Production and Activity of Cristazarin in the Lichen-Forming Fungus Cladonia metacorallifera
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Isolate
2.2. Media and Culture Conditions
2.3. Analysis of Secondary Metabolites Using Thin Layer Chromatography (TLC) and High Performance Liquid Chromatography (HPLC)
2.4. Purification of Cristazarin Peak (tR = 3.4 min) by Prep-HPLC
2.5. NMR Spectroscopy Analysis
2.6. Genome Information and PKS Gene Annotation
2.7. Analysis of Transcript Levels
2.8. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Assay
3. Results
3.1. Characterization of C. metacorallifera
3.2. Effects of the Carbon Source on the Production of Secondary Metabolites
3.3. Identification of Secondary Metabolites via NMR
3.4. Production of Cristazarin in a Liquid-Based System
3.5. Expression Analysis of 30 Putative PKS Genes in C. metacorallifera
3.6. Expression Analysis of White Collar-1 and White Collar-2 in C. metacorallifera
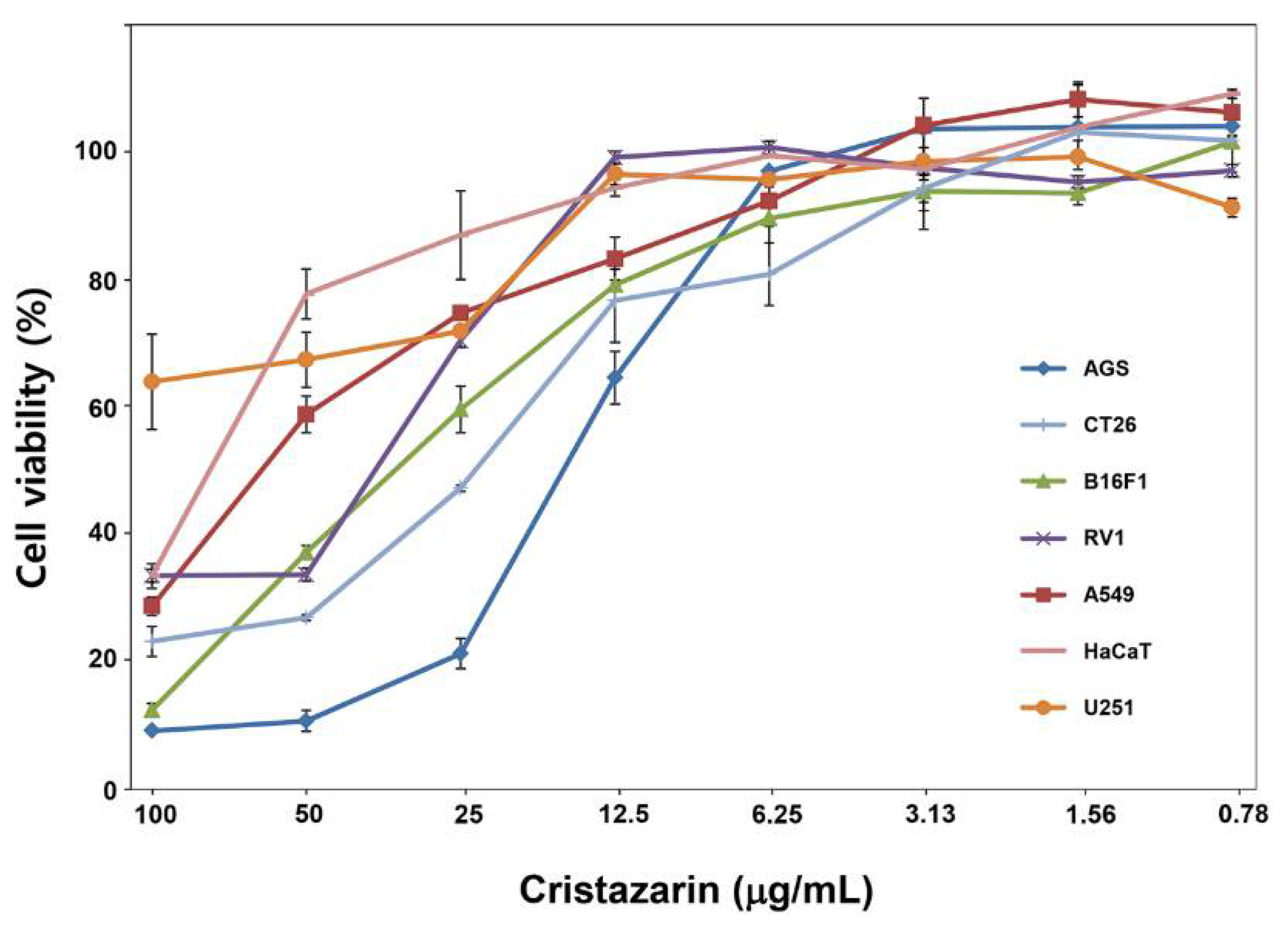
3.7. Biological Activity of Cristazarin Produced by C. metacorallifera
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Castle, H.; Kubsch, F. The production of usnic, didymic, and rhodocladonic acids by the fungal component of the lichen Cladonia cristatella. Arch. Biochem. 1949, 23, 158–160. [Google Scholar]
- Ejiri, H.; Sankawa, U. Graciliformin and its acetates in Cladonia graciliformis. Phytochemistry 1975, 14, 277–279. [Google Scholar] [CrossRef]
- Nakano, H.; Komiya, T.; Shibata, S. Anthraquinones of the lichens of Xanthoria and Caloplaca and their cultivated mycobionts. Phytochemistry 1972, 11, 3505–3508. [Google Scholar] [CrossRef]
- Yoshimura, I.; Kurokawa, T.; Kinoshita, K.; Yamamoto, Y.; Miyawaki, H. Lichen substances in cultured lichens. J. Hatt. Bot. Lab. 1994, 76, 249–261. [Google Scholar] [CrossRef]
- Kinoshita, Y.; Yamamoto, Y.; Yoshimura, I.; Kurokawa, T.; Yamada, Y. Production of usnic acid in cultured Usnea hirta. Lichenol 1993, 53, 137–145. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Matsubara, H.; Kinoshita, Y.; Kinoshita, K.; Koyama, K.; Takahashi, K.; Ahmadjiam, V.; Kurokawa, T.; Yoshimura, I. Naphthazarin derivatives from cultures of the lichen Cladonia cristatella. Phytochemistry 1996, 43, 1239–1242. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kinoshita, Y.; Matsubara, K.; Kinoshita, K.; Koyama, K.; Takahashi, K.; Kurokawa, T.; Yoshimura, I. Screening of biological activities and isolation of biological-active compounds from lichens. Recent Res. Devel. Phytochem. 1998, 2, 23–34. [Google Scholar]
- Brunauer, G.; Hager, A.; Grube, M.; Turk, R.; Stocker-Worgotter, E. Alterations in secondary metabolism of aposymbiotically grown mycobionts of Xanthoria elegans and cultured resynthesis stages. Plant Physiol. Biochem. 2007, 45, 146–151. [Google Scholar] [CrossRef]
