Comparative Genomics and Gene Pool Analysis Reveal the Decrease of Genome Diversity and Gene Number in Rice Blast Fungi by Stable Adaption with Rice
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Sequences
2.2. Single-Nucleotide Polymorphism Discovery and Population Genetics
2.3. Gene Prediction, Annotation, and Secreted Proteins Prediction
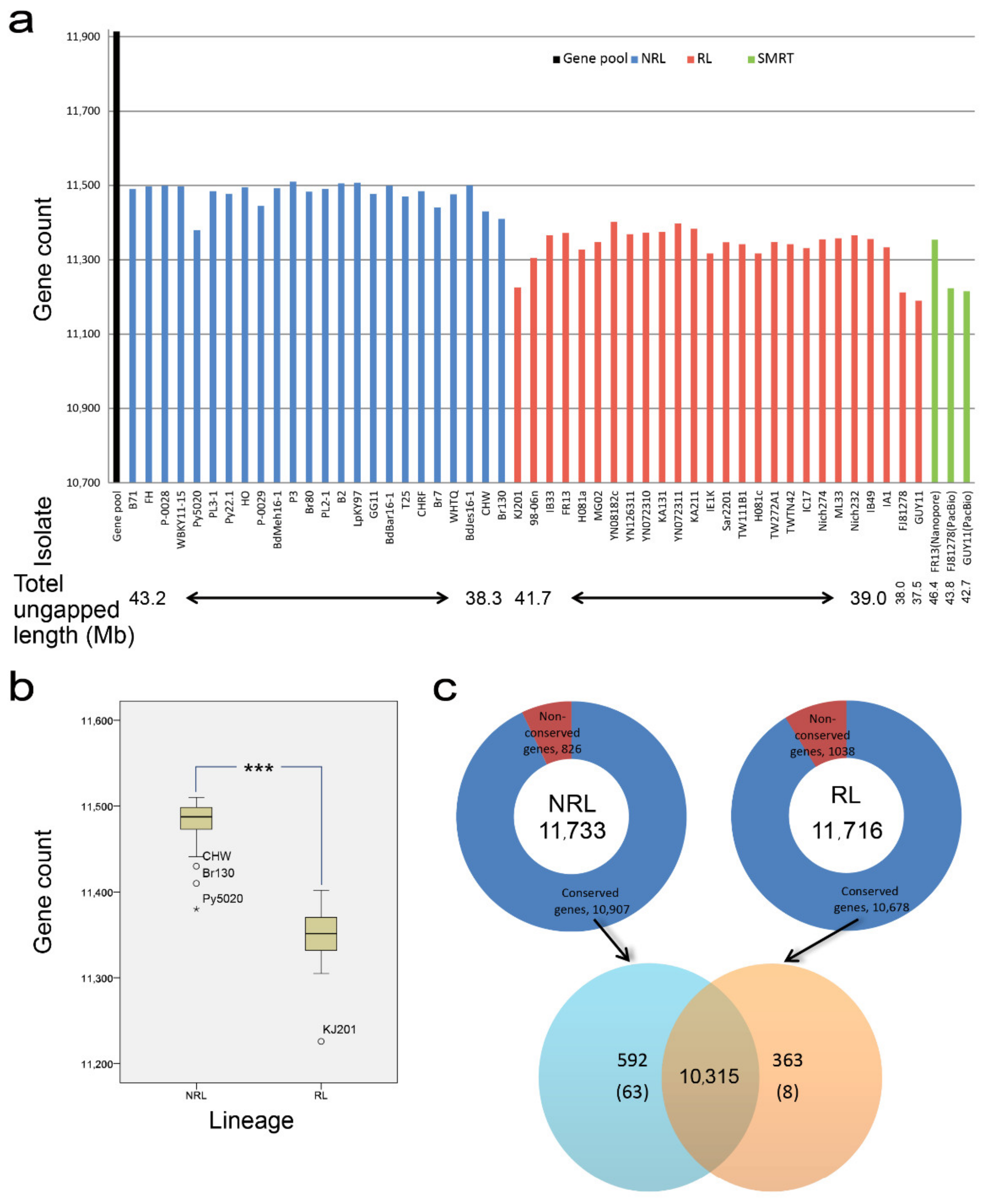
2.4. Gene Pool Analysis
2.5. Indentification of Expressed Genes
2.6. Statistical Analysis
2.7. Data Availability
3. Results
3.1. Population Genomic Analysis of Rice- and Non-Rice-Infecting Isolates
3.2. SNPs with Host Selection
3.3. Highly Polymorphic Distribution of 11 Cloned AVR Genes in Two Groups
3.4. Gene Pool Analysis between RL and NRL
3.5. Genes Associated with Host Adaptation of M. oryzae Identified by PAV Calling
4. Discussion
4.1. Comparative Genomes Analyses Present the Genome Regions Involving Host Selection between RL and NRL
4.2. Gene Pool Analysis Deepen the PAV Genes Promoting Host Specification of Rice Infecting Isolates
4.3. Functions of the Genes Related to Host Adaptation
4.4. The Gain or Loss of AVR and Effector Affects the Host Differentiation of M. oryzae
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kato, H.; Yamamoto, M.; Yamaguchi-Ozaki, T.; Kadouchi, H.; Iwamoto, Y.; Nakayashiki, H.; Tosa, Y.; Mori, S.M. Pathogenicity, mating ability and DNA restriction fragment length polymorphisms of Pyricularia populations isolated from Gramineae, Bambusideae and Zingiberaceae plants. J. Gen. Plant Pathol. 2000, 66, 30–47. [Google Scholar] [CrossRef]
- Couch, B.C.; Kohn, L.M. A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia 2002, 94, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Couch, B.C.; Fudal, I.; Lebrun, M.H.; Tharreau, D.; Valent, B.; van Kim, P.; Notteghem, J.L.; Kohn, L.M. Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics 2005, 170, 613–630. [Google Scholar] [CrossRef]
- Murakami, J.; Tosa, Y.; Kataoka, T.; Tomita, R.; Kawasaki, J.; Chuma, I.; Sesumi, Y.; Kusaba, M.; Nakayashiki, H.; Mayama, S. Analysis of host species specificity of Magnaporthe grisea toward wheat using a genetic cross between isolates from wheat and foxtail millet. Phytopathology 2000, 90, 1060–1067. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.S.; Tosa, Y.; Takabayashi, N.; Nakagawa, S.; Tomita, R.; Don, L.D.; Kusaba, M.; Nakayashiki, H.; Mayama, S. Characterization of an Avena isolate of Magnaporthe grisea and identification of a locus conditioning its specificity on oat. Can. J. Bot. 2002, 80, 1088–1095. [Google Scholar] [CrossRef]
- Tosa, Y.; Hirata, K.; Tamba, H.; Nakagawa, S.; Chuma, I.; Isobe, C.; Osue, J.; Urashima, A.S.; Don, L.D.; Kusaba, M.; et al. Genetic constitution and pathogenicity of Lolium isolates of Magnaporthe oryzae in comparison with host species-specific pathotypes of the blast fungus. Phytopathology 2004, 94, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.H. Rice Diseases; Kew Commonwealth Mycological Institute: Kew, UK, 1985. [Google Scholar]
