Strategies for Efficient Expression of Heterologous Monosaccharide Transporters in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media and Growth Conditions
2.2. Molecular Biology Techniques
2.3. Transport Assays
2.4. Analytical Methods
2.5. Prediction of Lysine Residues with Ubiquitinylation Potential
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goldemberg, J. Ethanol for a sustainable energy future. Science 2007, 315, 808–810. [Google Scholar] [CrossRef]
- Jaiswal, D.; de Souza, A.; Larsen, S.; Lebauer, D.S.; Miguez, F.E.; Sparovek, G.; Bollero, G.; Buckeridge, M.S.; Long, S.P. Brazilian sugarcane ethanol as an expandable green alternative to crude oil use. Nat. Clim. Chang. 2017, 7, 788–792. [Google Scholar] [CrossRef]
- Liu, Y.; Cruz-Morales, P.; Zargar, A.; Belcher, M.S.; Pang, B.; Englund, E.; Dan, Q.; Yin, K.; Keasling, J.D. Biofuels for a sustainable future. Cell 2021, 184, 1636–1647. [Google Scholar] [CrossRef]
- Jacobus, A.P.; Gross, J.; Evans, J.H.; Ceccato-Antonini, S.R.; Gombert, A.K. Saccharomyces cerevisiae strains used industrially for bioethanol production. Essays Biochem. 2021, 65, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Trichez, D.; Bergmann, J.C.; Calsing, L.C.G.; Cançado, L.J. How many bioethanol generations can we have? In Ethanol as a Green Alternative Fuel: Insight and Perspectives, 1st ed.; Treichel, H., Alves, S.L., Jr., Fongaro, G., Müller, C., Eds.; Nova Science Publishers Inc.: Hauppauge, NY, USA, 2019; pp. 21–55. [Google Scholar]
- dos Santos, L.V.; de Barros Grassi, M.C.; Gallardo, J.C.M.; Pirolla, R.A.S.; Calderón, L.L.; de Carvalho-Netto, O.V.; Parreiras, L.S.; Camargo, E.L.O.; Drezza, A.L.; Missawa, S.K.; et al. Second-generation ethanol: The need is becoming a reality. Ind. Biotechnol. 2016, 12, 40–57. [Google Scholar] [CrossRef]
- Gao, M.; Ploessl, D.; Shao, Z. Enhancing the co-utilization of biomass-derived mixed sugars by yeasts. Front. Microbiol. 2019, 9, 3264. [Google Scholar] [CrossRef]
- Kim, J.; Hwang, S.; Lee, S.M. Metabolic engineering for the utilization of carbohydrate portions of lignocellulosic biomass. Metab. Eng. 2021, in press. [Google Scholar] [CrossRef]
- Gírio, F.M.; Fonseca, C.; Carvalheiro, F.; Duarte, L.C.; Marques, S.; Bogel-Łukasik, R. Hemicelluloses for fuel ethanol: A review. Bioresour. Technol. 2010, 101, 4775–4800. [Google Scholar] [CrossRef]
- da Silva, A.S.; Espinheira, R.P.; Teixeira, R.S.S.; de Souza, M.F.; Ferreira-Leitão, V.; Bon, E.P.S. Constraints and advances in high-solids enzymatic hydrolysis of lignocellulosic biomass: A critical review. Biotechnol. Biofuels 2020, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Stambuk, B.U. Yeasts: The leading figures on bioethanol production. In Ethanol as a Green Alternative Fuel: Insight and Perspectives, 1st ed.; Treichel, H., Alves, S.L., Jr., Fongaro, G., Müller, C., Eds.; Nova Science Publishers Inc.: Hauppauge, NY, USA, 2019; pp. 57–91. [Google Scholar]
