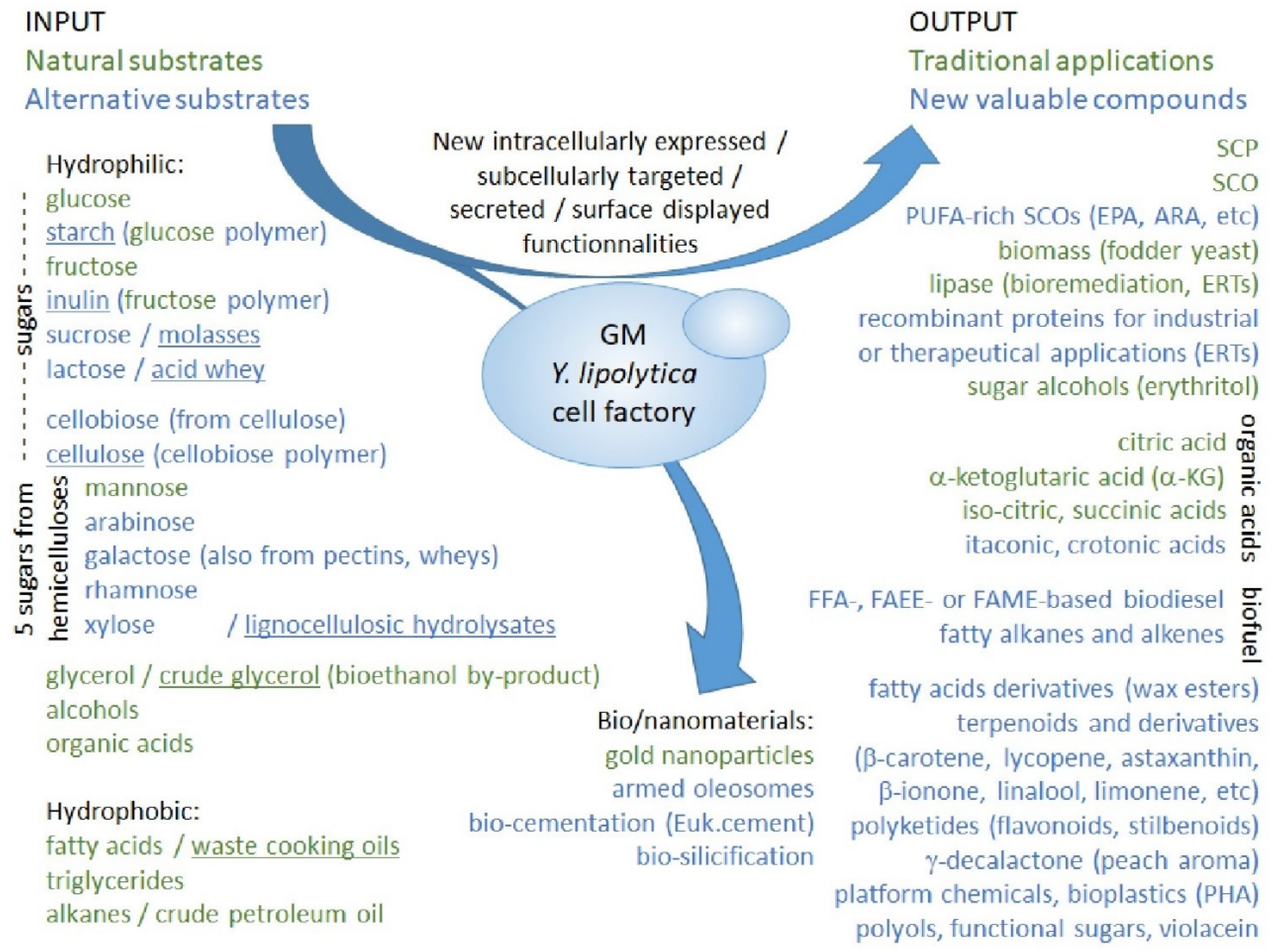
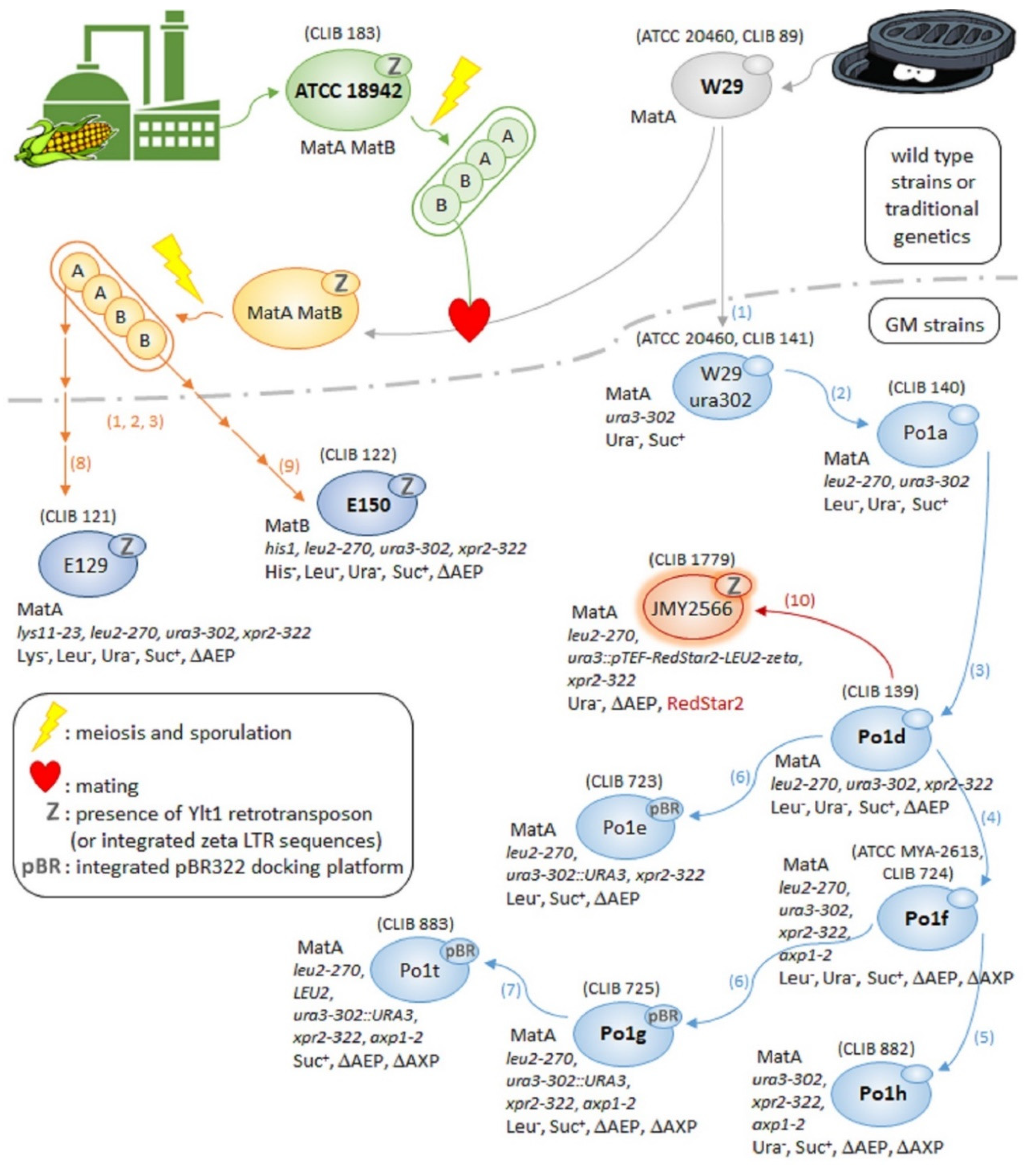
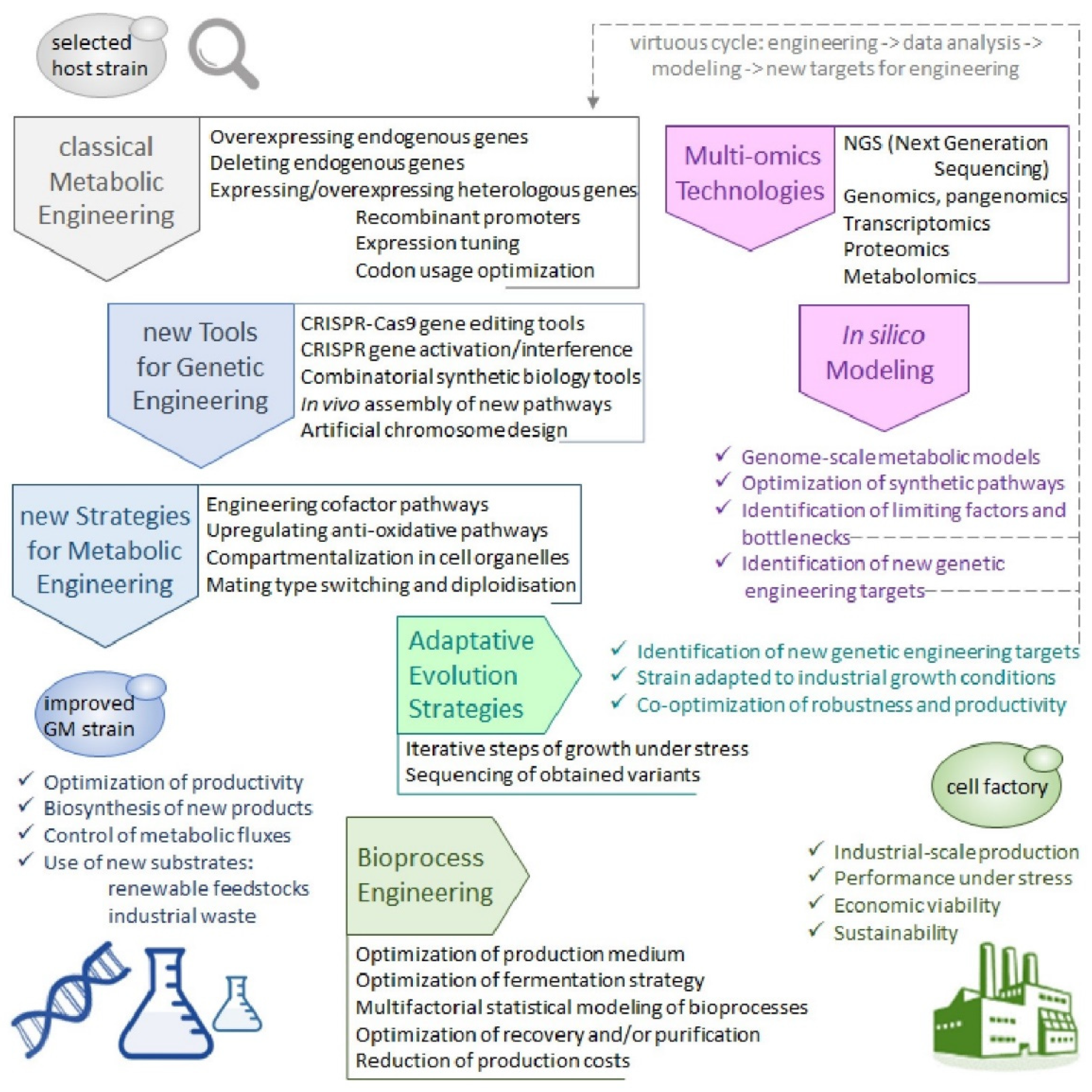
| Strain Usual Name/Origin (Reference Number in Yeast Collections—cf. Table 2) | Genotype Phenotype | Remarkable Characteristics | Usual Applications |
|---|---|---|---|
| A-101/carwash effluents, Poland [123] not publicly available (a) | ND wild-type prototroph | robust growth on oil [123], high citric acid production [124], sequenced strain [125] | in situ soil bioremediation [51,52], citric acid production [124], UV-mutagenesis and metabolic engineering host for design of improved strains producing citrate or erythritol [70,126,127,128] |
| ACA-DC 50109 [aka LGAM S(7)1]/Greece [129] (b) | ND wild-type prototroph | very high lipid content and productivity [129,130], robust growth on crude glycerol, simultaneous high lipid and citric acid yields [131] | production of organic acids (notably citric acid) and SCO [129,130,131,132,133,134], including cocoa butter substitute [31,32], parent of evolved strain with increase oleaginicity [135], metabolic engineering host for design of improved GM strains [133,134] |
| ACA-DC 5033 [aka ACA-YC 5033]/acid sourdough, Greece [136] | ND wild-type prototroph | robust growth on crude glycerol, simultaneous high lipid and citric acid yields [24] | SCO, citric acid and polyol production [24,137] |
| ATCC 18942 [aka YB-423]/corn-processing plant, USA [138] (CBS 6124, CLIB 183, JCM 2320 and 8060, MUCL 29853, NBRC 1548, NRRL YB-423) | MatA MatB wild-type prototroph | diploid type strain [138], robust growth [139] | yeast biomass production [139] |
| ATCC 20362 [aka 2002]/USA | ND wild-type prototroph | robust growth, high lipid content and productivity [57] | degradation of petroleum crude oil (US patent 3856667A), metabolic engineering host for design of Dupont GM PUFA-producing platform (cf. Section 2.3.2) [57,58] |
| ATCC 48436/soil, Japan [140] (CBS 6303, CLIB 703, JCM 8054, NBRC 10073) | MatA wild-type prototroph | produces lipase activators [140] | lipase production [140], parent strain for Artechno highly lipolytic mutants used for bioremediation (cf. Section 2.3.1) |