- Maciel, J.L.; Ceresini, P.C.; Castroagudin, V.L.; Zala, M.; Kema, G.H.; McDonald, B.A. Population structure and pathotype diversity of the wheat blast pathogen Magnaporthe oryzae 25 years after its emergence in Brazil. Phytopathology 2014, 104, 95–107. [Google Scholar] [CrossRef]
- Cruz, C.D.; Valent, B. Wheat blast disease: Danger on the move. Trop. Plant Pathol. 2017, 42, 210–222. [Google Scholar] [CrossRef]
- Farman, M.; Peterson, G.; Chen, L.; Starnes, J.; Valent, B.; Bachi, P.; Murdock, L.; Hershman, D.; Pedley, K.; Fernandes, J.M.; et al. The Lolium pathotype of Magnaporthe oryzae recovered from a single blasted wheat plant in the United States. Plant Dis. 2017, 101, 684–692. [Google Scholar] [CrossRef]
- Malaker, P.K.; Barma, N.; Tewari, T.P.; Collis, W.J.; Valent, B. First report of wheat blast caused by Magnaporthe oryzae pathotype triticum in Bangladesh. Plant Dis. 2016, 100, 2330. [Google Scholar] [CrossRef]
- Islam, M.T.; Croll, D.; Gladieux, P.; Soanes, D.M.; Persoons, A.; Bhattacharjee, P.; Hossain, M.S.; Gupta, D.R.; Rahman, M.M.; Mahboob, M.G.; et al. Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol. 2016, 14, 84. [Google Scholar] [CrossRef] [PubMed]
- Gladieux, P.; Condon, A.B.; Ravel, B.S.; Soanes, A.D.; Maciel, C.J.L.N. Gene flow between divergent cereal- and grass-specific lineages of the rice blast fungus Magnaporthe oryzae. mBio 2018, 9, e01219-17. [Google Scholar] [CrossRef] [PubMed]
- Saleh, D.; Milazzo, J.; Adreit, H.; Fournier, E.; Tharreau, D. South-East Asia is the center of origin, diversity and dispersion of the rice blast fungus, Magnaporthe oryzae. New Phytol. 2014, 201, 1440–1456. [Google Scholar] [CrossRef] [PubMed]
- Gladieux, P.; Ravel, S.; Rieux, A.; Cros-Arteil, S.; Adreit, H.; Milazzo, J.; Thierry, M.; Fournier, E.; Terauchi, R.; Tharreau, D. Coexistence of multiple endemic and pandemic lineages of the rice blast pathogen. mBio 2018, 9, e01806–e01817. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Norvienyeku, J.; Chen, M.; Bao, J.; Lin, L.; Chen, L.; Lin, Y.; Wu, X.; Cai, Z.; Zhang, Q.; et al. Directional selection from host plants is a major force driving host specificity in Magnaporthe Species. Sci. Rep. 2016, 6, 25591. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M.; Haigh, J. The hitch-hiking effect of a favourable gene. Genet. Res. 1974, 23, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Liberles, D. Detecting and understanding natural selection. In Codon Evolution: Mechanisms and Models; Cannarozzi, G.M., Schneider, A., Eds.; Oxford University Press: Oxford, UK, 2012. [Google Scholar]
- Kojima, K.-I. Is there a constant fitness value for a given genotype? NO! Evolution 1971, 25, 281–285. [Google Scholar] [CrossRef]