- Patiño, M.A.; Ortiz, J.P.; Velásquez, M.; Stambuk, B.U. d-Xylose consumption by nonrecombinant Saccharomyces cerevisiae: A review. Yeast 2019, 36, 541–556. [Google Scholar] [CrossRef]
- Ye, S.; Kim, J.W.; Kim, S.R. Metabolic engineering for improved fermentation of L-arabinose. J. Microbiol. Biotechnol. 2019, 29, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Arora, A. Tracking strategic developments for conferring xylose utilization/fermentation by Saccharomyces cerevisiae. Ann. Microbiol. 2020, 70, 50. [Google Scholar] [CrossRef]
- Jin, Y.S.; Lee, T.H.; Choi, Y.D.; Ryu, Y.W.; Seo, J.H. Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae containing genes for xylose reductase and xylitol dehydrogenase from Pichia stipitis. J. Microb. Biotechnol. 2000, 10, 564–567. [Google Scholar]
- Cadete, R.M.; De Las Heras, A.M.; Sandström, A.G.; Ferreira, C.; Gírio, F.; Gorwa-Grauslund, M.F.; Rosa, C.A.; Fonseca, C. Exploring xylose metabolism in Spathaspora species: XYL1.2 from Spathaspora passalidarum as the key for efficient anaerobic xylose fermentation in metabolic engineered Saccharomyces cerevisiae. Biotechnol. Biofuels 2016, 9, 167. [Google Scholar] [CrossRef]
- Mouro, A.; dos Santos, A.A.; Agnolo, D.D.; Gubert, G.F.; Bon, E.P.S.; Rosa, C.A.; Fonseca, C.; Stambuk, B.U. Combining xylose reductase from Spathaspora arborariae with xylitol dehydrogenase from Spathaspora passalidarum to promote xylose consumption and fermentation into xylitol by Saccharomyces cerevisiae. Fermentation 2020, 6, 72. [Google Scholar] [CrossRef]
- Brat, D.; Boles, E.; Wiedemann, B. Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2009, 75, 2304–2311. [Google Scholar] [CrossRef]
- Cunha, J.T.; Soares, P.O.; Romaní, A.; Thevelein, J.M.; Domingues, L. Xylose fermentation efficiency of industrial Saccharomyces cerevisiae yeast with separate or combined xylose reductase/xylitol dehydrogenase and xylose isomerase pathways. Biotechnol. Biofuels 2019, 12, 20. [Google Scholar] [CrossRef]
- Jeong, D.; Oh, E.J.; Ko, J.K.; Nam, J.-O.; Park, H.S.; Jin, Y.-S.; Lee, E.J.; Kim, S.R. Metabolic engineering considerations for the heterologous expression of xylose-catabolic pathways in Saccharomyces cerevisiae. PLoS ONE 2020, 15, e0236294. [Google Scholar] [CrossRef]
- Gonçalves, D.L.; Matsushika, A.; de Sales, B.B.; Goshima, T.; Bon, E.P.S.; Stambuk, B.U. Xylose and xylose/glucose co-fermentation by recombinant Saccharomyces cerevisiae strains expressing individual hexose transporters. Enzyme Microb. Technol. 2014, 63, 13–20. [Google Scholar] [CrossRef]
- Knoshaug, E.P.; Franden, M.A.; Stambuk, B.U.; Zhang, M.; Singh, A. Utilization and transport of l-arabinose by non-Saccharomyces yeasts. Cellulose 2009, 16, 729–741. [Google Scholar] [CrossRef]
