Virulence Traits and Population Genomics of the Black Yeast Aureobasidium melanogenum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virulence Factors
2.1.1. Strains and Growth Conditions
2.1.2. Growth at Human Body Temperature
2.1.3. Siderophore Production
2.1.4. Hemolytic Assay
2.1.5. Assimilation of Hydrocarbons
2.1.6. Assimilation of Neurotransmitters
2.2. Genome Sequencing and Population Genomics
2.2.1. Culture, Medium and Growth Conditions
2.2.2. DNA Isolation
2.2.3. Genome Sequencing
2.2.4. Variant Calling
2.2.5. Assembly and Annotation
2.2.6. Variant-Based Analysis
2.2.7. Phylogenetic Analyses
2.2.8. Identification of Individual Genes
3. Results
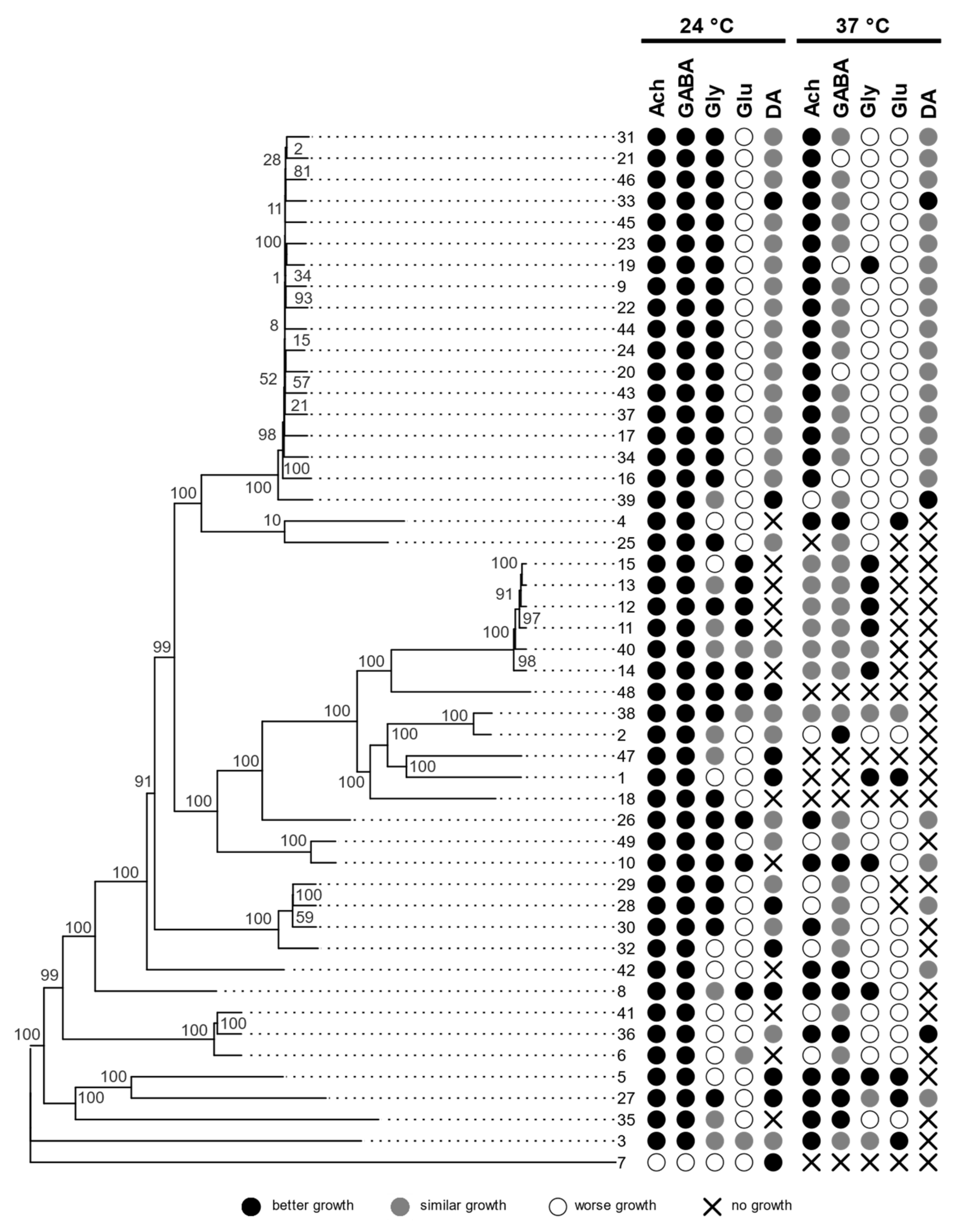
3.1. Growth at Human Body Temperature
3.2. Siderophore Production
3.3. Hemolytic Assay
3.4. Assimilation of Hydrocarbons
3.5. Assimilation of Neurotransmitters
3.6. Genomics
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chan, G.F.; Puad, M.S.A.; Chin, C.F.; Rashid, N.A.A. Emergence of Aureobasidium pullulans as human fungal pathogen and molecular assay for future medical diagnosis. Folia Microbiol. 2011, 56, 459–467. [Google Scholar] [CrossRef]
- Shiomi, N.; Yasuda, T.; Inoue, Y.; Kusumoto, N.; Iwasaki, S.; Katsuda, T.; Katoh, S. Characteristics of Neutralization of Acids by Newly Isolated Fungal Cells. J. Biosci. Bioeng. 2004, 97, 54–58. [Google Scholar] [CrossRef]
- Zajc, J.; Gostinčar, C.; Černoša, A.; Gunde-Cimerman, N. Stress-tolerant yeasts: Opportunistic pathogenicity versus biocontrol potential. Genes 2019, 10, 42. [Google Scholar] [CrossRef]
- Zalar, P.; Gostinčar, C.; de Hoog, G.S.; Uršič, V.; Sudhadham, M.; Gunde-Cimerman, N. Redefinition of Aureobasidium pullulans and its varieties. Stud. Mycol. 2008, 61, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Gostinčar, C.; Ohm, R.A.; Kogej, T.; Sonjak, S.; Turk, M.; Zajc, J.; Zalar, P.; Grube, M.; Sun, H.; Han, J.; et al. Genome sequencing of four Aureobasidium pullulans varieties: Biotechnological potential, stress tolerance, and description of new species. BMC Genom. 2014, 15, 549. [Google Scholar] [CrossRef] [PubMed]
- Prasongsuk, S.; Lotrakul, P.; Ali, I.; Bankeeree, W.; Punnapayak, H. The current status of Aureobasidium pullulans in biotechnology. Folia Microbiol. 2018, 63, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Diguță, C.F.; Proca, I.G.; Jurcoane, Ș.; Matei, F. Molecular characterization by PCR-RFLP of indigenous fungal isolates from hypersaline stream water in România. Folia Microbiol. 2018, 64, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Perini, L.; Mogrovejo, D.C.; Tomazin, R.; Gostinčar, C.; Brill, F.H.H.; Gunde-Cimerman, N. Phenotypes associated with pathogenicity: Their expression in arctic fungal isolates. Microorganisms 2019, 7, 600. [Google Scholar] [CrossRef]
- Ma, Z.C.; Fu, W.J.; Liu, G.L.; Wang, Z.P.; Chi, Z.M. High-level pullulan production by Aureobasidium pullulans var. melanogenium P16 isolated from mangrove system. Appl. Microbiol. Biotechnol. 2014, 98, 4865–4873. [Google Scholar] [CrossRef] [PubMed]