- Hamada, N. Induction of the production of lichen substances by non-metabolites. Bryologist 1996, 99, 68–70. [Google Scholar] [CrossRef]
- Hamada, N.; Miyagawa, H. Secondary metabolites from isolated lichen mycobionts cultured under different osmotic conditions. Lichenologist 1995, 27, 201–205. [Google Scholar] [CrossRef]
- Hamada, N.; Miyagawa, H.; Miyawaki, H.; Inoue, M. Lichen substances in mycobionts of crustose lichens cultured on media with extra sucrose. Bryologist 1996, 99, 71–74. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kinoshita, Y.; Kurokawa, T.; Yoshimura, I.; Ahmadjiam, V.; Yamada, Y. Cell growth and pigment production in suspension cultures of a mycobiont isolated from the lichen Cladonia cristatella. Can. J. Bot. 1995, 73, 590–594. [Google Scholar] [CrossRef]
- Stocker-Worgotter, E. Metabolic diversity of lichen-forming ascomycetous fungi: Culturing, polyketide and shikimate metabolite production, and PKS genes. Nat. Prod. Rep. 2008, 25, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Fahselt, D. Secondary biochemistry of lichens. Symbiosis 1994, 16, 134–140. [Google Scholar]
- O’Hagan, D. The Polyketide Metabolites; Ellis Horwood: Chichester, UK, 1991. [Google Scholar]
- Anand, S.; Prasad, M.V.; Yadav, G.; Kumar, N.; Shehara, J.; Ansari, M.Z.; Mohanty, D. SBSPKS: Structure based sequence analysis of polyketide synthases. Nucleic Acids Res. 2010, 38, W487–W496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ansari, M.Z.; Yadav, G.; Gokhale, R.S.; Mohanty, D. NRPS-PKS: A knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 2004, 32, W405–W413. [Google Scholar] [CrossRef]
- Bachmann, B.O.; Ravel, J. Chapter 8. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 2009, 458, 181–217. [Google Scholar] [CrossRef]
- Jenke-Kodama, H.; Dittmann, E. Bioinformatic perspectives on NRPS/PKS megasynthases: Advances and challenges. Nat. Prod. Rep. 2009, 26, 874–883. [Google Scholar] [CrossRef]
- Li, M.H.; Ung, P.M.; Zajkowski, J.; Garneau-Tsodikova, S.; Sherman, D.H. Automated genome mining for natural products. BMC Bioinform. 2009, 10, 185. [Google Scholar] [CrossRef] [Green Version]
- Starcevic, A.; Zucko, J.; Simunkovic, J.; Long, P.F.; Cullum, J.; Hranueli, D. ClustScan: An integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 2008, 36, 6882–6892. [Google Scholar] [CrossRef] [Green Version]
- Tae, H.; Kong, E.B.; Park, K. ASMPKS: An analysis system for modular polyketide synthases. BMC Bioinform. 2007, 8, 327. [Google Scholar] [CrossRef] [Green Version]
