Abstract
A major challenge in the research of transport proteins is to understand how single amino acid residues contribute to their structure and biological function. Amino acid substitutions that result in a selective advantage in adaptive laboratory evolution experiments can provide valuable hints at their role in transport proteins. In this study, we applied an evolutionary engineering strategy to alter the substrate specificity of the proton-coupled disaccharide transporter Mal11 in Saccharomyces cerevisiae, which has affinity for sucrose, maltose and glucose. The introduction of MAL11 in a strain devoid of all other sugar transporters and disaccharide hydrolases restored growth on glucose but rendered the strain highly sensitive to the presence of sucrose or maltose. Evolution in glucose-limited continuous cultures with pulse-wise addition of a concentrated sucrose solution at increasing frequency resulted in the enrichment of spontaneous mutant cells that were less sensitive to the presence of sucrose and maltose. Sequence analysis showed that in each of the two independent experiments, three mutations occurred in MAL11, which were found responsible for the disaccharide-insensitive phenotype via reverse engineering. Our work demonstrates how laboratory evolution with proton-motive force-driven uptake of a non-metabolizable substrate can be a powerful tool to provide novel insights into the role of specific amino acid residues in the transport function of Mal11.
1. Introduction
Over the course of evolution, a myriad of sugar transporters have emerged with diverse mechanisms, kinetics, regulation and substrate specificities [1,2]. Whereas some transporters are highly specific towards one sugar substrate, such as the fructose-specific Ffz1 from Zygosaccharomyces rouxii [3], others are able to mediate the uptake of multiple sugars, such as Mal11 from Saccharomyces cerevisiae [4,5,6]. Knowledge on the three-dimensional organization of amino acid residues and how they interact with each other to bind and translocate substrates is highly relevant to understand how transporters with broad substrate range evolved into specific transporters (or vice versa) in nature. Since sugars are important substrates of industrial biotechnology, their transporters are also relevant targets in engineering strategies for the improvement of microbial cell factories [7,8,9,10]. Ideally, transport proteins (and thus their encoding genes) can be designed at will, to obtain strains with unrestricted transport of the desired substrate without undesired inhibition or competition by other molecules. This scenario requires extensive knowledge on the structure and mechanism of transport proteins and the function of individual amino acid residues on their overall activity.
The steadily increasing number of resolved protein structures through methods such as X-ray crystallography has contributed to our understanding of the tertiary structure of membrane proteins [11,12]. Still, compared to soluble proteins, membrane protein structures are rather under-represented in the protein data bank, partially due to the difficulty of membrane protein crystallization [13]. In addition, computational methods for protein structure prediction, based on the crystal structure of a homologous protein, have provided valuable insights [14], and promising results are now obtained using the AI system AlphaFold [15]. Knowledge on the role of individual amino acid residues can be acquired by studying mutant transporter variants acquired via (targeted) mutagenesis [16,17] or adaptive laboratory evolution (ALE) [18,19,20]. For the latter approach, cultivation conditions are carefully designed to allow for the enrichment of mutants with a desired phenotype. By analyzing the genome of evolved strains, mutations can be identified that underlie the evolved phenotype, and (point) mutations in transport proteins hint at an important role of specific amino acid residues for transport function. This method is especially powerful when the investigated transport process is essential for growth [16,20]. For example, ALE has been applied to improve kinetics of proteins that mediate the uptake of an essential substrate by selecting for mutants with an improved growth rate through serial propagation in batch conditions [21,22,23,24]. Alternatively, transporters with improved affinity can be obtained by prolonged cultivation in substrate-limited continuous cultures [25,26]. Some evolutionary engineering approaches were aimed at specializing transporters with broad substrate affinity for one of its substrates. For example, Hxt transporters in S. cerevisiae, which transport glucose and xylose (albeit with moderate affinity and glucose inhibition), were specialized for xylose transport by evolving a hexokinase-deficient strain for growth on xylose in the presence of glucose [27,28,29]. The resulting mutant transporter variants were found to contain amino acid substitutions that decreased or abolished the affinity of the transporter for glucose but not for xylose. However, complete abolishment of glucose affinity was accompanied by a ~60% reduction of the Vmax for xylose.