- Majidi, Z.; Goudarzi, A.; Shamili, M.; Bagheri, A.; Seyahooei, M.A. High distribution rate of an emerging fungal pathogen on mango: A case study from southern Iran. Crop. Prot. 2021, 139, 105342. [Google Scholar] [CrossRef]
- Jiang, H.; Liu, N.N.; Liu, G.L.; Chi, Z.-M.; Wang, J.M.; Zhang, L.L.; Chi, Z.M. Melanin production by a yeast strain XJ5-1 of Aureobasidium melanogenum isolated from the Taklimakan desert and its role in the yeast survival in stress environments. Extremophiles 2016, 20, 567–577. [Google Scholar] [CrossRef]
- Novak Babič, M.; Zalar, P.; Ženko, B.; Schroers, H.-J.; Džeroski, S.; Gunde-Cimerman, N. Candida and Fusarium species known as opportunistic human pathogens from customer-accessible parts of residential washing machines. Fungal Biol. 2015, 119, 95–113. [Google Scholar] [CrossRef] [PubMed]
- Zupančič, J.; Babič, M.N.; Zalar, P.; Gunde-Cimerman, N. The black yeast Exophiala dermatitidis and other selected opportunistic human fungal pathogens spread from dishwashers to kitchens. PLoS ONE 2016, 11, e0148166. [Google Scholar] [CrossRef]
- Novak Babič, M.; Zalar, P.; Ženko, B.; Džeroski, S.; Gunde-Cimerman, N. Yeasts and yeast-like fungi in tap water and groundwater, and their transmission to household appliances. Fungal Ecol. 2016, 20, 30–39. [Google Scholar] [CrossRef]
- Humphries, Z.; Seifert, K.A.; Hirooka, Y.; Visagie, C.M. A new family and genus in Dothideales for Aureobasidium-like species isolated from house dust. IMA Fungus 2017, 8, 299–315. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Xue, S.J.; Li, Y.F.; Liu, G.L.; Chi, Z.M.; Hu, Z.; Chi, Z. Efficient transformation of sucrose into high pullulan concentrations by Aureobasidium melanogenum TN1-2 isolated from a natural honey. Food Chem. 2018, 257, 29–35. [Google Scholar] [CrossRef]
- Gostinčar, C.; Zajc, J.; Lenassi, M.; Plemenitaš, A.; de Hoog, S.; Al-Hatmi, A.M.S.; Gunde-Cimerman, N. Fungi between extremotolerance and opportunistic pathogenicity on humans. Fungal Divers. 2018, 93, 195–213. [Google Scholar] [CrossRef]
- Gostinčar, C.; Grube, M.; Gunde-Cimerman, N. Evolution of Fungal Pathogens in Domestic Environments? Fungal Biol. 2011, 115, 1008–1018. [Google Scholar] [CrossRef]
- Bolignano, G.; Criseo, G. Disseminated nosocomial fungal infection by Aureobasidium pullulans var. melanigenum: A case report. J. Clin. Microbiol. 2003, 41, 4483–4485. [Google Scholar] [CrossRef]
- Chen, W.T.; Tu, M.E.; Sun, P.L. Superficial Phaeohyphomycosis Caused by Aureobasidium melanogenum Mimicking Tinea Nigra in an Immunocompetent Patient and Review of Published Reports. Mycopathologia 2016, 181, 555–560. [Google Scholar] [CrossRef]
- Mershon-Shier, K.L.; Deville, J.G.; Delair, S.; Fothergill, A.W.; Wickes, B.; De Hoog, G.S.; Sutton, D.A.; Lewinski, M.A. Aureobasidium pullulans var. melanigenum fungemia in a pediatric patient. Med. Mycol. 2010, 49, 80–83. [Google Scholar] [CrossRef]
- Wang, S.C.; Lo, H.J.; Lin, L.J.; Chen, C.H. Port catheter-associated Aureobasidium melanigenum fungemia. J. Formos. Med. Assoc. 2018, 117, 346–347. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, K.; Joest, M.; Turan, N.; Schmidt, D.; Rath, P.M.; Steinmann, J. Hypersensitivity pneumonitis of a bagpipe player: Fungal antigens as trigger? Med. Mycol. Case Rep. 2019, 24, 44–47. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, M.; Rennie, R.; Sand, C.; Vaudry, W. Aureobasidium pullulans infection: Fungemia in an infant and a review of human cases. Diagn. Microbiol. Infect. Dis. 2005, 51, 209–213. [Google Scholar] [CrossRef]