- Weber, T.; Rausch, C.; Lopez, P.; Hoof, I.; Gaykova, V.; Huson, D.H.; Wohlleben, W. CLUSEAN: A computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J. Biotechnol. 2009, 140, 13–17. [Google Scholar] [CrossRef]
- Yadav, G.; Gokhale, R.S.; Mohanty, D. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 2003, 328, 335–363. [Google Scholar] [CrossRef]
- Avalos, J.; Estrada, A.F. Regulation by light in Fusarium. Fungal Genet. Biol. 2010, 47, 930–938. [Google Scholar] [CrossRef]
- Bayram, O.; Braus, G.H.; Fischer, R.; Rodriguez-Romero, J. Spotlight on Aspergillus nidulans photosensory systems. Fungal Genet. Biol. 2010, 47, 900–908. [Google Scholar] [CrossRef]
- Estrada, A.F.; Avalos, J. The White Collar protein WcoA of Fusarium fujikuroi is not essential for photocarotenogenesis, but is involved in the regulation of secondary metabolism and conidiation. Fungal Genet. Biol. 2008, 45, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Fuller, K.K.; Ringelberg, C.S.; Loros, J.J.; Dunlap, J.C. The fungal pathogen Aspergillus fumigatus regulates growth, metabolism, and stress resistance in response to light. mBio 2013, 4, e00142-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Ridenour, J.B.; Dunkle, L.D.; Bluhm, B.H. Regulation of stomatal tropism and infection by light in Cercospora zeae-maydis: Evidence for coordinated host/pathogen responses to photoperiod? PLoS Pathog. 2011, 7, e1002113. [Google Scholar] [CrossRef]
- Kim, H.; Son, H.; Lee, Y.W. Effects of light on secondary metabolism and fungal development of Fusarium graminearum. J. Appl. Microbiol. 2014, 116, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Hitzenhammer, E.; Buschl, C.; Sulyok, M.; Schuhmacher, R.; Kluger, B.; Wischnitzki, E.; Schmoll, M. YPR2 is a regulator of light modulated carbon and secondary metabolism in Trichoderma reesei. BMC Genome 2019, 20, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, G.J.; Pimentel-Elardo, S.; Nodwell, J.R. Dual-PKS cluster for biosynthesis of a light-induced secondary metabolite found from genome sequencing of Hyphodiscus hymeniophilus fungus. ChemBio Chem. 2020, 21, 2116–2120. [Google Scholar] [CrossRef]
- BeGora, M.D.; Fahselt, D. Usnic acid and atranorin concentrations in lichens in relation to bands of UV irradiance. Bryologist 2001, 104, 1687–1692. [Google Scholar] [CrossRef]
- Park, S.Y.; Choi, J.; Lee, G.W.; Kim, J.A.; Oh, S.O.; Jeong, M.H.; Yu, N.H.; Kim, S.; Lee, Y.H.; Hur, J.S. Draft genome sequence of lichen-forming fungus Cladonia metacorallifera strain KoLRI002260. Genome Announc. 2014, 2, e01065-13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stenroos, S.; Hyvonen, J.; Myllys, L.; Thell, A.; Ahti, T. Phylogeny of the genus Cladonia s.lat. (Cladoniaceae, Ascomycetes) inferred from molecular, morphological, and chemical data. Cladistics 2002, 18, 237–278. [Google Scholar] [CrossRef]
