Priming of Plant Defenses against Ophiostoma novo-ulmi by Elm (Ulmus minor Mill.) Fungal Endophytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant and Fungal Material
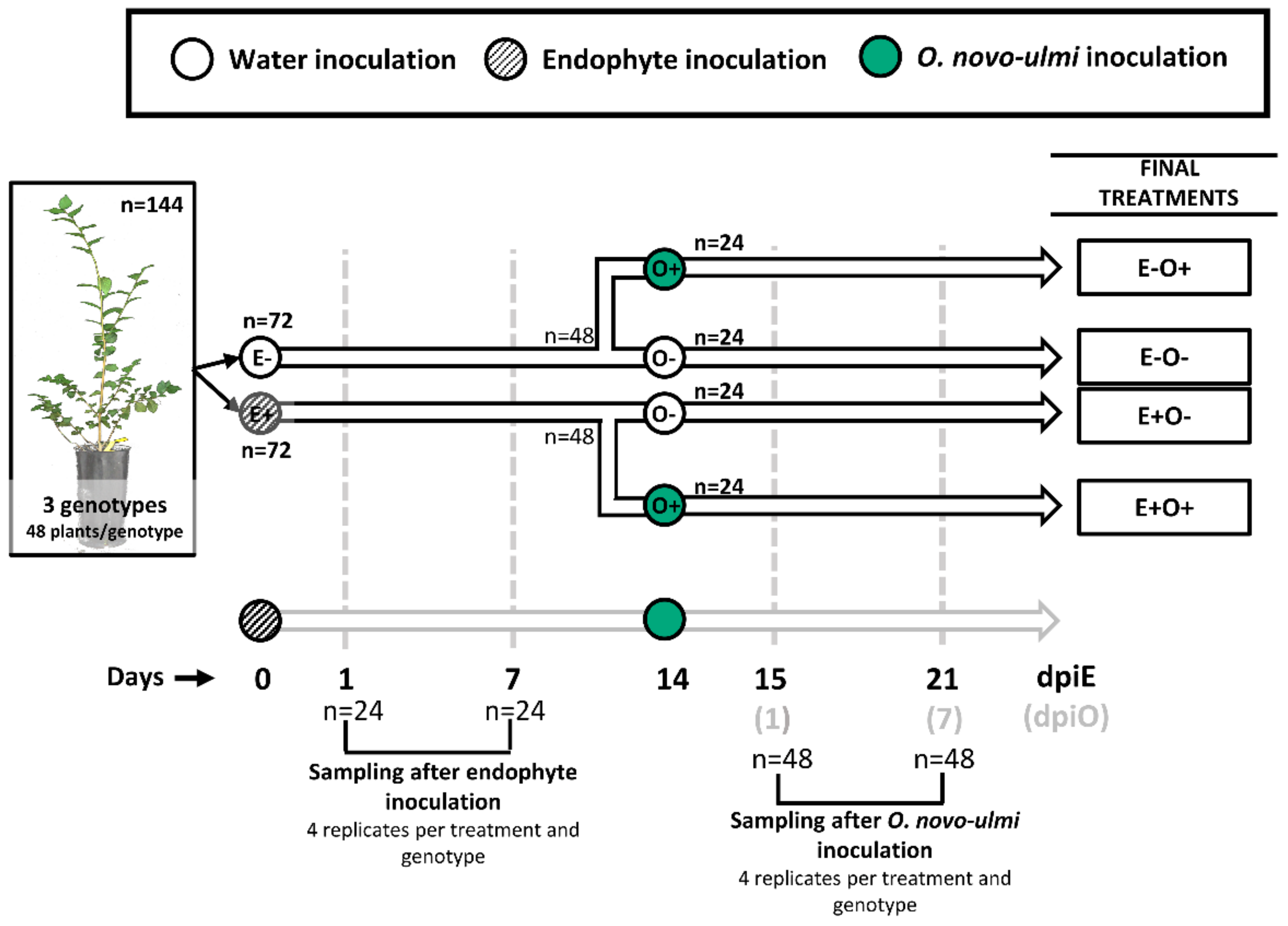
2.2. Experimental Design
2.3. O. novo-ulmi Impact and Spread in Inoculated Plants
2.4. Pathogen and Endophyte Quantification in Plant Tissues
2.5. Gene-Expression Analysis
2.6. Antioxidant Enzyme Activities
2.7. Statistical Analysis
3. Results
3.1. Plant Phenotypic Traits
3.2. Pathogen Quantification in Plant Tissues
3.3. Endophyte Quantification in Plant Tissues
3.4. Gene-Expression Analysis
3.4.1. U. minor Response to O. novo-ulmi Inoculation
3.4.2. U. minor Response to Endophyte Inoculation
3.4.3. U. minor Response to O. novo-ulmi in Plants Previously Inoculated with Endophytes
3.5. Antioxidant Enzyme Activities
4. Discussion
4.1. Different Activation of SA Genes in Response to O. novo-ulmi across U. minor Genotypes
4.2. Fungal Endophytes Activate a Transient SA and JA Response and Act as Priming Stimulators
4.3. A Rapid and Strong Defense Activation against O. novo-ulmi Is Triggered in Endophyte-Primed Plants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boyd, I.L.; Freer-Smith, P.H.; Gilligan, C.A.; Godfray, H.C.J. The Consequence of Tree Pests and Diseases for Ecosystem Services. Science 2013, 342, 1235773. [Google Scholar] [CrossRef]
- Santini, A.; Ghelardini, L.; De Pace, C.; Desprez-Loustau, M.L.; Capretti, P.; Chandelier, A.; Cech, T.; Chira, D.; Diamandis, S.; Gaitniekis, T.; et al. Biogeographical patterns and determinants of invasion by forest pathogens in Europe. New Phytol. 2013, 197, 238–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovalchuk, A.; Keriö, S.; Oghenekaro, A.O.; Jaber, E.; Raffaello, T.; Asiegbu, F.O. Antimicrobial defenses and resistance in forest trees: Challenges and perspectives in a genomic era. Annu. Rev. Phytopathol. 2013, 51, 221–244. [Google Scholar] [CrossRef]
- Tobias, P.A.; Guest, D.I. Tree immunity: Growing old without antibodies. Trends Plant Sci. 2014, 19, 367–370. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caarls, L.; Pieterse, C.M.J.; Van Wees, S.C.M. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 2015, 6, 170. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C.M. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal Modulation of Plant Immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.Y.; Wang, H.; Hu, P.; Hamby, R.; Jin, H. Small RNAs—Big Players in Plant-Microbe Interactions. Cell Host Microbe 2019, 2, 173–182. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Fang, Y.; Chen, L.; Wang, J.; Chen, X. Role of non-coding RNAs in plant immunity. Plant Commun. 2021, 2, 100180. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cañizares, C.; Jorrín, B.; Poole, P.S.; Tkacz, A. Understanding the holobiont: The interdependence of plants and their microbiome. Curr. Opin. Microbiol. 2017, 38, 188–196. [Google Scholar] [CrossRef]