- Slepecky, R.A.; Starmer, W.T. Phenotypic plasticity in fungi: A review with observations on Aureobasidium pullulans. Mycologia 2009, 101, 823–832. [Google Scholar] [CrossRef]
- Gauthier, G.M. Dimorphism in Fungal Pathogens of Mammals, Plants, and Insects. PLOS Pathog. 2015, 11, e1004608. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, X.; Liu, H.; Xi, L.; Cooper, C.R. Increased virulence of albino mutant of Fonsecaea monophora in Galleria mellonella. Med. Mycol. 2019, 57, 1018–1023. [Google Scholar] [CrossRef]
- Eisenman, H.C.; Greer, E.M.; McGrail, C.W. The role of melanins in melanotic fungi for pathogenesis and environmental survival. Appl. Microbiol. Biotechnol. 2020, 104, 4247–4257. [Google Scholar] [CrossRef]
- Gonçalves, S.M.; Duarte-Oliveira, C.; Campos, C.F.; Aimanianda, V.; ter Horst, R.; Leite, L.; Mercier, T.; Pereira, P.; Fernández-García, M.; Antunes, D.; et al. Phagosomal removal of fungal melanin reprograms macrophage metabolism to promote antifungal immunity. Nat. Commun. 2020, 11, 2282. [Google Scholar] [CrossRef] [PubMed]
- Lavrin, T.; Konte, T.; Kostanjšek, R.; Sitar, S.; Sepčič, K.; Prpar Mihevc, S.; Žagar, E.; Župunski, V.; Lenassi, M.; Rogelj, B.; et al. The Neurotropic Black Yeast Exophiala dermatitidis Induces Neurocytotoxicity in Neuroblastoma Cells and Progressive Cell Death. Cells 2020, 9, 963. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L. Iron and siderophores in fungal-host interactions. Mycol. Res. 2008, 112, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Schaible, U.E.; Kaufmann, S.H.E. Iron and microbial infection. Nat. Rev. Microbiol. 2004, 2, 946–953. [Google Scholar] [CrossRef]
- Prenafeta-Boldú, F.X.; Summerbell, R.; Sybren De Hoog, G. Fungi growing on aromatic hydrocarbons: Biotechnology’s unexpected encounter with biohazard? FEMS Microbiol. Rev. 2006, 30, 109–130. [Google Scholar] [CrossRef]
- Najafzadeh, M.J.; Sutton, D.A.; Keisari, M.S.; Zarrinfar, H.; de Hoog, G.S.; Chowdhary, A.; Meis, J.F. In Vitro Activities of Eight Antifungal Drugs against 104 Environmental and Clinical Isolates of Aureobasidium pullulans. Antimicrob. Agents Chemother. 2014, 58, 5629–5631. [Google Scholar] [CrossRef]
- Chen, L.; Chi, Z.; Liu, G.L.; Xue, S.J.; Wang, Z.P.; Hu, Z.; Chi, Z.M. Improved pullulan production by a mutant of Aureobasidium melanogenum TN3-1 from a natural honey and capsule shell preparation. Int. J. Biol. Macromol. 2019, 141, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, G.; Jiang, H.; Chi, Z.; Chi, Z. An insight into the iron acquisition and homeostasis in Aureobasidium melanogenum HN6.2 strain through genome mining and transcriptome analysis. Funct. Integr. Genom. 2019, 19, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Gostinčar, C.; Turk, M.; Zajc, J.; Gunde-Cimerman, N. Fifty Aureobasidium pullulans genomes reveal a recombining polyextremotolerant generalist. Environ. Microbiol. 2019, 21, 3638–3652. [Google Scholar] [CrossRef] [PubMed]