| D 1805/France [141] (ATCC 20390) | MatA MatB wild-type prototroph | non-sporulating diploid, robust growth [141,142], self-cycling fermentation [143] | organic acid production [143] |
| H222/soil, Germany [10,144] (CLIB 80) | MatA wild-type prototroph | better fructose assimilation, high citric acid production [144], sequenced strain [34,37] | organic acid production, metabolic engineering host for design of improved GM strains [145,146] |
| NCIM 3589/marine waters, India [147] | ND wild-type prototroph | biofilm formation [148] emulsifier production [149] | gold nanoparticle production [102] |
| SWJ-1b/marine fish gut, China [4] (MCCC 2E00068) | ND wild-type prototroph | high level of crude protein [4] | citric acid and SCP production [150], metabolic engineering host for design of improved GM strains [151,152] ARTP-mutagenesis host for design of improved strains producing erythritol [153] |
| W29/sewage water *, France [11,154] (ATCC 20460, CBS 7504, CICC 1778, CLIB 89, NBRC 113670, NRLL Y-3178, VKPM Y-3178) | MatA wild-type prototroph | high secretion level of proteins [10,155], sequenced strain [36,156] | organic acid production [11], basis for the Po1 series of heterologous protein-producing GM strains and the JMY2566 GM strain for high-throughput applications (cf. Figure 2), basis for GM obese strains (cf. Section 3.1.4) |
| E129/GM from a W29 and ATCC 18942 crossing [10,157] (CLIB 121) (cf. Figure 2) | MatA, lys11-23, leu2-270, ura3-302, xpr2-322 Lys−, Leu−, Ura−, Suc+, ΔAEP | able to grow on sucrose [10,15] deleted for alkaline extracellular protease [10] | heterologous protein production [155] |
| E150/GM from a W29 and ATCC 18942 crossing [10,157] (CLIB 122) (cf. Figure 2) | MatB, his1, leu2-270, ura3-302, xpr2-322 His−, Leu−, Ura−, Suc+, ΔAEP | able to grow on sucrose [10,15] deleted for alkaline extracellular protease [10], reference sequenced strain [33,34] | reference for assembling and annotating genomes [33,34] |