- Friesen, M.L.; Porter, S.S.; Stark, S.C.; von Wettberg, E.J.; Sachs, J.L.; Martinez-Romero, E. Microbially Mediated Plant Functional Traits. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 23–46. [Google Scholar] [CrossRef] [Green Version]
- Ferrol, N.; Azcón-Aguilar, C.; Pérez-Tienda, J. Review: Arbuscular mycorrhizas as key players in sustainable plant phosphorus acquisition: An overview on the mechanisms involved. Plant Sci. 2019, 280, 441–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zanne, A.E.; Abarenkov, K.; Afkhami, M.E.; Aguilar-Trigueros, C.A.; Bates, S.; Bhatnagar, J.M.; Busby, P.E.; Christian, N.; Cornwell, W.K.; Crowther, T.W.; et al. Fungal functional ecology: Bringing a trait-based approach to plant-associated fungi. Biol. Rev. 2020, 95, 409–433. [Google Scholar] [CrossRef]
- Porter, S.S.; Bantay, R.; Friel, C.A.; Garoutte, A.; Gdanetz, K.; Ibarreta, K.; Moore, B.M.; Shetty, P.; Siler, E.; Friesen, M.L. Beneficial microbes ameliorate abiotic and biotic sources of stress on plants. Funct. Ecol. 2020, 34, 2075–2086. [Google Scholar] [CrossRef] [Green Version]
- Latz, M.A.C.; Jensen, B.; Collinge, D.B.; Jørgensen, H.J.L. Endophytic fungi as biocontrol agents: Elucidating mechanisms in disease suppression. Plant Ecol. Divers. 2018, 11, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Berthelot, C.; Leyval, C.; Foulon, J.; Chalot, M.; Blaudez, D. Plant growth promotion, metabolite production and metal tolerance of dark septate endophytes isolated from metal-polluted poplar phytomanagement sites. FEMS Microbiol. Ecol. 2016, 92, fiw144. [Google Scholar] [CrossRef] [Green Version]
- Shoresh, M.; Harman, G.E.; Mastouri, F. Induced Systemic Resistance and Plant Responses to Fungal Biocontrol Agents. Annu. Rev. Phytopathol. 2010, 48, 21–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, J.F.; Torres, M.S. Is plant endophyte-mediated defensive mutualism the result of oxidative stress protection? Physiol. Plant. 2010, 138, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, C.E.; Gundel, P.E.; Helander, M.; Saikkonen, K. Endophytic mediation of reactive oxygen species and antioxidant activity in plants: A review. Fungal Divers. 2012, 54, 1–10. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A.H.M. Induced Systemic Resistance by Beneficial Microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Wees, S.C.; Van der Ent, S.; Pieterse, C.M. Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 2008, 11, 443–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Contreras-Cornejo, H.A.; Macías-Rodríguez, L.; Beltrán-Peña, E.; Herrera-Estrella, A.; López-Bucio, J. Trichoderma-induced plant immunity likely involves both hormonal- and camalexin-dependent mechanisms in Arabidopsis thaliana and confers resistance against necrotrophic fungi Botrytis cinerea. Plant Signal. Behav. 2011, 6, 1554–1563. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Medina, A.; Fernández, I.; Sánchez-Guzmán, M.J.; Jung, S.C.; Pascual, J.A.; Pozo, M.J. Deciphering the hormonal signaling network behind the systemic resistance induced by Trichoderma harzianum in tomato. Front. Plant Sci. 2013, 21, 818–822. [Google Scholar]