- Gladieux, P.; De Bellis, F.; Hann-Soden, C.; Svedberg, J.; Johannesson, H.; Taylor, J.W. Neurospora from Natural Populations: Population Genomics Insights into the Life History of a Model Microbial Eukaryote. In Statistical Population Genomics. Methods in Molecular Biology; Dutheil, J.Y., Ed.; Humana: New York, NY, USA, 2020; Volume 2090, pp. 313–336. ISBN 9781071601990. [Google Scholar]
- Milagres, A.M.F.; Machuca, A.; Napoleão, D. Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. J. Microbiol. Methods 1999, 37, 1–6. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Buxton, R. Blood Agar Plates and Hemolysis Protocols. Am. Soc. Microbiol. 2005, 1–9. Available online: https://www.asm.org/getattachment/7ec0de2b-bb16-4f6e-ba07-2aea25a43e76/protocol-2885.pdf (accessed on 14 January 2021).
- Satow, M.M.; Attili-Angelis, D.; de Hoog, G.S.; Angelis, D.F.; Vicente, V.A. Selective factors involved in oil flotation isolation of black yeasts from the environment. Stud. Mycol. 2008, 61, 157–163. [Google Scholar] [CrossRef]
- Harwood, C.R.; Cutting, S.M. (Eds.) Molecular Biological Methods for Bacillus; Wiley: Chichester, UK, 1990; ISBN 9780471923930. [Google Scholar]
- Fang, C.; Zhong, H.; Lin, Y.; Chen, B.; Han, M.; Ren, H.; Lu, H.; Luber, J.M.; Xia, M.; Li, W.; et al. Assessment of the cPAS-based BGISEQ-500 platform for metagenomic sequencing. Gigascience 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Alkan, C.; Coe, B.P.; Eichler, E.E. GATK toolkit. Nat. Rev. Genet. 2011, 12, 363–376. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Morgenstern, B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005, 33, 465–467. [Google Scholar] [CrossRef] [PubMed]
- Hoff, K.J.; Lomsadze, A.; Borodovsky, M.; Stanke, M. Whole-genome annotation with BRAKER. Methods Mol. Biol. 2019, 1962, 65–95. [Google Scholar] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simão, F.A.; Zdobnov, E.M. OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019, 47, D807–D811. [Google Scholar] [CrossRef]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Kishino, H.; Yano, T.A. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Schliep, K.; Potts, A.J.; Morrison, D.A.; Grimm, G.W. Intertwining phylogenetic trees and networks. Methods Ecol. Evol. 2017, 8, 1212–1220. [Google Scholar] [CrossRef]
- Revell, L.J. Bioinformatics-dendextend-an R package for visualizing, adjusting and comapring trees of hierachical clutering. Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Brooks, J.C.; Grünwald, N.J. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 2015, 6, 208. [Google Scholar] [CrossRef]
- Gostinčar, C. Towards Genomic Criteria for Delineating Fungal Species. J. Fungi 2020, 6, 246. [Google Scholar] [CrossRef]
- Baker, D.N.; Langmead, B. Dashing: Fast and accurate genomic distances with HyperLogLog. Genome Biol. 2019, 20, 265. [Google Scholar] [CrossRef] [PubMed]
- Moolhuijzen, P.; See, P.T.; Hane, J.K.; Shi, G.; Liu, Z.; Oliver, R.P.; Moffat, C.S. Comparative genomics of the wheat fungal pathogen Pyrenophora tritici-repentis reveals chromosomal variations and genome plasticity. BMC Genom. 2018, 19, 279. [Google Scholar] [CrossRef]