- Kim, J.A.; Hong, S.G.; Cheong, Y.H.; Koh, Y.J.; Hur, J.S. A new reducing polyketide synthase gene from the lichen-forming fungus Cladonia metacorallifera. Mycologia 2012, 104, 362–370. [Google Scholar] [CrossRef]
- Lilly, V.G.; Barnett, H.L. Physiology of the Fungi; McGraw-Hill, Inc.: New York, NY, USA, 1951. [Google Scholar]
- Culberson, C.F. Improved conditions and new data for the identification of lichen products by a standardized thin-layer chromatographic method. J. Chromatogr. 1972, 72, 113–125. [Google Scholar] [CrossRef]
- Park, S.Y.; Choi, J.; Lim, S.E.; Lee, G.W.; Park, J.; Kim, Y.; Kong, S.; Kim, S.R.; Rho, H.S.; Jeon, J.; et al. Global expression profiling of transcription factor genes provides new insights into pathogenicity and stress responses in the rice blast fungus. PLoS Pathog. 2013, 9, e1003350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Stenroos, S. Taxonomy of the Cladonia coccifera group II. Ann. Bot. Fenn. 1989, 26, 307–317. [Google Scholar]
- Honegger, R.; Kutasi, V. Anthraquinone Production in Three Aposymbiotically Cultured Teloschistalean Lichen Mycobionts: The Role of the Carbon Source; Institut National de la Recherche Agronomique: Paris, France, 1990. [Google Scholar]
- Schlotter, Y.M.; Veenhof, E.Z.; Brinkhof, B.; Rutten, V.P.; Spee, B.; Willemse, T.; Penning, L.C. A GeNorm algorithm-based selection of reference genes for quantitative real-time PCR in skin biopsies of healthy dogs and dogs with atopic dermatitis. Vet. Immunol. Immunopathol. 2009, 129, 115–118. [Google Scholar] [CrossRef] [Green Version]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [Green Version]
- Palmqvist, K.; Dahlman, L.; Jonsson, A.; Nash, T.H. The carbon economy of lichens. In Lichen Biology, 2nd ed.; Nash, T.H., III, Ed.; Cambridge University Press: Cambridge, UK, 2008; pp. 182–215. [Google Scholar]
- Wastlhuber, R.; Loos, E. Differences between cultured and freshly isolated cyanobiont from Peltigera-is there symbiosis-specific regulation of a glucose carrier? Lichenologist 2007, 28, 67–78. [Google Scholar] [CrossRef]
- Cornejo, C.; Nelson, P.R.; Stepanchikova, I.; Himelbrant, D.; JØRgensen, P.-M.; Scheidegger, C. Contrasting pattern of photobiont diversity in the Atlantic and Pacific populations of Erioderma pedicellatum (Pannariaceae). Lichenologist 2016, 48, 275–291. [Google Scholar] [CrossRef]
- Gustavs, L.; Schiefelbein, U.; Tatyana Darienko, T.; Proschold, T. Symbioses of the green algal genera Coccomyxa and Elliptochloris (Trebouxiophyceae, Chlorophyta). In Algal and Cyanobacteria Symbioses; Grobe, M., Seckbach, J., Mugga, L., Eds.; World Scientific Publishing Company: Singapore, 2017; pp. 169–208. [Google Scholar] [CrossRef]