A more stringent selection for specialized transporters could be achieved if transport of the undesired substrate can be made toxic to the cells (counter-selection), while transport of the desired substrate provides an evolutionary benefit. Transport of a metabolizable sugar is usually not toxic, but exceptions can be found when sugar uptake is coupled to the uptake of a proton (proton symport). Uptake via such a proton-symporter is not only driven by the concentration gradient of the sugar, but also by the chemical and electrical potential difference of the proton-motive force (pmf). At typical values of the pmf in S. cerevisiae (−150 to −200 mV [30]), this driving force could enable a 1000-fold accumulation of sugar inside the cell. Such accumulation would lead to osmotic bursting of cells, and therefore excessive substrate accumulation is prevented by regulation of transporters and enzymes that catalyze the intracellular degradation of the sugar. The importance of this regulation is demonstrated by the phenomenon referred to as ‘substrate-accelerated cell death’, which has for example been observed when a pulse of maltose was administered to prolonged, maltose-limited S. cerevisiae chemostat cultures [26,31]. The sudden switch from maltose-limited to maltose-excess conditions decreased culture viability by up to 70%, which was hypothesized to be due to uncontrolled, sudden and fast uptake of maltose and protons into the cells, leading to rapid acidification and an osmotic burst.
In this study, we investigated whether the proton-motive force could be employed to create a selective pressure against substrate uptake by proton-coupled transporters. To this end, we introduced MAL11, which encodes a proton-symporter with affinity for sucrose, maltose and glucose [4,5,6] as the sole sugar transporter gene in an S. cerevisiae strain that is not able to hydrolyze sucrose or maltose. Since these disaccharides are not metabolized, we expected that cells would suffer negative effects from their intracellular accumulation, which is ‘forced’ by the proton-motive force due to proton-coupled uptake. By evolving this strain in glucose-limited chemostats with increasing addition of sucrose, we selected for mutants that were less sensitive to the presence of sucrose, while maintaining their ability to take up glucose. This evolution was followed by an analysis of mutations and growth characterization of evolved and reverse engineered strains in medium with glucose, supplemented with various concentrations of sucrose or maltose.
2. Materials and Methods
2.1. Strains and Maintenance
All Saccharomyces cerevisiae strains used in this study are derived from the CEN.PK lineage [32]. For long-term storage, glycerol was added to cells that were grown until late exponential phase, to obtain a final concentration of 30% (v/v) glycerol, after which 1 mL aliquots were stored at −80 °C. Plasmids were propagated in Escherichia coli XL1-Blue cells (Agilent, Santa Clara, CA, USA), which were also stored at −80 °C after addition of glycerol to a final concentration of 25% (v/v) to overnight cultures.
2.2. Molecular Biology Techniques
Plasmids were isolated from yeast strains using the Zymoprep Yeast Plasmid Miniprep II kit (Baseclear, Leiden, The Netherlands), after which 1 µL of this mixture was transformed, according to the manufacturer’s protocol, into XL1-Blue chemically competent E. coli cells (Agilent) for plasmid propagation. Plasmids were isolated from E. coli cells using the GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific, Waltham, MA, USA). Transformation of S. cerevisiae strains was performed using LiAc/ssDNA/PEG, as previously described [33]. After transformation, single colonies were re-streaked three consecutive times to ensure an isogenic single cell line. For whole-genome sequencing, genomic DNA was isolated from stationary S. cerevisiae cultures using the QIAGEN Blood & Cell Culture DNA Kit with 100/G Genomics-tips (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Whole-genome sequencing of these DNA samples was performed by Genomescan (Leiden, The Netherlands). Then, 350 bp insert libraries were constructed, which were paired-end sequenced (150 bp reads) with an Illumina HiSeq X sequencer. The data analysis was performed as described previously [34]. Sanger sequencing of genes was performed using the Macrogen (Amsterdam, the Netherlands) EZ-seq sequencing service according to the provided instructions.