| Po1d/GM from W29 [158] (CLIB 139) (cf. Figure 2) | MatA, leu2-270, ura3-302, xpr2-322 Leu−, Ura−, Suc+, ΔAEP | able to grow on sucrose [15] deleted for alkaline extracellular protease [158] | heterologous protein production [19,20,158], metabolic engineering host for design of GM strains for multiple applications [19,20] metabolic engineering host for design of Oxyrane ERT-producing platform (cf. Section 2.3.2) [61] |
| Po1f/GM from W29 [159] (ATCC MYA-2613, CLIB 724, VKPM Y-3155) (cf. Figure 2) | MatA, leu2-270, ura3-302, xpr2-322, axp1-2 Leu−, Ura−, Suc+, ΔAEP, ΔAXP | able to grow on sucrose [15] deleted for both extracellular proteases [159], sequenced strain [160,161] | heterologous protein production [19,20,159], metabolic engineering host for design of GM strains for multiple applications [19,20] |
| Po1g/GM from W29 [159] (CLIB 725) (cf. Figure 2) | MatA, leu2-270, ura3-302::URA3, xpr2-322, axp1-2 Leu−, Suc+, ΔAEP, ΔAXP | able to grow on sucrose [15] deleted for both extracellular proteases, carry a pBR322 docking platform [159] | heterologous protein production [19,20,159], included in the YLEX kit for expression/secretion of heterologous proteins [19,20] (cf. Section 2.3.2) |
| Po1h/GM from W29 [18,43] (CLIB 882) (cf. Figure 2) | MatA, ura3-302, xpr2-322, axp1-2 Ura−, Suc+, ΔAEP, ΔAXP | able to grow on sucrose [15] deleted for both extracellular proteases [18,43] | heterologous protein production [18,19,20,43], metabolic engineering host for design of GM strains for multiple applications [19,20] |
| Po1t/GM from W29 [18,43] (CLIB 883) (cf. Figure 2) | MatA, leu2-270, LEU2, ura3-302::URA3, xpr2-322, axp1-2 Suc+, ΔAEP, ΔAXP | able to grow on sucrose [15] deleted for both extracellular proteases, carry a pBR322 docking platform, GM prototroph [18,43] | negative control for heterologous protein production by other Po1 strains [18,43] |
| JMY2566/GM from W29 [162] (CLIB 1779) (cf. Figure 2) | MatA, leu2-270, ura3::pTEF-RedStar2-LEU2-zeta, xpr2-322 Ura−, ΔAEP, RedStar2 | deleted for alkaline extracellular protease, fluorescent (red) strain, carry a zeta LTR sequences docking platform [162] | high-throughput mutant library screening [162] |
| VKM Y-2373/Russia (Fed) VKM Y-2412/Russia (Fed) not publicly available (c) | ND wild-type prototroph | natural overproducers of, respectively, (iso)citric acid [163,164] and KGA [165] | organic acid production [163,164,165], basis for traditionally obtained 704-UV4-A/NG50 mutant for improved (iso)citric acid production [163] |