- Rech, G.E.; Sanz-Martin, J.M.; Anisimova, M.; Sukno, S.A.; Thon, M.R. Natural selection on coding and noncoding DNA sequences is associated with virulence genes in a plant pathogenic fungus. Genome Biol. Evol. 2014, 6, 2368–2379. [Google Scholar] [CrossRef]
- Ye, W.; Wang, Y.; Tyler, B.M.; Wang, Y. Comparative genomic analysis among four representative isolates of Phytophthora sojae reveals genes under evolutionary selection. Front Microbiol. 2016, 7, 1547. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Dong, F.; Stavrinides, J.; Guttman, D.S. Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2007, 2, e209. [Google Scholar] [CrossRef]
- Charlesworth, B.; Nordborg, M.; Charlesworth, D. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet. Res. 1997, 70, 155–174. [Google Scholar] [CrossRef]
- Majewski, J.; Cohan, F.M. Adapt globally, act locally: The effect of selective sweeps on bacterial sequence diversity. Genetics 1999, 152, 1459–1474. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M.; Wiehe, T. Genetic hitch-hiking in a subdivided population. Genet. Res. 1998, 71, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, R. Molecular signatures of natural selection. Annu. Rev. Genet. 2005, 39, 197–218. [Google Scholar] [CrossRef] [PubMed]
- Mohd-Assaad, N.; McDonald, B.A.; Croll, D. Genome–wide detection of genes under positive selection in worldwide populations of the barley scald pathogen. Genome Biol. Evol. 2018, 10, 1315–1332. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Kurata, N.; Wei, X.; Wang, Z.X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W.; et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef]
- Pieck, M.L.; Ruck, A.; Farman, M.L.; Peterson, G.L.; Stack, J.P.; Valent, B.; Pedley, K.F. Genomics-based marker discovery and diagnostic assay development for wheat blast. Plant Dis. 2017, 101, 103–109. [Google Scholar] [CrossRef]
- Park, S.; Kim, S.; Koh, H.; Lee, Y. A blast lesion mimic mutant of rice. In Advances in Rice Blast; Tharreau, D., Lebrun, M.H., Talbot, N.J., Notteghem, J.L., Eds.; Kluwer Academic Press: Dordrecht, The Netherlands, 2000. [Google Scholar]
- Dong, Y.; Li, Y.; Zhao, M.; Jing, M.; Liu, X.; Liu, M.; Guo, X.; Zhang, X.; Chen, Y.; Liu, Y.; et al. Global genome and transcriptome analyses of Magnaporthe oryzae epidemic isolate 98-06 uncover novel effectors and pathogenicity-related genes, revealing gene gain and lose dynamics in genome evolution. PLoS Pathog. 2015, 11, e1004801. [Google Scholar] [CrossRef] [PubMed]