- Conrath, U.; Beckers, G.J.M.; Flors, V.; García-Agustín, P.; Jakab, G.; Mauch, F.; Newman, M.-A.; Pieterse, C.M.J.; Poinssot, B.; Pozo, M.J.; et al. Priming: Getting Ready for Battle. Mol. Plant-Microbe Interact. 2006, 19, 1062–1071. [Google Scholar] [CrossRef] [Green Version]
- Conrath, U.; Beckers, G.J.M.; Langenbach, C.J.G.; Jaskiewicz, M.R. Priming for Enhanced Defense. Annu. Rev. Phytopathol. 2015, 53, 97–119. [Google Scholar] [CrossRef]
- Martinez-Medina, A.; Flors, V.; Heil, M.; Mauch-Mani, B.; Pieterse, C.M.J.; Pozo, M.J.; Ton, J.; van Dam, N.M.; Conrath, U. Recognizing Plant Defense Priming. Trends Plant Sci. 2016, 21, 818–822. [Google Scholar] [CrossRef] [Green Version]
- Romeralo, C.; Witzell, J.; Romeralo-Tapia, R.; Botella, L.; Diez, J.J. Antagonistic activity of fungal endophyte filtrates against Gremmeniella abietina infections on Aleppo pine seedlings. Eur. J. Plant Pathol. 2015, 80, 30–39. [Google Scholar] [CrossRef]
- Martínez-Álvarez, P.; Fernández-González, R.A.; Sanz-Ros, A.V.; Pando, V.; Diez, J.J. Two fungal endophytes reduce the severity of pitch canker disease in Pinus radiata seedlings. Biol. Control 2016, 94, 1–10. [Google Scholar] [CrossRef]
- Schlegel, M.; Dubach, V.; von Buol, L.; Sieber, T.N. Effects of endophytic fungi on the ash dieback pathogen. FEMS Microbiol. Ecol. 2016, 92, 1–8. [Google Scholar] [CrossRef]
- Christian, N.; Herre, E.A.; Mejia, L.C.; Clay, K. Exposure to the leaf litter microbiome of healthy adults protects seedlings from pathogen damage. Proc. R. Soc. B Biol. Sci. 2017, 284, 20170641. [Google Scholar] [CrossRef]
- Martínez-Arias, C.; Macaya-Sanz, D.; Witzell, J.; Martín, J.A. Enhancement of Populus alba tolerance to Venturia tremulae upon inoculation with endophytes showing in vitro biocontrol potential. Eur. J. Plant Pathol. 2019, 153, 1031–1042. [Google Scholar] [CrossRef]
- Mejía, L.C.; Herre, E.A.; Sparks, J.P.; Winter, K.; García, M.N.; Van Bael, S.A.; Stitt, J.; Shi, Z.; Zhang, Y.; Guiltinan, M.J.; et al. Pervasive effects of a dominant foliar endophytic fungus on host genetic and phenotypic expression in a tropical tree. Front. Microbiol. 2014, 5, 419. [Google Scholar] [CrossRef]
- Liao, H.-L.; Bonito, G.; Rojas, J.A.; Hameed, K.; Wu, S.; Schadt, C.W.; Labbé, J.; Tuskan, G.A.; Martin, F.; Grigoriev, I.V.; et al. Fungal Endophytes of Populus trichocarpa Alter Host Phenotype, Gene Expression, and Rhizobiome Composition. Mol. Plant-Microbe Interact. 2019, 32, 853–864. [Google Scholar] [CrossRef]
- Santini, A.; Faccoli, M. Dutch elm disease and elm bark beetles: A century of association. iForest-Biogeosci. For. 2015, 8, 126–134. [Google Scholar] [CrossRef] [Green Version]
- Martín, J.A.; Sobrino-Plata, J.; Rodríguez-Calcerrada, J.; Collada, C.; Gil, L. Breeding and scientific advances in the fight against Dutch elm disease: Will they allow the use of elms in forest restoration? New For. 2019, 50, 183–215. [Google Scholar] [CrossRef] [Green Version]
- Martín, J.A.; Domínguez, J.; Solla, A.; Brasier, C.M.; Webber, J.F.; Santini, A.; Martínez-Arias, C.; Bernier, L.; Gil, L. Complexities underlying the breeding and deployment of Dutch elm disease resistant elms. New For. 2021, 1–36. [Google Scholar] [CrossRef]