- Fonseca, C.; Olofsson, K.; Ferreira, C.; Runquist, D.; Fonseca, L.L.; Hahn-Hägerdal, B.; Lidén, G. The glucose/xylose facilitator Gxf1 from Candida intermedia expressed in a xylose-fermenting industrial strain of Saccharomyces cerevisiae increases xylose uptake in SSCF of wheat straw. Enzyme Microb. Technol. 2011, 48, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Young, E.; Poucher, A.; Comer, A.; Bailey, A.; Alper, H. Functional survey for heterologous sugar transport proteins, using Saccharomyces cerevisiae as a host. Appl. Environ. Microbiol. 2011, 77, 3311–3319. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Liu, Z.L.; Ma, M.; Slininger, P.J. New genotypes of industrial yeast Saccharomyces cerevisiae engineered with YXI and heterologous xilose transporters improve xylose utilization and ethanol production. Biocatal. Agricult. Biotechnol. 2013, 2, 247–254. [Google Scholar] [CrossRef]
- Knoshaug, E.P.; Vidgren, V.; Magalhães, F.; Jarvis, E.E.; Franden, M.A.; Zhang, M.; Singh, A. Novel transporters from Kluyveromyces marxianus and Pichia guilliermondii expressed in Saccharomyces cerevisiae enable growth on l-arabinose and d-xylose. Yeast 2015, 32, 615–628. [Google Scholar] [CrossRef] [PubMed]
- de Sales, B.B.; Scheid, B.; Gonçalves, D.L.; Knychala, M.M.; Matsushika, A.; Bon, E.P.; Stambuk, B.U. Cloning novel sugar transporters from Scheffersomyces (Pichia) stipitis allowing d-xylose fermentation by recombinant Saccharomyces cerevisiae. Biotechnol. Lett. 2015, 37, 1973–1982. [Google Scholar] [CrossRef]
- Jojima, T.; Omumasaba, C.A.; Inui, M.; Yukawa, H. Sugar transporters in efficient utilization of mixed sugar substrates: Current knowledge and outlook. Appl. Microbiol. Biotechnol. 2010, 85, 471–480. [Google Scholar] [CrossRef]
- Hara, K.Y.; Kobayashi, J.; Yamada, R.; Sasaki, D.; Kuriya, Y.; Hirono-Hara, Y.; Ishii, J.; Araki, M.; Kondo, A. Transporter engineering in biomass utilization by yeast. FEMS Yeast Res. 2017, 17, fox061. [Google Scholar] [CrossRef]
- Sharma, N.K.; Behera, S.; Arora, R.; Kumar, S.; Sani, R.K. Xylose transport in yeast for lignocellulosic ethanol production: Current status. J. Biosci. Bioeng. 2018, 125, 259–267. [Google Scholar] [CrossRef]
- Nijland, J.G.; Driessen, A.J.M. Engineering of pentose transport in Saccharomyces cerevisiae for biotechnological applications. Front. Bioeng. Biotechnol. 2020, 7, 464. [Google Scholar] [CrossRef]
- Wieczorke, R.; Krampe, S.; Weierstall, T.; Freidel, K.; Hollenberg, C.P.; Boles, E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999, 464, 123–128. [Google Scholar] [CrossRef]
- Boles, E.; Oreb, M. A growth-based screening system for hexose transporters in yeast. Methods Mol. Biol. 2018, 1713, 123–135. [Google Scholar] [CrossRef]
- Hamacher, T.; Becker, J.; Gárdonyi, M.; Hahn-Hägerdal, B.; Boles, E. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 2002, 148, 2783–2788. [Google Scholar] [CrossRef] [PubMed]