- Chiapello, H.; Mallet, L.; Guerin, C.; Aguileta, G.; Amselem, J.; Kroj, T.; Ortega-Abboud, E.; Lebrun, M.H.; Henrissat, B.; Gendrault, A.; et al. Deciphering genome content and evolutionary relationships of isolates from the fungus Magnaporthe oryzae attacking different host plants. Genome Biol. Evol. 2015, 7, 2896–2912. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Q.; Meng, F.Z.; Zhang, M.M.; Yin, L.F.; Yin, W.X.; Lin, Y.; Hsiang, T.; Peng, Y.L.; Wang, Z.H.; Luo, C.X. A putative Zn2Cys6 transcription factor is associated with isoprothiolane resistance in Magnaporthe oryzae. Front. Microbiol. 2018, 9, 2608. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Chen, M.; Lin, L.; Han, Y.; Bao, J.; Tang, W.; Lin, L.; Lin, Y.; Somai, R.; Lu, L.; et al. Population genomic analysis of the rice blast fungus reveals specific events associated with expansion of three main clades. ISME J. 2018, 12, 1867–1878. [Google Scholar] [CrossRef] [PubMed]
- Shirke, M.D.; Mahesh, H.B.; Gowda, M. Genome-wide comparison of Magnaporthe species reveals a host-specific pattern of secretory proteins and transposable elements. PLoS ONE 2016, 11, e0162458. [Google Scholar] [CrossRef]
- Chen, C.; Lian, B.; Hu, J.; Zhai, H.; Wang, X.; Venu, R.C.; Liu, E.; Wang, Z.; Chen, M.; Wang, B.; et al. Genome comparison of two Magnaporthe oryzae field isolates reveals genome variations and potential virulence effectors. BMC Genom. 2013, 14, 887. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Singh, P.K.; Gupta, D.K.; Mahato, A.K.; Sarkar, C.; Rathour, R.; Singh, N.K.; Sharma, T.R. Analysis of Magnaporthe oryzae genome reveals a fungal effector, which is able to induce resistance response in transgenic rice line containing resistance gene, Pi54. Front. Plant Sci. 2016, 7, 1140. [Google Scholar] [CrossRef]
- Zhu, K.; Bao, J.; Zhang, L.; Yang, X.; Li, Y.; Zhu, M.; Lin, Q.; Zhao, Z.; Zhou, B.; Lu, G. Comparative analysis of the genome of the field isolate V86010 of the rice blast fungus Magnaporthe oryzae from Philippines. J. Integr. Agric. 2017, 16, 2222–2230. [Google Scholar] [CrossRef]
- Gowda, M.; Shirke, M.D.; Mahesh, H.B.; Chandarana, P.; Rajamani, A.; Chattoo, B.B. Genome analysis of rice-blast fungus Magnaporthe oryzae field isolates from southern India. Genome Data 2015, 5, 284–291. [Google Scholar] [CrossRef][Green Version]
- Bao, J.; Chen, M.; Zhong, Z.; Tang, W.; Lin, L.; Zhang, X.; Jiang, H.; Zhang, D.; Miao, C.; Tang, H.; et al. PacBio sequencing reveals transposable elements as a key contributor to genomic plasticity and virulence variation in Magnaporthe oryzae. Mol. Plant 2017, 10, 1465–1468. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [PubMed]
- Vilella, A.J.; Blanco-Garcia, A.; Hutter, S.; Rozas, J. VariScan: Analysis of evolutionary patterns from large-scale DNA sequence polymorphism data. Bioinformatics 2005, 21, 2791–2793. [Google Scholar] [CrossRef] [PubMed]