- Wu, Y.; Ma, X.; Pan, Z.; Kale, S.D.; Song, Y.; King, H.; Zhang, Q.; Presley, C.; Deng, X.; Wei, C.I.; et al. Comparative genome analyses reveal sequence features reflecting distinct modes of host-adaptation between dicot and monocot powdery mildew 06 Biological Sciences 0604 Genetics. BMC Genom. 2018, 19, 705. [Google Scholar] [CrossRef] [PubMed]
- Janbon, G.; Ormerod, K.L.; Paulet, D.; Byrnes, E.J.; Yadav, V.; Chatterjee, G.; Mullapudi, N.; Hon, C.C.; Billmyre, R.B.; Brunel, F.; et al. Analysis of the Genome and Transcriptome of Cryptococcus neoformans var. grubii Reveals Complex RNA Expression and Microevolution Leading to Virulence Attenuation. PLoS Genet. 2014, 10, e1004261. [Google Scholar] [CrossRef]
- Floudas, D.; Held, B.W.; Riley, R.; Nagy, L.G.; Koehler, G.; Ransdell, A.S.; Younus, H.; Chow, J.; Chiniquy, J.; Lipzen, A.; et al. Evolution of novel wood decay mechanisms in Agaricales revealed by the genome sequences of Fistulina hepatica and Cylindrobasidium torrendii. Fungal Genet. Biol. 2015, 76, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Spanu, P.D.; Abbott, J.C.; Amselem, J.; Burgis, T.A.; Soanes, D.M.; Stüber, K.; Van Themaat, E.V.L.; Brown, J.K.M.; Butcher, S.A.; Gurr, S.J.; et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 2010, 330, 1543–1546. [Google Scholar] [CrossRef]
- Zhu, S.; Cao, Y.Z.; Jiang, C.; Tan, B.Y.; Wang, Z.; Feng, S.; Zhang, L.; Su, X.H.; Brejova, B.; Vinar, T.; et al. Sequencing the genome of Marssonina brunnea reveals fungus-poplar co-evolution. BMC Genom. 2012, 13, 382. [Google Scholar] [CrossRef]
- Kim, H.-S.; Lohmar, J.M.; Busman, M.; Brown, D.W.; Naumann, T.A.; Divon, H.H.; Lysøe, E.; Uhlig, S.; Proctor, R.H. Identification and distribution of gene clusters required for synthesis of sphingolipid metabolism inhibitors in diverse species of the filamentous fungus Fusarium. BMC Genom. 2020, 21, 510. [Google Scholar] [CrossRef]
- Huo, J.; Wang, Y.; Hao, Y.; Yao, Y.; Wang, Y.; Zhang, K.; Tan, X.; Li, Z.; Wang, W. Genome sequence resource for colletotrichum scovillei, the cause of anthracnose disease of chili. Mol. Plant. Microbe Interact. 2021, 34, 122–126. [Google Scholar] [CrossRef]
- Lin, R.; Zhang, X.; Xin, B.; Zou, M.; Gao, Y.; Qin, F.; Hu, Q.; Xie, B.; Cheng, X. Genome sequence of Isaria javanica and comparative genome analysis insights into family S53 peptidase evolution in fungal entomopathogens. Appl. Microbiol. Biotechnol. 2019, 103, 7111–7128. [Google Scholar] [CrossRef]
- Proctor, R.H.; McCormick, S.P.; Kim, H.S.; Cardoza, R.E.; Stanley, A.M.; Lindo, L.; Kelly, A.; Brown, D.W.; Lee, T.; Vaughan, M.M.; et al. Evolution of structural diversity of trichothecenes, a family of toxins produced by plant pathogenic and entomopathogenic fungi. PLoS Pathog. 2018, 14, e1006946. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Liu, Z.; Lin, R.; Li, E.; Mao, Z.; Ling, J.; Yang, Y.; Yin, W.B.; Xie, B. Biosynthesis of Antibiotic Leucinostatins in Bio-control Fungus Purpureocillium lilacinum and Their Inhibition on Phytophthora Revealed by Genome Mining. PLoS Pathog. 2016, 12, e1005685. [Google Scholar] [CrossRef] [PubMed]