- Brasier, C.M. Ophiostoma novo-ulmi sp. nov., causative agent of current Dutch elm disease pandemics. Mycopathologia 1991, 115, 151–161. [Google Scholar] [CrossRef]
- Brasier, C.M. Intercontinental Spread and Continuing Evolution of the Dutch Elm Disease Pathogens. In The Elms; Springer: Boston, MA, USA, 2000; pp. 61–72. [Google Scholar]
- Martín, J.A.; Witzell, J.; Blumenstein, K.; Rozpedowska, E.; Helander, M.; Sieber, T.N.; Gil, L. Resistance to Dutch Elm Disease Reduces Presence of Xylem Endophytic Fungi in Elms (Ulmus spp.). PLoS ONE 2013, 8, e56987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín, J.A.; Macaya-Sanz, D.; Witzell, J.; Blumenstein, K.; Gil, L. Strong in vitro antagonism by elm xylem endophytes is not accompanied by temporally stable in planta protection against a vascular pathogen under field conditions. Eur. J. Plant Pathol. 2015, 142, 185–196. [Google Scholar] [CrossRef]
- Blumenstein, K.; Albrectsen, B.R.; Martín, J.A.; Hultberg, M.; Sieber, T.N.; Helander, M.; Witzell, J. Nutritional niche overlap potentiates the use of endophytes in biocontrol of a tree disease. BioControl 2015, 60, 655–667. [Google Scholar] [CrossRef]
- Macaya-Sanz, D.; Witzell, J.; Collada, C.; Gil, L.; Martin, J.A. Structure of core fungal endobiome in Ulmus minor: Patterns within the tree and across genotypes differing in tolerance to Dutch elm disease. bioRxiv 2020. [Google Scholar] [CrossRef]
- Martínez-Arias, C.; Sobrino-Plata, J.; Ormeño-Moncalvillo, S.; Gil, L.; Rodríguez-Calcerrada, J.; Martín, J.A. Endophyte inoculation enhances Ulmus minor resistance to Dutch elm disease. Fungal Ecol. 2021, 50, 101024. [Google Scholar] [CrossRef]
- Martínez-Arias, C.; Sobrino-Plata, J.; Medel, D.; Gil, L.; Martín, J.A.; Rodríguez-Calcerrada, J. Stem endophytes increase root development, photosynthesis, and survival of elm plantlets (Ulmus minor Mill.). J. Plant Physiol. 2021, 261, 153420. [Google Scholar] [CrossRef]
- Perdiguero, P.; Venturas, M.; Cervera, M.T.; Gil, L.; Collada, C. Massive sequencing of Ulmus minor’s transcriptome provides new molecular tools for a genus under the constant threat of Dutch elm disease. Front. Plant Sci. 2015, 6, 541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín, J.; Solla, A.; Venturas, M.; Collada, C.; Domínguez, J.; Miranda, E.; Fuentes, P.; Burón, M.; Iglesias, S.; Gil, L. Seven Ulmus minor clones tolerant to Ophiostoma novo-ulmi registered as forest reproductive material in Spain. iForest-Biogeosci. For. 2015, 8, 172–180. [Google Scholar] [CrossRef] [Green Version]
- Martín, J.A.; Sobrino-Plata, J.; Coira, B.; Medel, D.; Collada, C.; Gil, L. Growth resilience and oxidative burst control as tolerance factors to Ophiostoma novo-ulmi in Ulmus minor. Tree Physiol. 2019, 39, 1512–1524. [Google Scholar] [CrossRef]
- Tchernoff, V. Methods for screening and for the rapid selection of elms for resistance to dutch elm disease. Acta Bot. Neerl. 1965, 14, 409–452. [Google Scholar] [CrossRef]