- Rousset, F. genepop’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 2008, 8, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Keller, O.; Kollmar, M.; Stanke, M.; Waack, S. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 2011, 27, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Nielsen, H.; von Heijne, G.; Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Yang, J.; Li, Z.; Hu, S.; Yao, N.; Dean, R.A.; Zhao, W.; Shen, M.; Zhang, H.; Li, C.; et al. Comparative analysis of the genomes of two field isolates of the rice blast fungus Magnaporthe oryzae. PLoS Genet. 2012, 8, e1002869. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Saunders, D.G.; Mitsuoka, C.; Natsume, S.; Kosugi, S.; Saitoh, H.; Inoue, Y.; Chuma, I.; Tosa, Y.; Cano, L.M.; et al. Host specialization of the blast fungus Magnaporthe oryzae is associated with dynamic gain and loss of genes linked to transposable elements. BMC Genom. 2016, 17, 370. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Populations: Experimental Results and Evolutionary Deductions; University of Chicago Press: Chicago, IL, USA, 1977; Volume 3. [Google Scholar]
- Kang, S.; Sweigard, J.A.; Valent, B. The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact 1995, 8, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Sweigard, J.A.; Carroll, A.M.; Kang, S.; Farrall, L.; Chumley, F.G.; Valent, B. Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell 1995, 7, 1221–1233. [Google Scholar] [CrossRef] [PubMed]
- Orbach, M.J.; Farrall, L.; Sweigard, J.A.; Chumley, F.G.; Valent, B. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 2000, 12, 2019–2032. [Google Scholar] [CrossRef] [PubMed]
- Bohnert, H.U.; Fudal, I.; Dioh, W.; Tharreau, D.; Notteghem, J.L.; Lebrun, M.H. A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 2004, 16, 2499–2513. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Saitoh, H.; Fujisawa, S.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Tosa, Y.; Chuma, I.; Takano, Y.; Win, J.; et al. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 2009, 21, 1573–1591. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, B.; Wu, J.; Lu, G.; Hu, Y.; Zhang, X.; Zhang, Z.; Zhao, Q.; Feng, Q.; Zhang, H.; et al. The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant Microbe Interact 2009, 22, 411–420. [Google Scholar] [CrossRef]
- Zheng, Y.; Zheng, W.; Lin, F.; Zhang, Y.; Yi, Y.; Wang, B.; Lu, G.; Wang, Z.; Wu, W. AVR1-CO39 is a predominant locus governing the broad avirulence of Magnaporthe oryzae 2539 on cultivated rice (Oryza sativa L.). Mol. Plant Microbe Interact. 2011, 24, 13–17. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, L.; Wu, W.; He, L.; Yang, X.; Pan, Q. Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Sci. Rep. 2015, 5, 11642. [Google Scholar] [CrossRef]
- Wu, J.; Kou, Y.; Bao, J.; Li, Y.; Tang, M.; Zhu, X.; Ponaya, A.; Xiao, G.; Li, J.; Li, C.; et al. Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9-mediated blast resistance in rice. New Phytol. 2015, 206, 1463–1475. [Google Scholar] [CrossRef] [PubMed]