- Kono, M.; Tanabe, H.; Ohmura, Y.; Satta, Y.; Terai, Y. Physical contact and carbon transfer between a lichen-forming Trebouxia alga and a novel Alphaproteobacterium. Microbiology 2017, 163, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Lines, C.E.M.; Ratcliffe, R.G.; Rees, T.A.V.; Southon, T.E. A 13C NMR study of photosynthate transport and metabolism in the Lichen Xanthoria calcicola Oxner. New Phytol. 1989, 111, 447–456. [Google Scholar] [CrossRef]
- Elshobary, M.E.; Osman, M.E.; Abo-Shady, A.M.; Komatsu, E.; Perreault, H.; Sorensen, J.; Piercey-Normore, M.D. Algal carbohydrates affect polyketide synthesis of the lichen-forming fungus Cladonia rangiferina. Mycologia 2016, 108, 646–656. [Google Scholar] [CrossRef] [Green Version]
- Elix, J.A.; Stocker-Wörgötter, E. Biochemistry and secondary metabolites. In Lichen Biology, 2nd ed.; Nash, H., III, Ed.; Cambridge University Press: Cambridge, UK, 2008; pp. 104–133. [Google Scholar]
- Boustie, J.; Grube, M. Lichens-a promising source of bioactive secondary metabolites. Plant Genet. Resour. 2005, 3, 273–287. [Google Scholar] [CrossRef] [Green Version]
- Seliwanoff, T. Notiz über eine Fruchtzuckerreaction. Ber. Dtsch. Chem. Ges. 1887, 20, 181–182. [Google Scholar] [CrossRef] [Green Version]
- Raharjo, T.J.; Chang, W.-T.; Choi, Y.H.; Peltenburg-Looman, A.M.G.; Verpoorte, R. Olivetol as product of a polyketide synthase in Cannabis sativa L. Plant Sci. 2004, 166, 381–385. [Google Scholar] [CrossRef]
- Kozaki, A.; Sasaki, Y. Light-dependent changes in redox status of the plastidic acetyl-CoA carboxylase and its regulatory component. Biochem. J. 1999, 339 Pt 3, 541–546. [Google Scholar] [CrossRef]





| Target Genes * | Forward Primer (5’ → 3’) | Reverse Primer (5’ → 3’) |
|---|---|---|
| β-tubulin | TGAGGCACTTTACGACATCTG | GAAGTGAAGACGAGGGAAAGG |
| α-tubulin | TCGTCTCTTCAATCACTGCC | GAATTGGATTCATGTGCAGCC |
| EF1β | TCTACAGGGCAAGTTCAATCG | GCAAAGTAGAGGATCAGGAGTG |
| UEP1 | CCTCCATCGCATTACCCTTAG | CTTCCCTGCGAAAATCAAACG |
| Actin2 | CACAGCAACCCTATACTCCTTC | CTATTCTCACTATCACACCATCCC |
| GAPDH | AAGACGCTGAATGGGATATGG | TCTTATGCAAGCTTAGCCCTC |
| CmPKS1 | GCTGTTTTTGCGGGCATGGA | CATACGGACGGCTTGATGT |
| CmPKS2 | ACCAGTTCGGATCACTTC | CGTAGCAATATCTGTTCG |
| CmPKS3 | CAAGGCTTCCACTTCTCA | CGGAAGATGTGTAACCTC |
| CmPKS4 | TGCGATTCACGCTCCATA | TGACATATCTCTGGCACG |
| CmPKS5 | GTCGAACGTATCATTCAT | GATCGATATGAGTGTGCA |
| CmPKS6 | TCGAAGAGGTGCAAGCAA | GTTGACCAGCTGTCGAAG |
| CmPKS7 | TATCGAGTACACCAGTGC | GCACAGTGTTCTGATCGT |
| CmPKS8 | GTCTCATTAGCTATGTAC | ACGGACCAGCTCTCTTGG |
| CmPKS9 | AGAGTGCAGCAGAGCTGT | CGTAGTGTCTCTTGGTGC |