- Solis-Escalante, D.; van den Broek, M.; Kuijpers, N.G.; Pronk, J.T.; Boles, E.; Daran, J.M.; Daran-Lapujade, P. The genome sequence of the popular hexose-transport-deficient Saccharomyces cerevisiae strain EBY.VW4000 reveals LoxP/Cre-induced translocations and gene loss. FEMS Yeast Res. 2015, 15, fou004. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wijsman, M.; Swiat, M.A.; Marques, W.L.; Hettinga, J.K.; van den Broek, M.; Torre Cortés, P.; Mans, R.; Pronk, J.T.; Daran, J.M.; Daran-Lapujade, P. A toolkit for rapid CRISPR-SpCas9 assisted construction of hexose-transport-deficient Saccharomyces cerevisiae strains. FEMS Yeast Res. 2019, 19, foy107. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Suh, S.O.; Marshall, C.J.; Blackwell, M. Morphological and ecological similarities: Wood-boring beetles associated with novel xylose-fermenting yeasts, Spathaspora passalidarum gen. sp. nov. and Candida jeffriesii sp. nov. Mycol. Res. 2006, 110, 1232–1241. [Google Scholar] [CrossRef]
- Wohlbach, D.J.; Kuo, A.; Sato, T.K.; Potts, K.M.; Salamov, A.A.; Labutti, K.M.; Sun, H.; Clum, A.; Pangilinan, J.L.; Lindquist, E.A.; et al. Comparative genomics of xylose-fermenting fungi for enhanced biofuel production. Proc. Natl. Acad. Sci. USA 2011, 108, 13212–13217. [Google Scholar] [CrossRef]
- Cadete, R.M.; Santos, R.O.; Melo, M.A.; Mouro, A.; Gonçalves, D.L.; Stambuk, B.U.; Rosa, C.A. Spathaspora arborariae sp. nov., a d-xylose fermenting yeast species isolated from rotting wood in Brazil. FEMS Yeast Res. 2009, 9, 1338–1342. [Google Scholar] [CrossRef] [PubMed]
- Lobo, F.P.; Gonçalves, D.L.; Alves, S.L., Jr.; Gerber, A.L.; Vasconcelos, A.T.R.; Basso, L.C.; Franco, G.R.; Soares, M.A.; Cadete, R.M.; Rosa, C.A.; et al. Draft genome sequence of the d-xylose-fermenting yeast Spathaspora arborariae UFMG-HM19.1AT. Genome Announc. 2014, 2, e01163-13. [Google Scholar] [CrossRef]
- Kruckeberg, A.L.; Ye, L.; Berden, J.A.; van Dam, K. Functional expression, quantification and cellular localization of the Hxt2 hexose transporter of Saccharomyces cerevisiae tagged with the green fluorescent protein. Biochem. J. 1999, 339, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Matsushika, A.; Inoue, H.; Murakami, K.; Takimura, O.; Sawayama, S. Bioethanol production performance of five recombinant strains of laboratory and industrial xylose-fermenting Saccharomyces cerevisiae. Bioresour. Technol. 2009, 100, 2392–2398. [Google Scholar] [CrossRef]
- Kasahara, M.; Maeda, M. Contribution to substrate recognition of two aromatic amino acid residues in putative transmembrane segment 10 of the yeast sugar transporters Gal2 and Hxt2. J. Biol. Chem. 1998, 273, 29106–29112. [Google Scholar] [CrossRef]