- Farman, M.L.; Eto, Y.; Nakao, T.; Tosa, Y.; Nakayashiki, H.; Mayama, S.; Leong, S.A. Analysis of the structure of the AVR1-CO39 avirulence locus in virulent rice-infecting isolates of Magnaporthe grisea. Mol. Plant Microbe Interact. 2002, 15, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Vy, T.T.P.; Yoshida, K.; Asano, H.; Mitsuoka, C.; Asuke, S.; Anh, V.L.; Cumagun, C.J.R.; Chuma, I.; Terauchi, R.; et al. Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science 2017, 357, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Okagaki, L.H.; Sailsbery, J.K.; Eyre, A.W.; Dean, R.A. Comparative genome analysis and genome evolution of members of the magnaporthaceae family of fungi. BMC Genom. 2016, 17, 135. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.W.; Branco, S.; Gao, C.; Hann-Soden, C.; Montoya, L.; Sylvain, I.; Gladieux, P. Sources of fungal genetic variation and associating it with phenotypic diversity. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Ellison, C.E.; Hall, C.; Kowbel, D.; Welch, J.; Brem, R.B.; Glass, N.L.; Taylor, J.W. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc. Natl. Acad. Sci. USA 2011, 108, 2831–2836. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Gonda, I.; Sun, H.; Ma, Q.; Bao, K.; Tieman, D.M.; Burzynski-Chang, E.A.; Fish, T.L.; Stromberg, K.A.; Sacks, G.L.; et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 2019, 51, 1044–1051. [Google Scholar] [CrossRef]
- Zhao, J.; Sauvage, C.; Zhao, J.; Bitton, F.; Bauchet, G.; Liu, D.; Huang, S.; Tieman, D.M.; Klee, H.J.; Causse, M. Meta-analysis of genome-wide association studies provides insights into genetic control of tomato flavor. Nat. Commun. 2019, 10, 1534. [Google Scholar] [CrossRef]
- Ceresini, P.C.; Castroagudin, V.L.; Rodrigues, F.A.; Rios, J.A.; Eduardo Aucique-Perez, C.; Moreira, S.I.; Alves, E.; Croll, D.; Maciel, J.L.N. Wheat blast: Past, present, and future. Annu. Rev. Phytopathol. 2018, 56, 427–456. [Google Scholar] [CrossRef]
- Peter, J.; De Chiara, M.; Friedrich, A.; Yue, J.X.; Pflieger, D.; Bergstrom, A.; Sigwalt, A.; Barre, B.; Freel, K.; Llored, A.; et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 2018, 556, 339–344. [Google Scholar] [CrossRef]
- Dunn, B.; Richter, C.; Kvitek, D.J.; Pugh, T.; Sherlock, G. Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res. 2012, 22, 908–924. [Google Scholar] [CrossRef]
- Sharma, R.; Mishra, B.; Runge, F.; Thines, M. Gene loss rather than gene gain is associated with a host jump from monocots to dicots in the Smut Fungus Melanopsichium pennsylvanicum. Genome Biol. Evol. 2014, 6, 2034–2049. [Google Scholar] [CrossRef]
- Gabriel, G.; Klingel, K.; Otte, A.; Thiele, S.; Hudjetz, B.; Arman-Kalcek, G.; Sauter, M.; Shmidt, T.; Rother, F.; Baumgarte, S.; et al. Differential use of importin-α isoforms governs cell tropism and host adaptation of influenza virus. Nat. Commun. 2011, 2, 156. [Google Scholar] [CrossRef]
- Yang, X.; Ding, F.; Zhang, L.; Sheng, Y.; Zheng, X.; Wang, Y. The importin α subunit PsIMPA1 mediates the oxidative stress response and is required for the pathogenicity of Phytophthora sojae. Fungal Genet. Biol. 2015, 82, 108–115. [Google Scholar] [CrossRef]