- Solla, A.; Martín, J.A.; Ouellette, G.B.; Gil, L. Influence of Plant Age on Symptom Development in Ulmus minor Following Inoculation by Ophiostoma novo-ulmi. Plant Dis. 2005, 89, 1035–1040. [Google Scholar] [CrossRef] [Green Version]
- Pita, P.; Rodríguez-Calcerrada, J.; Medel, D.; Gil, L. Further insights into the components of resistance to Ophiostoma novo-ulmi in Ulmus minor: Hydraulic conductance, stomatal sensitivity and bark dehydration. Tree Physiol. 2017, 38, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Duchesne, L.; Jeng, R.S.; Hubbes, M.; Sticklen, M.B. Accumulation of mansonones E and F in seedlings of Ulmus americana in response to inoculation with Ophiostoma ulmi. Trees 1990, 4, 187–190. [Google Scholar] [CrossRef]
- Del Sorbo, G.; Scala, F.; Parrella, G.; Lorito, M.; Comparini, C.; Ruocco, M.; Scala, A. Functional Expression of the Gene cu, Encoding the Phytotoxic Hydrophobin Cerato-ulmin, Enables Ophiostoma quercus, a Nonpathogen on Elm, to Cause Symptoms of Dutch Elm Disease. Mol. Plant-Microbe Interact. 2000, 13, 43–53. [Google Scholar] [CrossRef] [Green Version]
- Perdiguero, P.; Sobrino-Plata, J.; Venturas, M.; Martín, J.A.; Gil, L.; Collada, C.; Cervera, M.T.; Gil, L.; Collada, C. Gene expression trade-offs between defence and growth in English elm induced by Ophiostoma novo-ulmi. Plant Cell Environ. 2018, 41, 198–214. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Ding, P.; Wang, D.; Cheng, Y.T.; He, J.; Gao, M.; Xu, F.; Li, Y.; Zhu, Z.; et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18220–18225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogt, T. Phenylpropanoid Biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shim, J.S.; Jung, C.; Lee, S.; Min, K.; Lee, Y.-W.; Choi, Y.; Lee, J.S.; Song, J.T.; Kim, J.-K.; Choi, Y.D. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 2013, 73, 483–495. [Google Scholar] [CrossRef]
- Kim, D.S.; Hwang, B.K. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [CrossRef] [Green Version]
- Stintzi, A.; Browse, J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 10625–10630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breeze, E. Master MYCs: MYC2, the Jasmonate Signaling “Master Switch”. Plant Cell 2019, 31, 9–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Xue, L.; Chintamanani, S.; Germain, H.; Lin, H.; Cui, H.; Cai, R.; Zuo, J.; Tang, X.; Li, X.; et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 Repress SALICYLIC ACID INDUCTION DEFICIENT2 Expression to Negatively Regulate Plant Innate Immunity in Arabidopsis. Plant Cell 2009, 21, 2527–2540. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.C.; Hong, J.K.; Kim, Y.J.; Hwang, B.K. Pepper gene encoding thionin is differentially induced by pathogens, ethylene and methyl jasmonate. Physiol. Mol. Plant Pathol. 2000, 56, 207–216. [Google Scholar] [CrossRef]