- Kasahara, T.; Maeda, M.; Ishiguro, M.; Kasahara, M. Identification by comprehensive chimeric analysis of a key residue responsible for high affinity glucose transport by yeast HXT2. J. Biol. Chem. 2007, 282, 13146–13150. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, T.; Kasahara, M. Identification of a key residue determining substrate affinity in the yeast glucose transporter Hxt7: A two-dimensional comprehensive study. J. Biol. Chem. 2010, 285, 26263–26268. [Google Scholar] [CrossRef] [PubMed]
- Batista, A.S.; Miletti, L.C.; Stambuk, B.U. Sucrose fermentation by Saccharomyces cerevisiae lacking hexose transport. J. Mol. Microbiol. Biotechnol. 2004, 8, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Alves-Jr, S.L.; Thevelein, J.M.; Stambuk, B.U. Extracellular maltotriose hydrolysis by Saccharomyces cerevisiae cells lacking the AGT1 permease. Lett. Appl. Microbiol. 2018, 67, 377–383. [Google Scholar] [CrossRef]
- Sen, A.; Acosta-Sampson, L.; Alvaro, C.G.; Ahn, J.S.; Cate, J.H.; Thorner, J. Internalization of heterologous sugar transporters by endogenous α-arrestins in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2016, 82, 7074–7085. [Google Scholar] [CrossRef]
- Petracek, M.E.; Longtine, M.S. PCR-based engineering of yeast genome. Methods Enzymol. 2002, 350, 445–469. [Google Scholar]
- Kang, Y.S.; Kane, J.; Kurjan, K.; Stadel, J.M.; Tipper, D.J. Effects of expression of mammalian G alpha and hybrid mammalian-yeast G alpha proteins on the yeast pheromone response signal transduction pathway. Mol. Cell. Biol. 1990, 10, 2582–2590. [Google Scholar]
- Mumberg, D.; Müller, R.; Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 1995, 156, 119–122. [Google Scholar] [CrossRef]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Short Protocols in Molecular Biology, 3rd ed.; John Wiley & Sons: New York, NY, USA, 1995. [Google Scholar]
- Gietz, D.; St Jean, A.; Woods, R.A.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1425. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov (accessed on 21 October 2016).
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Pirovano, W.; Feenstra, K.A.; Heringa, J. PRALINETM: A strategy for improved multiple alignment of transmembrane proteins. Bioinformatics 2008, 24, 492–497. [Google Scholar] [CrossRef]
- PRALINE multiple sequence alignment. Available online: https://www.ibi.vu.nl/programs/pralinewww (accessed on 22 February 2017).
- Radivojac, P.; Vacic, V.; Haynes, C.; Cocklin, R.R.; Mohan, A.; Heyen, J.W.; Goebl, M.G.; Iakoucheva, L.M. Identification, analysis, and prediction of protein ubiquitination sites. Proteins 2010, 78, 365–380. [Google Scholar] [CrossRef] [PubMed]
- UbPred. Available online: http://www.ubpred.org (accessed on 22 February 2017).
- Horák, J. The role of ubiquitin in down-regulation and intracellular sorting of membrane proteins: Insights from yeast. Biochim. Biophys. Acta. 2003, 1614, 139–155. [Google Scholar] [CrossRef]
- Kahlhofer, J.; Leon, S.; Teis, D.; Schmidt, O. The α-arrestin family of ubiquitin ligase adaptors links metabolism with selective endocytosis. Biol. Cell. 2021, 113, 183–219. [Google Scholar] [CrossRef]