| WSH-Z06/oil-polluted soil (refinery), China [166] not publicly available (d) | ND wild-type prototroph | thiamine-auxotrophic natural overproducer of KGA [166,167], sequenced strain [168] | KGA and keto acids production [167], basis for traditionally obtained hyper-producer mutants [168], metabolic engineering host for design of improved GM strains [169,170] |
| YB-392/gluten settler, USA YB-419/maize fiber tailings, USA (NRRL YB-392 and NRRL YB-419) YB-420, YB-566 and YB-567 (not publicly available, do not appear on online catalog) | ND wild-type prototrophs | biomass hydrolysate consumption, inhibitor tolerance, high lipid/fatty acid or sugar alcohol production (cf. Section 3.2) [118], sequenced strains [171] | five strains selected as promising candidates for industrial biocatalysis [118,171] |
| Country (Per Alphabetic Order) | Acronym of the Collection (WDCM Number) | Full Name of the Culture Collection | Website ISO Standard | Number of Strains of the Yarrowia Genus or Clade (C. for Candida) | Remarkable Yarrowia lipolytica Strains (Haploid, Unless Specified) |
|---|---|---|---|---|---|
| Belgium | BCCM/MUCL (WDCM 308) | Belgian Coordinated Collections of Microorganisms/MUCL Agro-food and Environmental Fungal Collection | http://bccm.belspo.be/about-us/bccm-mucl (accessed on 3 June 2021) ISO 9001:2015 | 29 Y. lipolytica + 3 C. (Y.) alimentaria 5 Y. deformans 1 C. (Y.) galli 1 C. hispaniensis 2 C. (Y.) hollandica 1 C. (Y.) osloensis * 4 Y. yakushimensis (including type strain for all) 2 Yarrowia sp. | MUCL 29853: diploid type strain (ATCC 18942) |
| China (PR) | CICC (WDCM 582) | China Center of Industrial Culture Collection | http://www.china-cicc.org (accessed on 3 June 2021) http://www.english.china-cicc.org (accessed on 3 June 2021) ISO 9001:2008 ISO 17025:2016; ISO 17034:2018 | 34 Y. lipolytica (with intended biotechnological applications indicated) 1 Y. brassicae (type strain) | W29 (CICC 1778) CICC 33063 for erythritol production CICC 31268 and 32291 edible and feed yeasts |