- Huang, H.; Gao, H.; Liu, B.; Fan, M.; Wang, J.; Wang, C.; Tian, H.; Wang, L.; Xie, C.; Wu, D.; et al. BHLH13 Regulates Jasmonate-Mediated Defense Responses and Growth. Evol. Bioinform. 2018, 14, 117693431879026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, S.; Wang, X.; Chen, D.; Yang, X.; Wang, M.; Turrà, D.; Di Pietro, A.; Zhang, W. The Secreted Peptide PIP1 Amplifies Immunity through Receptor-Like Kinase 7. PLoS Pathog. 2014, 10, e1004331. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Sobrino-Plata, J.; Ortega-Villasante, C.; Laura Flores-Cáceres, M.; Escobar, C.; Del Campo, F.F.; Hernández, L.E. Differential alterations of antioxidant defenses as bioindicators of mercury and cadmium toxicity in alfalfa. Chemosphere 2009, 77, 946–954. [Google Scholar] [CrossRef]
- Martín, J.A.; Solla, A.; García-Vallejo, M.C.; Gil, L. Chemical changes in Ulmus minor xylem tissue after salicylic acid or carvacrol treatments are associated with enhanced resistance to Ophiostoma novo-ulmi. Phytochemistry 2012, 83, 104–109. [Google Scholar] [CrossRef]
- Sherif, S.M.; Shukla, M.R.; Murch, S.J.; Bernier, L.; Saxena, P.K. Simultaneous induction of jasmonic acid and disease-responsive genes signifies tolerance of American elm to Dutch elm disease. Sci. Rep. 2016, 6, 21934. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Zheng, Z.; Huang, J.; Lai, Z.; Fan, B. Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 2009, 4, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tsuda, K.; Truman, W.; Sato, M.; Nguyen, L.V.; Katagiri, F.; Glazebrook, J. CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J. 2011, 67, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Dong, Q.; Yu, D. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 2012, 185–186, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.A.; Solla, A.; Esteban, L.G.; de Palacios, P.; Gil, L. Bordered pit and ray morphology involvement in elm resistance to Ophiostoma novo-ulmi. Can. J. For. Res. 2009, 39, 420–429. [Google Scholar] [CrossRef]
- Martín, J.A.; Solla, A.; Ruiz-Villar, M.; Gil, L. Vessel length and conductivity of Ulmus branches: Ontogenetic changes and relation to resistance to Dutch elm disease. Trees-Struct. Funct. 2013, 27, 1239–1248. [Google Scholar] [CrossRef]
- Venturas, M.; López, R.; Martín, J.A.; Gascó, A.; Gil, L. Heritability of Ulmus minor resistance to Dutch elm disease and its relationship to vessel size, but not to xylem vulnerability to drought. Plant Pathol. 2014, 63, 500–509. [Google Scholar] [CrossRef]
- Valverde, P.; Trapero, C.; Arquero, O.; Serrano, N.; Barranco, D.; Muñoz Díez, C.; López-Escudero, F.J. Highly infested soils undermine the use of resistant olive rootstocks as a control method of verticillium wilt. Plant Pathol. 2021, 70, 144–153. [Google Scholar] [CrossRef]
- Petkovsek, M.; Slatnar, A.; Stampar, F.; Veberic, R. Phenolic compounds in apple leaves after infection with apple scab. Biol. Plant 2011, 55, 725. [Google Scholar] [CrossRef]
- Wasternack, C.; Strnad, M. Jasmonate signaling in plant stress responses and development—Active and inactive compounds. New Biotechnol. 2016, 33, 604–613. [Google Scholar] [CrossRef]
- Korolev, N.; Rav David, D.; Elad, Y. The role of phytohormones in basal resistance and Trichoderma-induced systemic resistance to Botrytis cinerea in Arabidopsis thaliana. BioControl 2008, 53, 667–683. [Google Scholar] [CrossRef]