- Barata-Antunes, C.; Alves, R.; Talaia, G.; Casal, M.; Gerós, H.; Mans, R.; Paiva, S. Endocytosis of nutrient transporters in fungi: The ART of connecting signaling and trafficking. Comput. Struct. Biotechnol. J. 2021, 19, 1713–1737. [Google Scholar] [CrossRef]
- Nikko, E.; Pelham, H.R. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 2009, 10, 1856–1867. [Google Scholar] [CrossRef]
- Llopis-Torregrosa, V.; Ferri-Blázquez, A.; Adam-Artigues, A.; Deffontaines, E.; van Heusden, G.P.; Yenush, L. Regulation of the yeast Hxt6 hexose transporter by the Rod1 α-arrestin, the Snf1 protein kinase, and the Bmh2 14-3-3 protein. J. Biol. Chem. 2016, 291, 14973–14985. [Google Scholar] [CrossRef] [PubMed]
- Savocco, J.; Nootens, S.; Afokpa, W.; Bausart, M.; Chen, X.; Villers, J.; Renard, H.F.; Prévost, M.; Wattiez, R.; Morsomme, P. Yeast α-arrestin Art2 is the key regulator of ubiquitylation-dependent endocytosis of plasma membrane vitamin B1 transporters. PLoS Biol. 2019, 17, e3000512. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, A.F.; McCartney, R.R.; Chandrashekarappa, D.G.; Zhang, B.B.; Thorner, J.; Schmidt, M.C. 2-Deoxyglucose impairs Saccharomyces cerevisiae growth by stimulating Snf1-regulated and α-arrestin-mediated trafficking of hexose transporters 1 and 3. Mol. Cell. Biol. 2015, 35, 939–955. [Google Scholar] [CrossRef]
- Tamayo Rojas, S.A.; Schmidl, S.; Boles, E.; Oreb, M. Glucose-induced internalization of the S. cerevisiae galactose permease Gal2 is dependent on phosphorylation and ubiquitination of its aminoterminal cytoplasmic tail. FEMS Yeast Res. 2021, 21, foab019. [Google Scholar] [CrossRef]
- Roy, A.; Kim, Y.B.; Cho, K.H.; Kim, J.H. Glucose starvation-induced turnover of the yeast glucose transporter Hxt1. Biochim. Biophys. Acta 2014, 1840, 2878–2885. [Google Scholar] [CrossRef]
- Hovsepian, J.; Defenouillère, Q.; Albanèse, V.; Váchová, L.; Garcia, C.; Palková, Z.; Léon, S. Multilevel regulation of an α-arrestin by glucose depletion controls hexose transporter endocytosis. J. Cell. Biol. 2017, 216, 1811–1831. [Google Scholar] [CrossRef] [PubMed]
- Lazar, Z.; Neuvéglise, C.; Rossignol, T.; Devillers, H.; Morin, N.; Robak, M.; Nicaud, J.M.; Crutz-Le Coq, A.M. Characterization of hexose transporters in Yarrowia lipolytica reveals new groups of Sugar Porters involved in yeast growth. Fungal Genet. Biol. 2017, 100, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Liu, D.; Lu, X.; Zong, H.; Song, J.; Zhuge, B. Identification and characterization from Candida glycerinogenes of hexose transporters having high efficiency at high glucose concentrations. Appl. Microbiol. Biotechnol. 2018, 102, 5557–5567. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Shen, Y.; Gu, L.; Wang, Z.; Su, N.; Niu, K.; Guo, W.; Hou, S.; Bao, X.; Tian, C.; et al. Identification and characterization of an efficient d-xylose transporter in Saccharomyces cerevisiae. J. Agric. Food Chem. 2020, 68, 2702–2710. [Google Scholar] [CrossRef]