| CmPKS10 | GCTCCGCAAGAACAGCAA | TGGAATGCGGCAGTGATC |
| CmPKS11 | ACTGATGCTTCATGTCGA | TGAGCCGTTGCACCAACA |
| CmPKS12 | CAGGTCTTGGAGACTACT | GGTCGACATTCCTGTCTT |
| CmPKS13 | ACTAGACAGGCTCCAGAG | TTCATTGTGTCGAAGGTC |
| CmPKS14 | TCAGCATACTAACACTGC | TCAGACACCTGAAGACCT |
| CmPKS15 | AGGTACATAATCCAGAGA | AGTATACCATTCGCATCG |
| CmPKS16 | ATTGGCCTCTTGAGCGCT | GATGTACGTATCCGGATA |
| CmPKS17 | ATGGCTGAAGAGGCGGAT | GCGCGACCAATTGAATCA |
| CmPKS18 | ATGGAGATGGCGATACGA | TGAGAGTACCAGCCGCAT |
| CmPKS19 | TGTGGATGTTGCCTGTCA | GACAGCATCTTCGAGACG |
| CmPKS20 | GAGGAGAAGTGGCATCTA | GCATTCTCAAGTCCTTCA |
| CmPKS21 | TGGCTTCTGATTATACCACG | GCTAACGTTCGGAGACGATG |
| CmPKS22 | ATGGAGCTCTGCATGGTA | TCATCGGCATTGTGAATC |
| CmPKS23 | CAGCATACCTGCCGAGAG | ACACCGCATTCTCAAGT |
| CmPKS24 | ACACATAATGAAGACATC | GTCGATAATGTTCTGGAG |
| CmPKS25 | TCATCGGCACAGTGCACA | GTATAGCAATGCGATATC |
| CmPKS26 | ACATCGTGGAGCAGAAGG | TGTGATGCCAGCGTCTTCT |
| CmPKS27 | CACAGTGGTACGAAGACA | CGATGTAGCATGAGGTAT |
| CmPKS28 | GCGACGTAGATGGATATG | GATGATTGGTTGCTGGAC |
| CmPKS29 | GGCGAGACGATATTGATCCAT | CTTGGCCAGTTGAACCG |
| CmPKS30 | TGTTAGACAAGCTCACTT | TGAATATGATGCTATCGT |
| Cmwc1 | TGCTAATTGCCATACCCG | CCGTAGCTGAATTGTGTGAG |
| Cmwc2 | TCGCTTCTTCTACCGCTT | CTTGGTTAGGCGCTCATT |
| C No. | 13C a | 1H (Number of Protons, Multiplicity) b (j-Hz) c |
|---|---|---|
| 1 | 176.99 | |
| 2 | 155.83 | |
| 3 | 128.74 | |
| 4 | 181.97 | |
| 4a | 105.02 | |
| 5 | 166.69 | |
| 6 | 109.19 | 6.62 (1H, s) |
| 7 | 158.51 | |
| 8 | 158.61 | |
| 8a | 111.70 | |
| 9 | 17.10 | 2.61 (2H, q, 7.5) |
| 10 | 12.99 | 1.11 (3H, t, 7.5) |
| 11 | 57.14 | 3.93 (3H, s) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, M.-H.; Park, C.-H.; Kim, J.A.; Choi, E.D.; Kim, S.; Hur, J.-S.; Park, S.-Y. Production and Activity of Cristazarin in the Lichen-Forming Fungus Cladonia metacorallifera. J. Fungi 2021, 7, 601. https://doi.org/10.3390/jof7080601
Jeong M-H, Park C-H, Kim JA, Choi ED, Kim S, Hur J-S, Park S-Y. Production and Activity of Cristazarin in the Lichen-Forming Fungus Cladonia metacorallifera. Journal of Fungi. 2021; 7(8):601. https://doi.org/10.3390/jof7080601
Chicago/Turabian StyleJeong, Min-Hye, Chan-Ho Park, Jung A Kim, Eu Ddeum Choi, Soonok Kim, Jae-Seoun Hur, and Sook-Young Park. 2021. "Production and Activity of Cristazarin in the Lichen-Forming Fungus Cladonia metacorallifera" Journal of Fungi 7, no. 8: 601. https://doi.org/10.3390/jof7080601
APA StyleJeong, M.-H., Park, C.-H., Kim, J. A., Choi, E. D., Kim, S., Hur, J.-S., & Park, S.-Y. (2021). Production and Activity of Cristazarin in the Lichen-Forming Fungus Cladonia metacorallifera. Journal of Fungi, 7(8), 601. https://doi.org/10.3390/jof7080601