- Gan, P.; Narusaka, M.; Kumakura, N.; Tsushima, A.; Takano, Y.; Narusaka, Y.; Shirasu, K. Genus-wide comparative genome analyses of colletotrichum species reveal specific gene family losses and gains during adaptation to specific infection lifestyles. Genome Biol. Evol. 2016, 8, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Kracher, B.; Hiruma, K.; Münch, P.C.; Garrido-Oter, R.; Thon, M.R.; Weimann, A.; Damm, U.; Dallery, J.F.; Hainaut, M.; et al. Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun. 2016, 7, 11362. [Google Scholar] [CrossRef]
- Robert, V.A.; Casadevall, A. Vertebrate endothermy restricts most fungi as potential pathogens. J. Infect. Dis. 2009, 200, 1623–1626. [Google Scholar] [CrossRef] [PubMed]
- Kitiyanant, V.; Lotrakul, P.; Kanchanabanca, C.; Padungros, P.; Punnapayak, H.; Prasongsuk, S.; Chanvorachote, P. Fusigen reduces intracellular reactive oxygen species and nitric oxide levels. In Vivo 2019, 33, 425–432. [Google Scholar] [CrossRef]
- Wang, W.; Chi, Z.M.; Chi, Z.; Li, J.; Wang, X.H. Siderophore production by the marine-derived Aureobasidium pullulans and its antimicrobial activity. Bioresour. Technol. 2009, 100, 2639–2641. [Google Scholar] [CrossRef]
- Wang, W.; Chi, Z.; Liu, G.; Buzdar, M.A.; Chi, Z.; Gu, Q. Chemical and biological characterization of siderophore produced by the marine-derived Aureobasidium pullulans HN6.2 and its antibacterial activity. BioMetals 2009, 22, 965–972. [Google Scholar] [CrossRef]
- Sepcic, K.; Zalar, P.; Gunde-Cimerman, N. Low water activity induces the production of bioactive metabolites in halophilic and halotolerant fungi. Mar. Drugs 2011, 9, 43–58. [Google Scholar] [CrossRef]
- Nayak, A.P.; Green, B.J.; Beezhold, D.H. Fungal hemolysins. Med. Mycol. 2013, 51, 1–16. [Google Scholar] [CrossRef]
- Blasi, B.; Poyntner, C.; Rudavsky, T.; Prenafeta-Boldú, F.X.; De Hoog, S.; Tafer, H.; Sterflinger, K. Pathogenic yet environmentally friendly? black fungal candidates for bioremediation of pollutants. Geomicrobiol. J. 2016, 33, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Restrepo-Flórez, J.M.; Bassi, A.; Thompson, M.R. Microbial degradation and deterioration of polyethylene—A review. Int. Biodeterior. Biodegrad. 2014, 88, 83–90. [Google Scholar] [CrossRef]
- Cappitelli, F.; Sorlini, C. Microorganisms Attack Synthetic Polymers in Items Representing Our Cultural Heritage. Appl. Environ. Microbiol. 2008, 74, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.A.; Hasan, F.; Hameed, A.; Ahmed, S. Biological degradation of plastics: A comprehensive review. Biotechnol. Adv. 2008, 26, 246–265. [Google Scholar] [CrossRef] [PubMed]
- Huttova, M.; Kralinsky, K.; Horn, J.; Marinova, I.; Iligova, K.; Fric, J.; Spanik, S.; Filka, J.; Uher, J.; Kurak, J.; et al. Prospective study of nosocomial fungal meningitis in children—Report of 10 cases. Scand. J. Infect. Dis. 1998, 30, 485–487. [Google Scholar] [CrossRef] [PubMed]
- Kutleša, M.; Mlinarić-Missoni, E.; Hatvani, L.; Voncina, D.; Simon, S.; Lepur, D.; Baršić, B. Chronic fungal meningitis caused by Aureobasidium proteae. Diagn. Microbiol. Infect. Dis. 2012, 73, 271–272. [Google Scholar] [CrossRef]