- Pereira, I.O.; dos Santos, A.A.; Gonçalves, D.L.; Purificação, M.; Guimarães, N.C.; Tramontina, R.; Coutouné, N.; Zanella, E.; Matsushika, A.; Stambuk, B.U.; et al. Comparison of Spathaspora passalidarum and recombinant Saccharomyces cerevisiae for integration of first- and second-generation ethanol production. FEMS Yeast Res. 2021, 21, foab048. [Google Scholar] [CrossRef]
- Nijland, J.G.; Vos, E.; Shin, H.Y.; de Waal, P.P.; Klaassen, P.; Driessen, A.J. Improving pentose fermentation by preventing ubiquitination of hexose transporters in Saccharomyces cerevisiae. Biotechnol. Biofuels 2016, 9, 158. [Google Scholar] [CrossRef]
- Leandro, M.J.; Gonçalves, P.; Spencer-Martins, I. Two glucose/xylose transporter genes from the yeast Candida intermedia: First molecular characterization of a yeast xylose-H+ symporter. Biochem. J. 2006, 395, 543–549. [Google Scholar] [CrossRef]
- Li, H.; Schmitz, O.; Alper, H.S. Enabling glucose/xylose co-transport in yeast through the directed evolution of a sugar transporter. Appl. Microbiol. Biotechnol. 2016, 100, 10215–10223. [Google Scholar] [CrossRef]



| Strains, Plasmids and Primers | Relevant Features, Genotype or Sequence | Source |
|---|---|---|
| Yeast strains: | ||
| Sp. arborariae UFMG-HM19.1AT | Isolated from rotting wood in Minas Gerais, Brazil | [39] |
| Sp. passalidarum UFMG-CM-Y474 | Isolated from rotting wood in Roraima, Brazil | [17] |
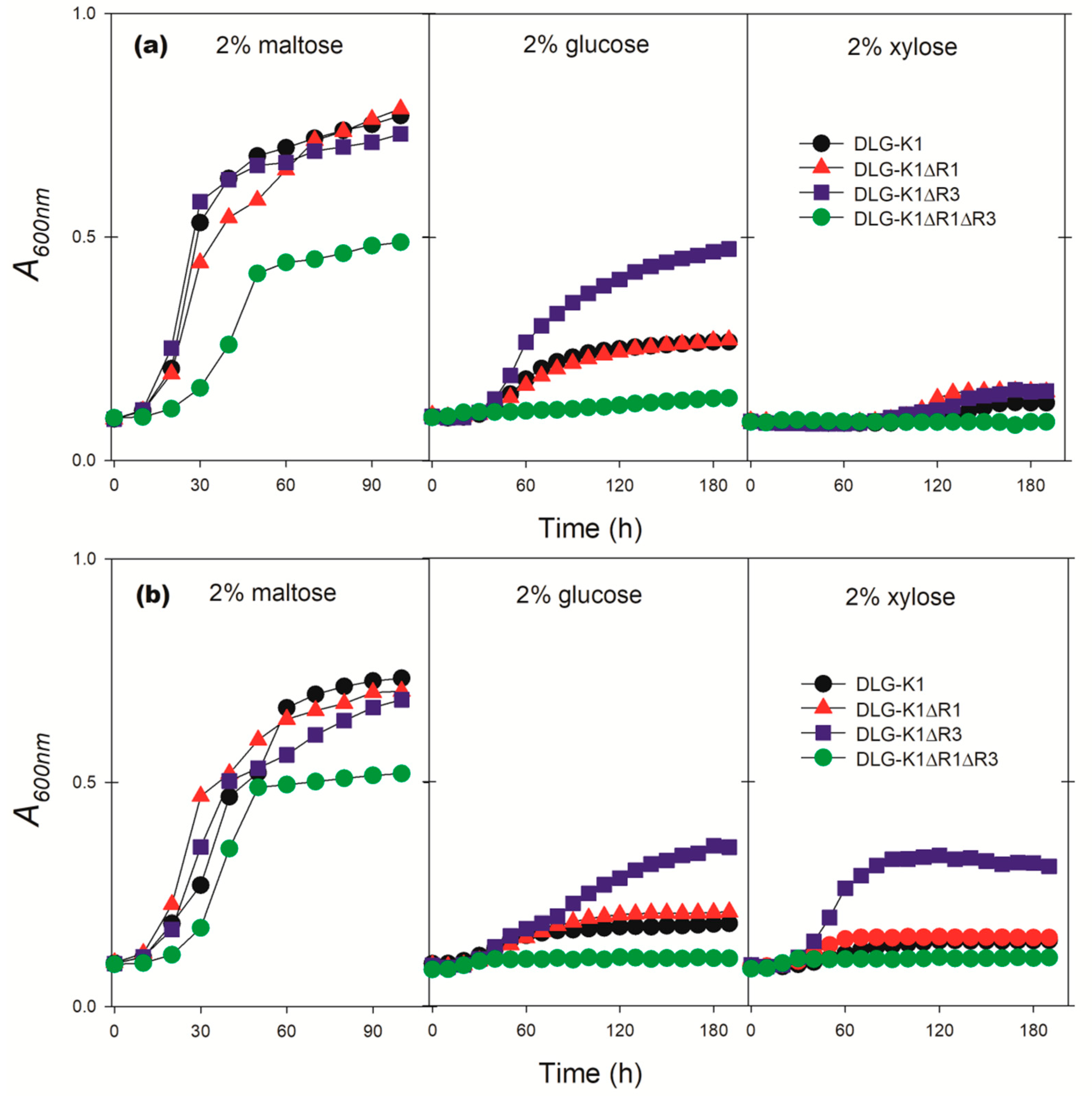
| S. cerevisiae DLG-K1 | MATa hxt1Δ::HIS3::Δhxt4 hxt2Δ::HIS3 hxt5::LEU2 hxt7::HIS3 hxt3Δ::LEU2::hxt6 gal2Δ::DR 1 ura3-52 his3-11,15 leu2-3,112 MAL2 SUC2AUR1::pAUR-XKXDHXR | [21] |
| S. cerevisiae DLG-K1∆R1 | Isogenic to DLG-K1, but rod1∆::LoxP-KanMX6-LoxP | This work |
| S. cerevisiae DLG-K1∆R3 | Isogenic to DLG-K1, but rog3∆::LoxP-BleR-LoxP | This work |
| S. cerevisiae DLG-K1∆R1∆R3 | Isogenic to DLG-K1, but rog3∆::LoxP-BleR-LoxP rod1∆::LoxP-KanMX6-LoxP | This work |
| Plasmids: | ||
| pUG6 | LoxP-PTEF-KanMX6-TTEF-LoxP | [49] |
| pUG66 | LoxP-PTEF-BleR-TTEF-LoxP | [49] |
| pPGK | 2 µ URA3 PPGK1-TPGK1 | [50] |
| pGPD-426 | 2 µ URA3 PTDH3-TCYC1 | [51] |
| pPGK-SsXUT1 | 2 µ URA3 PPGK1-SsXUT1-TPGK1 | [27] |
| pPGK-SpXUT1 | 2 µ URA3 PPGK1-SpXUT1-TPGK1 | This work |
| pPGK-SpXUT1ΔC | 2 µ URA3 PPGK1-SpXUT1ΔC-TPGK1 | This work |
| pGPD-SaXUT1 | 2 µ URA3 PTDH3-SaXUT1-TCYC1 | This work |
| pGPD-SaXUT1ΔNC | 2µ URA3 PTDH3-SaXUT1ΔNC-TCYC1 | This work |
| Primers: 2 | ||
| pPGK-SpXUT1-F | AGATCGGAATTCAAGCTTATGCACGGAGGTTCAGACG | This work |
| pPGK-SpXUT1-R | GCCGGATCCGGCTTAAGCACTGTCAGCATCAGC | This work |
| pPGK-SpXUT1ΔC-R | GGCGGATCCAAATTAGTCAGAGTCTAATTCTTCTCCGCC | This work |
| pGPD-SaXUT1-F | AGATCGGAATTCAAGCTTGGATCCATGCACGGAGGTTCAGATAGTAA | This work |
| pGPD-SaXUT1-R | GCCCTCGAGGTCGACCCCGGGGGCTTAATCAGCATCAGCAACCTTTTC | This work |