| CGMCC (WDCM 550) | China General Microbiological Culture Collection Center | http://www.cgmcc.net (accessed on 3 June 2021) http://www.cgmcc.net/english/ (accessed on 3 June 2021) ISO 9001:2010; ISO 14001:2010 | 113 Y. lipolytica | ||
| MCCC (WDCM 1051) | Marine Culture Collection of China | http://www.mccc.org.cn/ (accessed on 3 June 2021) ISO 9001:2011 | 135 Y. lipolytica + 1 Yarrowia sp. | SWJ-1b (MCCC 2E00068): marine strain, for citric acid and SCP production | |
| France | CIRM-Levures (WDCM 788) | Centre International de Ressources Microbiennes—Levures | https://www6.inrae.fr/cirm/Levures (accessed on 3 June 2021) https://www6.inrae.fr/cirm_eng/Yeasts (accessed on 3 June 2021) ISO 9001:2015 | 123 Y. lipolytica (including numerous GM laboratory strains) + 4 Y. deformans 1 C. hispaniensis (including type strain for both) 2 Yarrowia sp. | CLIB 183: diploid type strain (ATCC 18942) CLIB 703: type strain of Candida paralipolytica, for lipase production (ATCC 48436) E122 (CLIB 120): GM E129 (CLIB 121): GM E150 (CLIB 122): GM, sequenced (reference strain) H222 (CLIB 80): sequenced W29 (CLIB 89): sequenced W29 ura302 (CLIB 141): GM Po1a (CLIB 140): GM Po1d (CLIB 139): GM Po1e (CLIB 723): GM Po1f (CLIB 724): GM, sequenced Po1g (CLIB 725): GM Po1h (CLIB 882): GM Po1t (CLIB 883): GM |
| Germany | DSMZ (WDCM 274) | Leibniz-Institut DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH | http://www.dsmz.de/ (accessed on 3 June 2021) ISO 9001:2008 ISO 17025:2005; ISO Guide 34:2009 | 6 Y. lipolytica + 1 Y. deformans | |
| Greece | ACA-DC (WDCM 609) | Agricultural College of Athens-Dairy Collection | http://www.aca-dc.gr/ (accessed on 3 June 2021) | 16 Y. lipolytica (isolated mainly from food) | ACA-DC 50109 §, ACA-DC 5033: applied to citric acid and, respectively, SCO and polyol production |
| India | NCIM (WDCM 3) | National Collection of Industrial Microorganisms | https://www.ncl-india.org/files/NCIM/Default.aspx (accessed on 3 June 2021) | 6 Y. lipolytica (erroneously indicated as Y. lipolitica) | NCIM 3589: marine strain, for gold nanoparticle production |