- Ghassemi, S.; Lichius, A.; Bidard, F.; Lemoine, S.; Rossignol, M.N.; Herold, S.; Seidl-Seiboth, V.; Seiboth, B.; Espeso, E.A.; Margeot, A.; et al. The ss-importin KAP8 (Pse1/Kap121) is required for nuclear import of the cellulase transcriptional regulator XYR1, asexual sporulation and stress resistance in Trichoderma reesei. Mol. Microbiol. 2015, 96, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Van Vu, B.; Itoh, K.; Nguyen, Q.B.; Tosa, Y.; Nakayashiki, H. Cellulases belonging to glycoside hydrolase families 6 and 7 contribute to the virulence of Magnaporthe oryzae. Mol. Plant Microbe Interact. 2012, 25, 1135–1141. [Google Scholar] [CrossRef]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Mentlak, T.A.; Kombrink, A.; Shinya, T.; Ryder, L.S.; Otomo, I.; Saitoh, H.; Terauchi, R.; Nishizawa, Y.; Shibuya, N.; Thomma, B.P.; et al. Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 2012, 24, 322–335. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Yu, Y.; Huang, J.; Meng, F.; Pang, J.; Zhao, Q.; Islam, M.A.; Xu, N.; Tian, Y.; Liu, J. Binding of the Magnaporthe oryzae chitinase MoChia1 by a rice tetratricopeptide repeat protein allows free chitin to trigger immune responses. Plant Cell 2019, 31, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Song, L.; Peng, C.; Liu, X.; Liu, L.; Zhang, Y.; Wang, W.; Zhou, J.; Wang, S.; Ebbole, D.; et al. A Magnaporthe chitinase interacts with a rice jacalin-related lectin to promote host colonization. Plant Physiol. 2019, 179, 1416–1430. [Google Scholar] [CrossRef]
- Jashni, M.K.; Mehrabi, R.; Collemare, J.; Mesarich, C.H.; de Wit, P.J. The battle in the apoplast: Further insights into the roles of proteases and their inhibitors in plant-pathogen interactions. Front Plant Sci. 2015, 6, 584. [Google Scholar] [CrossRef]
- Schirrmann, M.K.; Zoller, S.; Croll, D.; Stukenbrock, E.H.; Leuchtmann, A.; Fior, S. Genomewide signatures of selection in Epichloë reveal candidate genes for host specialization. Mol. Ecol. 2018, 27, 3070–3086. [Google Scholar] [CrossRef]
- Gao, X.; Yin, C.; Liu, X.; Peng, J.; Chen, D.; He, D.; Shi, W.; Zhao, W.; Yang, J.; Peng, Y.L. A glycine-rich protein MoGrp1 functions as a novel splicing factor to regulate fungal virulence and growth in Magnaporthe oryzae. Phytopathol. Res. 2019, 1, 2. [Google Scholar] [CrossRef]
- Paoletti, M. Vegetative incompatibility in fungi: From recognition to cell death, whatever does the trick. Fungal Biol. Rev. 2016, 30, 152–162. [Google Scholar] [CrossRef]
- Kumar, S. Caspase function in programmed cell death. Cell Death Differ. 2007, 14, 32–43. [Google Scholar] [CrossRef]
- Zhang, N.; Cai, G.; Price, D.C.; Crouch, J.A.; Gladieux, P.; Hillman, B.; Khang, C.H.; LeBrun, M.H.; Lee, Y.H.; Luo, J.; et al. Genome wide analysis of the transition to pathogenic lifestyles in Magnaporthales fungi. Sci. Rep. 2018, 8, 5862. [Google Scholar] [CrossRef]