- Trupl, J.; Minarik, T.; Sufliarsky, J.; Spanik, S.; Kremery, V. Nosocomial bacterial and fungal meningitis in cancer patients. Support. Care Cancer 1995, 3, 425–427. [Google Scholar] [CrossRef]
- Gostinčar, C.; Stajich, J.E.; Zupančič, J.; Zalar, P.; Gunde-Cimerman, N. Genomic evidence for intraspecific hybridization in a clonal and extremely halotolerant yeast. BMC Genom. 2018, 19, 364. [Google Scholar] [CrossRef]
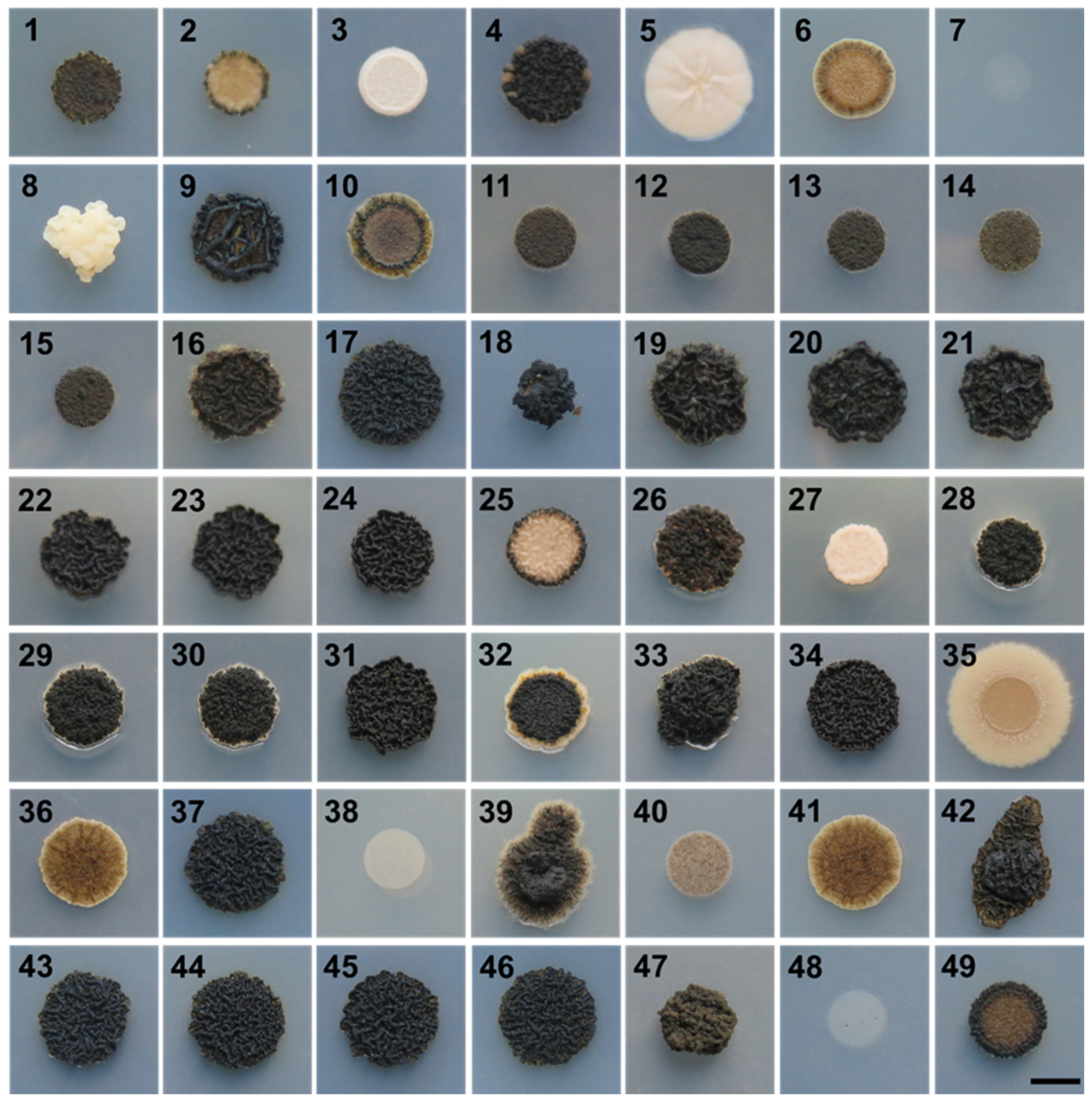





| Culture Collection Strain Number | Present Study Number | Isolation Habitat | Sampling Site Location |
|---|---|---|---|
| EXF-924 | 1 | Glacial: ponds on sea ice | Arctic; Svalbard, Ny Alesund |
| EXF-926 | 2 | Glacial: surface glacial ice | Arctic; Svalbard, Ny Alesund |
| EXF-3233 | 3 | Other: deep sea (4500 m b.s.l.) | Japan |
| EXF-3371 | 4 | Other: soil | Thailand |
| EXF-3378 | 5 | Water: public fountain | Thailand, Bangkok |
| EXF-3397 | 6 | Clinical: endoperitoneal fluid | Greece, Athens |
| EXF-3399 | 7 | Other: decomposing military textile | USA, Florida |
| EXF-4450 | 8 | Unknown: Iskra factory | Slovenia |
| EXF-5590 | 9 | Dishwasher: rubber seal | Slovenia |
| EXF-6171 | 10 | Glacial: black glacier | Argentina |
| EXF-7932 | 11 | Kitchen: metal drain on the kitchen sink | Sweden |
| EXF-7946 | 12 | Kitchen: metal holder for washed dishes | Sweden |
| EXF-8016 | 13 | Bathroom: between faucet and sink | Sweden |
| EXF-8022 | 14 | Refrigerator: inner surface | Sweden |
| EXF-8044 | 15 | Kitchen: metal holder for washed dishes | Sweden |
| EXF-8258 | 16 | Water: water from well | Slovenia |
| EXF-9877 | 17 | Tap water | Slovenia, Rodica |
| EXF-11403 | 18 | Refrigerator | Sweden |
| EXF-8492 | 19 | Water: water from well | Slovenia |
| EXF-8678 | 20 | Water: water from well | Slovenia, Šentvid |
| EXF-8689 | 21 | Water: water from well | Slovenia, Kleče |
| EXF-8695 | 22 | Water: water from well | Slovenia, Hrastje |
| EXF-8702 | 23 | Water: water from well | Slovenia, Brest |
| EXF-8986 | 24 | Saltern: fango mud from Sečovlje salterns | Slovenia, Sečovlje |
| EXF-9262 | 25 | Kitchen: rubber on kitchen drain | Slovenia, Gomilsko |
| EXF-9470 | 26 | Kitchen: counter above dishwasher | Slovenia, Gomilsko |
| EXF-9272 | 27 | Kitchen: strainer basket | Slovenia, Vojnik |
| EXF-9298 | 28 | Kitchen: plastic mesh on kitchen drain | Slovenia, Podlog v Savinjski dolini |
| EXF-9304 | 29 | Kitchen: strainer basket | Slovenia, Podlog v Savinjski dolini |
| EXF-9313 | 30 | Kitchen: sink | Slovenia, Podlog v Savinjski dolini |
| EXF-9454 | 31 | Tap water | Slovenia, Podlog v Savinjski dolini |
| EXF-9484 | 32 | Kitchen: counter above dishwasher | Slovenia, Velenje |
| EXF-9887 | 33 | Tap water | Slovenia, Velenje |
| EXF-9516 | 34 | Kitchen: sink drain | Slovenia, Zgornji Dolič |
| EXF-9539 | 35 | Kitchen: strainer basket | Slovenia, Lokovica |
| EXF-9540 | 36 | Dishwasher door | Slovenia, Celje |
| EXF-10064 | 37 | Tap water | Slovenia, Ormož |
| EXF-11060 | 38 | Other: ceiling | Slovenia, Celje |
| EXF-9875 | 39 | Tap water | Slovenia, Rodica |
| EXF-9906 | 40 | Saltern: Arthrocnemum sp. plant from Sečovlje saltern | Slovenia, Sečovlje |