| Japan | NBRC (WDCM 825) | NITE (National Institute of Technology and Evaluation) Biological Resource Center | https://www.nite.go.jp/en/nbrc/index.html (accessed on 3 June 2021) https://www.nite.go.jp/nbrc/catalogue/NBRCDispSearchServlet?lang=en (accessed on 3 June 2021) ISO 9001:2008 | 20 Y. lipolytica + 1 C. (Y.) phangngensis 1 Y. porcina (type strain for both) | NBRC 1548: diploid type strain (ATCC 18942) NBRC 10073: type strain of Candida paralipolytica, for lipase production (ATCC 48436) W29 (NBRC 113670) |
| JCM (WDCM 567) | Japan Collection of Microorganisms | https://jcm.brc.riken.jp/en/ (accessed on 3 June 2021) ISO 9001:2015 | 22 Y. lipolytica + 4 Y. deformans 1 Y. keelungensis 1 Y. yakushimensis (including type strain for all) 5 Yarrowia sp. | JCM 8057: type strain (ATCC 20177) JCM 2320 and 8060: diploid type strain (ATCC 18942) JCM 8054: type strain of Candida paralipolytica, for lipase production (ATCC 48436) | |
| Netherlands | CBS-KNAW (WDCM 133) | CBS Filamentous fungi and Yeast Collection-Westerdijk Fungal Biodiversity Institute | http://www.westerdijkinstitute.nl/ (accessed on 3 June 2021) https://wi.knaw.nl/page/Collection (accessed on 3 June 2021) https://theyeasts.org/ (accessed on 3 June 2021) ISO 9001:2007 | 38 Y. lipolytica + 3 C. (Y.) alimentaria 1 Y. brassicae 1 Y. bubula 18 Y. deformans 1 Y. divulgata 2 Y. galli 2 C. (Y.) hollandica 1 Y. keelungensis 4 Y. osloensis 1 Y. parophoni * 1 C. (Y.) phangngensis 2 Y. porcina 4 Y. yakushimensis 2 C. hispaniensis (including type strain for all) | CBS 8108: type strain (ATCC 20177) CBS 6124: diploid type strain (ATCC 18942) CBS 6303: type strain of Candida paralipolytica, for lipase production (ATCC 48436) W29 (CBS 7504) |
| Russia (Fed) | VKPM (WDCM 588) | Russian National Collection of Industrial Microorganisms | https://vkpm.genetika.ru/ (accessed on 3 June 2021) | 28 Y. lipolytica | W29 (Y-3178) Po1f (Y-3155): GM Po1f Ura+ (Y-3483): GM |
| VKM (WDCM 342) | All-Russian Collection of Microorganisms | http://www.vkm.ru/Catalogue.htm (accessed on 3 June 2021) | 17 Y. lipolytica + 1 Y. deformans (including type strain for all) | VKM Y-2373, VKM Y-2412: applied to organic acid production | |