- Kanzaki, H.; Yoshida, K.; Saitoh, H.; Fujisaki, K.; Hirabuchi, A.; Alaux, L.; Fournier, E.; Tharreau, D.; Terauchi, R. Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J. 2012, 72, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Huang, H.; Meusnier, I.; Adreit, H.; Ducasse, A.; Bonnot, F.; Pan, L.; He, X.; Kroj, T.; Fournier, E.; et al. Pathogen effectors and plant immunity determine specialization of the blast fungus to rice subspecies. eLife 2016, 5, e19377. [Google Scholar] [CrossRef]
- Wang, B.-h.; Ebbole, D.J.; Wang, Z.-h. The arms race between Magnaporthe oryzae and rice: Diversity and interaction of Avr and R genes. J. Integr. Agric. 2017, 16, 2746–2760. [Google Scholar] [CrossRef]
- Jones, J.D.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Li, J.; Zhang, X.; Zhang, Q.; Huang, J.; Chen, J.Q.; Hartl, D.L.; Tian, D. Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc. Natl. Acad. Sci. USA 2013, 110, 18572–18577. [Google Scholar] [CrossRef] [PubMed]





| Lineage | Intronic Regions | Intergenic Regions |
| RL (n = 24) | 0.0002665 | 0.0005304 |
| NRL (n = 24) | 0.0021783 | 0.0035670 |
| TL (n = 14) | 0.0021997 | 0.0033026 |
| LL (n = 10) | 0.0016510 | 0.0029569 |
| EL (n = 7) | 0.0032822 | 0.0050494 |
| Isolates | Features | Number of Genes | Hosts (Rice or Non-Rice) | |||||
| Spearman a | Spearman b | Mann-Whitney U Test c | ||||||
| ρ | Sig | ρ | Sig | U | Z | Sig | ||
| 48 isolates of RL and NRL | Total length | 0.626 ** | 0.000 | 0.767 ** | 0.000 | 33.000 | −5.258 | 0 ** |
| Total ungapped length | 0.693 ** | 0.000 | 0.752 ** | 0.000 | 38.000 | −5.155 | 0 ** | |
| Scaffold (contig) number | −0.079 | 0.595 | −0.244 | 0.095 | 207.000 | −1.670 | 0.095 | |
| N50 | −0.081 | 0.585 | −0.066 | 0.655 | 266.000 | −0.454 | 0.650 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Q.; Wang, Y.; Liu, L.-N.; Shi, K.; Li, C.-Y. Comparative Genomics and Gene Pool Analysis Reveal the Decrease of Genome Diversity and Gene Number in Rice Blast Fungi by Stable Adaption with Rice. J. Fungi 2022, 8, 5. https://doi.org/10.3390/jof8010005
Wu Q, Wang Y, Liu L-N, Shi K, Li C-Y. Comparative Genomics and Gene Pool Analysis Reveal the Decrease of Genome Diversity and Gene Number in Rice Blast Fungi by Stable Adaption with Rice. Journal of Fungi. 2022; 8(1):5. https://doi.org/10.3390/jof8010005
Chicago/Turabian StyleWu, Qi, Yi Wang, Li-Na Liu, Kai Shi, and Cheng-Yun Li. 2022. "Comparative Genomics and Gene Pool Analysis Reveal the Decrease of Genome Diversity and Gene Number in Rice Blast Fungi by Stable Adaption with Rice" Journal of Fungi 8, no. 1: 5. https://doi.org/10.3390/jof8010005
APA StyleWu, Q., Wang, Y., Liu, L.-N., Shi, K., & Li, C.-Y. (2022). Comparative Genomics and Gene Pool Analysis Reveal the Decrease of Genome Diversity and Gene Number in Rice Blast Fungi by Stable Adaption with Rice. Journal of Fungi, 8(1), 5. https://doi.org/10.3390/jof8010005