- Ryu, C.; Hu, C.; Reddy, M.S.; Kloepper, J.W. Different signaling pathways of induced resistance by rhizobacteria in Arabidopsis thaliana against two pathovars of Pseudomonas syringae. New Phytol. 2003, 160, 413–420. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; van Wees, S.C.M.; van Pelt, J.A.; Knoester, M.; Laan, R.; Gerrits, H.; Weisbeek, P.J.; van Loon, L.C. A Novel Signaling Pathway Controlling Induced Systemic Resistance in Arabidopsis. Plant Cell 1998, 10, 1571–1580. [Google Scholar] [CrossRef] [Green Version]
- Stein, E.; Molitor, A.; Kogel, K.-H.; Waller, F. Systemic Resistance in Arabidopsis Conferred by the Mycorrhizal Fungus Piriformospora indica Requires Jasmonic Acid Signaling and the Cytoplasmic Function of NPR1. Plant Cell Physiol. 2008, 49, 1747–1751. [Google Scholar] [CrossRef]
- Mathys, J.; De Cremer, K.; Timmermans, P.; Van Kerckhove, S.; Lievens, B.; Vanhaecke, M.; Cammue, B.P.A.; De Coninck, B. Genome-Wide Characterization of ISR Induced in Arabidopsis thaliana by Trichoderma hamatum T382 Against Botrytis cinerea Infection. Front. Plant Sci. 2012, 3, 108. [Google Scholar] [CrossRef] [Green Version]
- Niu, D.-D.; Liu, H.-X.; Jiang, C.-H.; Wang, Y.-P.; Wang, Q.-Y.; Jin, H.-L.; Guo, J.-H. The Plant Growth–Promoting Rhizobacterium Bacillus cereus AR156 Induces Systemic Resistance in Arabidopsis thaliana by Simultaneously Activating Salicylate- and Jasmonate/Ethylene-Dependent Signaling Pathways. Mol. Plant-Microbe Interact. 2011, 24, 533–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamioudis, C.; Pieterse, C.M.J. Modulation of Host Immunity by Beneficial Microbes. Mol. Plant-Microbe Interact. 2012, 25, 139–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, A.; Christensen, M.J.; Takemoto, D.; Park, P.; Scott, B. Reactive Oxygen Species Play a Role in Regulating a Fungus–Perennial Ryegrass Mutualistic Interaction. Plant Cell 2006, 18, 1052–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, A.; Takemoto, D.; Hyon, G.-S.; Park, P.; Scott, B. NoxA activation by the small GTPase RacA is required to maintain a mutualistic symbiotic association between Epichloë festucae and perennial ryegrass. Mol. Microbiol. 2008, 68, 1165–1178. [Google Scholar] [CrossRef]
- Plett, J.M.; Martin, F.M. Know your enemy, embrace your friend: Using omics to understand how plants respond differently to pathogenic and mutualistic microorganisms. Plant J. 2018, 93, 729–746. [Google Scholar] [CrossRef] [Green Version]
- Plett, J.M.; Martin, F. Blurred boundaries: Lifestyle lessons from ectomycorrhizal fungal genomes. Trends Genet. 2011, 27, 14–22. [Google Scholar] [CrossRef]