| USA | ATCC (WDCM 1) | American Type Culture Collection | http://www.atcc.org/ (accessed on 3 June 2021) ISO 9001:2015 ISO 13485:2016; ISO 17025:2017; ISO 17034:2016 | 132 Y. lipolytica + 1 Y. deformans 1 C. (Y.) phangngensis * (type strain for both) | ATCC 20177: type strain ATCC 18942: diploid type strain ATCC 48436: type strain of Candida paralipolytica, for lipase production D 1805 (ATCC 20390): non-sporulating diploid for organic acid production ATCC 20362: basis for Dupont PUFA-producing platform W29 (ATCC 20460) Po1f (ATCC MYA2613): GM |
| NRRL (WDCM 97) | Agricultural Research Service (ARS) Culture Collection | http://nrrl.ncaur.usda.gov/ (accessed on 3 June 2021) | 34 Y. lipolytica + 1 C. (Y.) alimentaria 1 Y. bubula 2 Y. deformans 1 Y. divulgata 1 C. (Y.) galli 1 C. (Y.) hollandica 1 Y. keelungensis 1 C. (Y.) osloensis 2 C. (Y.) phangngensis * 1 Y. porcina 1 Y. yakushimensis 2 C. hispaniensis (including type strain for all) | NRRL YB-423: diploid type strain (ATCC 18942) YB-392, YB-419, YB-420, YB-566 and YB-567: selected as promising candidate strains for industrial biocatalysis, all sequenced W29 (Y-63746) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madzak, C. Yarrowia lipolytica Strains and Their Biotechnological Applications: How Natural Biodiversity and Metabolic Engineering Could Contribute to Cell Factories Improvement. J. Fungi 2021, 7, 548. https://doi.org/10.3390/jof7070548
Madzak C. Yarrowia lipolytica Strains and Their Biotechnological Applications: How Natural Biodiversity and Metabolic Engineering Could Contribute to Cell Factories Improvement. Journal of Fungi. 2021; 7(7):548. https://doi.org/10.3390/jof7070548
Chicago/Turabian StyleMadzak, Catherine. 2021. "Yarrowia lipolytica Strains and Their Biotechnological Applications: How Natural Biodiversity and Metabolic Engineering Could Contribute to Cell Factories Improvement" Journal of Fungi 7, no. 7: 548. https://doi.org/10.3390/jof7070548
APA StyleMadzak, C. (2021). Yarrowia lipolytica Strains and Their Biotechnological Applications: How Natural Biodiversity and Metabolic Engineering Could Contribute to Cell Factories Improvement. Journal of Fungi, 7(7), 548. https://doi.org/10.3390/jof7070548