| EXF-9911 | 41 | Kitchen: sink drain | Slovenia, Ormož |
| EXF-9937 | 42 | Kitchen: rubber on kitchen drain | Slovenia, Ljutomer |
| EXF-10061 | 43 | Tap water | Slovenia, Trebnje |
| EXF-10062 | 44 | Tap water | Slovenia, Litija |
| EXF-10066 | 45 | Tap water | Slovenia, Planina pri Sevnici |
| EXF-10333 | 46 | Tap water | Slovenia, Ljubljana |
| EXF-10372 | 47 | Other: air in National Gallery restoration centre | Slovenia, Ljubljana |
| EXF-10726 | 48 | Other: integument of a male alate ant of Atta sexdens rubropilosa | Brazil, Sao Paolo |
| EXF-11028 | 49 | Aquarium water: Proteus anguinus | Slovenia, Ljubljana |
| Culture Collection Strain Number | Genus | Isolation Habitat | Sampling Site Location |
|---|---|---|---|
| EXF-2481 | Aureobasidium subglaciale | Glacial: subglacial ice from seawater | Arctic; Svalbard, Ny Alesund |
| EXF-4632 | Aureobasidium subglaciale | Plant: decaying leaves of Convallaria sp. | Slovenia |
| EXF-12298 | Aureobasidium subglaciale | Refrigerator | Sweden |
| EXF-150 | Aureobasidium pullulans | Saltern: hypersaline water, active saltpans Seča, Droga Portorož | Slovenia, Portorož |
| EXF-3670 | Aureobasidium pullulans | Glacial: ice at the edge of the glacier | Arctic; Svalbard, Ny Alesund |
| EXF-10629 | Aureobasidium pullulans | Other: car reservoir fuel | Slovenia |
| EXF-11318 | Aureobasidium pullulans | Plant: apple surface | Slovenia, Horjul |
| EXF-3398 | Aureobasidium namibiae | Other: dolomitic marble | Namibia, Namib desert |
| Minimum | Mean | Maximum | Standard Deviation | |
|---|---|---|---|---|
| Genome assembly size (Mbp) | 25.31 | 41.43 | 54.65 | 10.25 |
| GC content (%) | 49.14 | 50.04 | 51.79 | 0.46 |
| CDS total length (Mbp) | 12.41 | 22.01 | 30.21 | 5.52 |
| CDS total length (% of genome) | 46.00 | 53.18 | 57.62 | 2.47 |
| Gene models (n) | 7480 | 18745 | 27345 | 6405 |
| Gene average length (bp) | 1109 | 1378 | 1945 | 232 |
| Number of exons (n) | 20572 | 43523 | 56418 | 11369 |
| Exons per gene (average) | 2.05 | 2.42 | 2.90 | 0.30 |
| Number of introns (n) | 7480 | 18745 | 27345 | 6405 |
| Intron average length (bp) | 86 | 94 | 162 | 11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Černoša, A.; Sun, X.; Gostinčar, C.; Fang, C.; Gunde-Cimerman, N.; Song, Z. Virulence Traits and Population Genomics of the Black Yeast Aureobasidium melanogenum. J. Fungi 2021, 7, 665. https://doi.org/10.3390/jof7080665
Černoša A, Sun X, Gostinčar C, Fang C, Gunde-Cimerman N, Song Z. Virulence Traits and Population Genomics of the Black Yeast Aureobasidium melanogenum. Journal of Fungi. 2021; 7(8):665. https://doi.org/10.3390/jof7080665
Chicago/Turabian StyleČernoša, Anja, Xiaohuan Sun, Cene Gostinčar, Chao Fang, Nina Gunde-Cimerman, and Zewei Song. 2021. "Virulence Traits and Population Genomics of the Black Yeast Aureobasidium melanogenum" Journal of Fungi 7, no. 8: 665. https://doi.org/10.3390/jof7080665
APA StyleČernoša, A., Sun, X., Gostinčar, C., Fang, C., Gunde-Cimerman, N., & Song, Z. (2021). Virulence Traits and Population Genomics of the Black Yeast Aureobasidium melanogenum. Journal of Fungi, 7(8), 665. https://doi.org/10.3390/jof7080665