- Plett, J.M.; Daguerre, Y.; Wittulsky, S.; Vayssieres, A.; Deveau, A.; Melton, S.J.; Kohler, A.; Morrell-Falvey, J.L.; Brun, A.; Veneault-Fourrey, C.; et al. Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl. Acad. Sci. USA 2014, 111, 8299–8304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kloppholz, S.; Kuhn, H.; Requena, N. A Secreted Fungal Effector of Glomus intraradices Promotes Symbiotic Biotrophy. Curr. Biol. 2011, 21, 1204–1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimenez-Ibanez, S.; Chini, A.; Solano, R. How Microbes Twist Jasmonate Signaling around Their Little Fingers. Plants 2016, 5, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, O.P.; Sharma, P.; Gupta, R.K.; Sharma, I. Current status on role of miRNAs during plant-fungus interaction. Physiol. Mol. Plant Pathol. 2014, 85, 1–7. [Google Scholar] [CrossRef]
- Kandel, E.R.; Dudai, Y.; Mayford, M.R. The Molecular and Systems Biology of Memory. Cell 2014, 157, 163–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conrath, U. Molecular aspects of defence priming. Trends Plant Sci. 2011, 16, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Pastor, V.; Luna, E.; Mauch-Mani, B.; Ton, J.; Flors, V. Primed plants do not forget. Environ. Exp. Bot. 2013, 94, 46–56. [Google Scholar] [CrossRef]
- Amaral, J.; Ribeyre, Z.; Vigneaud, J.; Sow, M.D.; Fichot, R.; Messier, C.; Pinto, G.; Nolet, P.; Maury, S. Advances and Promises of Epigenetics for Forest Trees. Forests 2020, 11, 976. [Google Scholar] [CrossRef]
- Alizadeh, H.; Behboudi, K.; Ahmadzadeh, M.; Javan-Nikkhah, M.; Zamioudis, C.; Pieterse, C.M.J.; Bakker, P.A.H.M. Induced systemic resistance in cucumber and Arabidopsis thaliana by the combination of Trichoderma harzianum Tr6 and Pseudomonas sp. Ps14. Biol. Control 2013, 65, 14–23. [Google Scholar] [CrossRef]
- Sarma, B.K.; Yadav, S.K.; Singh, S.; Singh, H.B. Microbial consortium-mediated plant defense against phytopathogens: Readdressing for enhancing efficacy. Soil Biol. Biochem. 2015, 87, 25–33. [Google Scholar] [CrossRef]
- van Wees, S.C.M.; de Swart, E.A.M.; van Pelt, J.A.; van Loon, L.C.; Pieterse, C.M.J. Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2000, 97, 8711–8716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halim, V.A.; Altmann, S.; Ellinger, D.; Eschen-Lippold, L.; Miersch, O.; Scheel, D.; Rosahl, S. PAMP-induced defense responses in potato require both salicylic acid and jasmonic acid. Plant J. 2009, 57, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Derksen, H.; Rampitsch, C.; Daayf, F. Signaling cross-talk in plant disease resistance. Plant Sci. 2013, 207, 79–87. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Arias, C.; Sobrino-Plata, J.; Gil, L.; Rodríguez-Calcerrada, J.; Martín, J.A. Priming of Plant Defenses against Ophiostoma novo-ulmi by Elm (Ulmus minor Mill.) Fungal Endophytes. J. Fungi 2021, 7, 687. https://doi.org/10.3390/jof7090687
Martínez-Arias C, Sobrino-Plata J, Gil L, Rodríguez-Calcerrada J, Martín JA. Priming of Plant Defenses against Ophiostoma novo-ulmi by Elm (Ulmus minor Mill.) Fungal Endophytes. Journal of Fungi. 2021; 7(9):687. https://doi.org/10.3390/jof7090687
Chicago/Turabian StyleMartínez-Arias, Clara, Juan Sobrino-Plata, Luis Gil, Jesús Rodríguez-Calcerrada, and Juan Antonio Martín. 2021. "Priming of Plant Defenses against Ophiostoma novo-ulmi by Elm (Ulmus minor Mill.) Fungal Endophytes" Journal of Fungi 7, no. 9: 687. https://doi.org/10.3390/jof7090687