| pGPD-SaXUT1ΔNC-F | GCCGGATCCAAAATGCGTTTAGAAATCGCCGGTAAACC | This work |
| pGPD-SaXUT1ΔNC-R | GCCCTCGAGGTCGACTTAATCAGAATCTAAGTCTTCTAATCC | This work |
| ROD1Δ-F | ATGTTTTCATCATCATCTCGACCTTCAAAAGAGCCATTACCCAGCTGAAGCTTCGTACGC | This work |
| ROD1Δ-R | CTATGAGCGATCCCGTTTTGTGAACATCTCCATTAAATTAGCATAGGCCACTAGTGGATC | This work |
| ROG3Δ-F | GGCGTTGATAAAGAGCCAATATCTATTGTTGCTACATAGACCAGCTGAAGCTTCGTACGC | This work |
| ROG3Δ-R | CGACTATCGTTTGTTACCCTTTGATAGAAAACCTCCCATAGCATAGGCCACTAGTGGATC | This work |
| V-ROD1-F | AGTCGAGTCCCTTGGTACAT | This work |
| V-ROD1-INT-F | CTGCCGTCACTTATGCTCTG | This work |
| V-ROD1-R | CGAATGATGTCTGTGGGATC | This work |
| V-ROG3-F | GCAAGTACAGAGTCCTACCA | This work |
| V-ROG3-INT-F | CTGTGTGCAAGATTGTGATG | This work |
| V-ROG3-R | GCCAGTTAGAGTGCGTAAAT | This work |
| V-KanR-F | CCGGTTGCATTCGATTCC | This work |
| V-BleR-F | CCTTCTATGAAAGGTTGGGC | This work |
| Strain | Transport of 14C-Glucose | Transport of 14C-Xylose | ||
|---|---|---|---|---|
| Km (mM) | Vmax (mmol h−1 gDCW−1) | Km (mM) | Vmax (mmol h−1 gDCW−1) | |
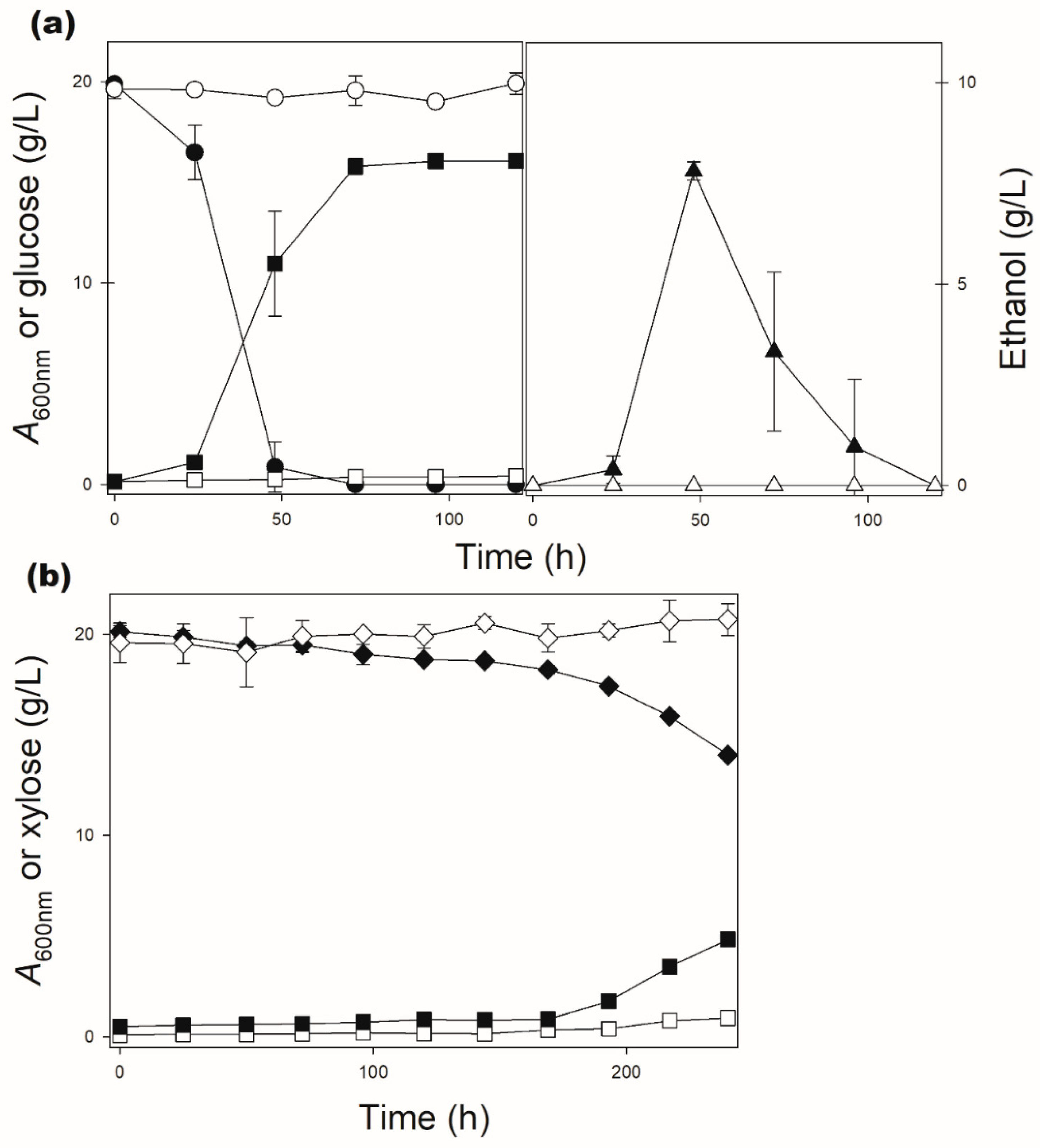
| DLG-K1 + pPGK-SsXUT1 | 24.5 ± 4.1 | 10.8 ± 1.8 | 417.7 ± 176 | 72.4 ± 27 |
| DLG-K1 + pPGK-SpXUT1 | 26.1 ± 11.5 | 4.3 ± 1.9 | 711 ± 550 | 72.3 ± 7.5 |
| Plasmid and Carbon Source | Ethanol Produced (g/L) by Strains: | |||
|---|---|---|---|---|
| DLG-K1 | DLG-K1∆R1 | DLG-K1∆R3 | DLG-K1∆R1R3 | |
| pPGK-SpXUT1 | ||||
| 2% maltose | 9.0 ± 0.2 | 9.3 ± 0.6 | 9.3 ± 0.5 | 7.6 ± 0,2 |
| 2% glucose | 0.0 | 0.0 | 7.1 ± 0.2 | 0.0 |
| pGPD-SaXUT1 | ||||
| 2% maltose | 8.3 ± 0.1 | 8.2 ± 0.1 | 7.9 ± 0.3 | 5.8 ± 0.1 |
| 2% glucose | 0.0 | 0.0 | 1.1 ± 0.1 | 0.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knychala, M.M.; dos Santos, A.A.; Kretzer, L.G.; Gelsleichter, F.; Leandro, M.J.; Fonseca, C.; Stambuk, B.U. Strategies for Efficient Expression of Heterologous Monosaccharide Transporters in Saccharomyces cerevisiae. J. Fungi 2022, 8, 84. https://doi.org/10.3390/jof8010084
Knychala MM, dos Santos AA, Kretzer LG, Gelsleichter F, Leandro MJ, Fonseca C, Stambuk BU. Strategies for Efficient Expression of Heterologous Monosaccharide Transporters in Saccharomyces cerevisiae. Journal of Fungi. 2022; 8(1):84. https://doi.org/10.3390/jof8010084
Chicago/Turabian StyleKnychala, Marilia M., Angela A. dos Santos, Leonardo G. Kretzer, Fernanda Gelsleichter, Maria José Leandro, César Fonseca, and Boris U. Stambuk. 2022. "Strategies for Efficient Expression of Heterologous Monosaccharide Transporters in Saccharomyces cerevisiae" Journal of Fungi 8, no. 1: 84. https://doi.org/10.3390/jof8010084
APA StyleKnychala, M. M., dos Santos, A. A., Kretzer, L. G., Gelsleichter, F., Leandro, M. J., Fonseca, C., & Stambuk, B. U. (2022). Strategies for Efficient Expression of Heterologous Monosaccharide Transporters in Saccharomyces cerevisiae. Journal of Fungi, 8(1), 84. https://doi.org/10.3390/jof8010084