2.3. Strain Construction
All S. cerevisiae strains used in this study are listed in Table 1.

Table 1.
S. cerevisiae strains used in this study. The prefix ‘pUDE’ signifies an episomal plasmid.
Plasmid pUDE432 [36] was introduced into strain IMK1010, resulting in strain IMZ786, which was subsequently evolved in duplicate using independent bioreactor cultures. After evolution, the evolved population from each bioreactor was stocked, resulting in IMS1225 and IMS1226. Both strains were plated on SMD with 5 g L−1 sucrose and re-streaked three consecutive times. The resulting single-colony isolates were grown in liquid SMD with 5 g L−1 sucrose and stocked as IMS1230 and IMS1231, respectively. The plasmids pUDE1222 and pUDE1223 were isolated from IMS1230 and IMS1231, respectively, and reintroduced in IMK1010, resulting in strains IME753 and IME754, respectively.
2.4. Media and Cultivation
E. coli cultures were grown at 37 °C in LB medium, supplemented with 100 μg mL−1 ampicillin for selection and maintenance of plasmids. Yeast strains were grown on synthetic medium (SM), which was heat sterilized for 20 min at 121 °C, after which a filter-sterilized vitamin solution and 20 g L−1 glucose was added (SMD) [37]. Medium for anaerobic cultivations was additionally supplemented with 10 mg L−1 ergosterol and 420 mg L−1 Tween 80, which were added in from a concentrated solution (800×) in absolute ethanol [38]. For preparation of solid medium plates, 2% (w/v) agar was added to the media prior to heat sterilization. Medium used in bioreactor cultivations was additionally supplemented with 0.2 g L−1 Antifoam C (Sigma Aldrich, Saint Louis, MO, USA). Aerobic shake flask cultures were grown in 500 mL round bottom flasks with 100 mL medium, which were incubated in an Innova orbital shaker (Eppendorf, Nijmegen, the Nederlands) at 200 rpm at 30 °C. Laboratory evolution of IMZ786 was conducted in 2 L laboratory bioreactors (Applikon, Delft, the Netherlands) with a 1 L working volume. Cultures were stirred at 800 rpm, the temperature was controlled at 30 °C and the pH was kept constant at 5.0 through automated addition of 2.0 M KOH. The cultures were sparged with 500 mL N2 min−1 (<5 ppm O2), and medium vessels were sparged with nitrogen as well. To enable evolution in continuous culture set-up, SMD medium pumps were switched on after glucose depletion in a preceding batch phase, to obtain a constant flowrate. The volume was kept constant at 1 L using an effluent pump that was controlled by an electric level sensor, resulting in a stable dilution rate. After a stable CO2 concentration in the reactor off-gas was observed, a filter-sterilized 500 g L−1 sucrose solution was added to the culture in one-second pulses via a separate pump. Based on observations on the online measurements of the CO2 concentration in the reactor off-gas, the frequency of these pulses was manually increased and decreased to exert a selective pressure on the culture but prevent culture washout.
Maximum specific growth rates were determined in a Growth-Profiler system (EnzyScreen, Heemstede, The Netherlands) equipped with 96-well plates in a culture volume of 250 μL, set at 250 rpm and 30 °C. The measurement interval was set at 20 min. Raw green values were corrected for well-to-well variation using measurements of a 96-well plate containing a culture with an externally determined optical density, measured at 660 nm using a Libra S11 spectrophotometer (Biochrom, Cambridge, United Kingdom), of 4.19 in all wells. Optical densities were calculated by converting green values (corrected for well-to-well variation) using a calibration curve that was determined by fitting a third-degree polynomial through 71 measurements of cultures with known OD values between 0.75 and 24.5 (Supplementary Data S2). Growth rates were determined using the calculated optical densities of at least 10 points (corresponding to at least 200 min) in the exponential phase. Exponential growth was assumed when an exponential curve could be fitted with an R2 of at least 0.985. To validate the experimental procedure and data analysis, the growth rates of laboratory strain CEN.PK113-7D were also determined in all conditions, which corresponded to within 5% of previously determined values for this strain grown aerobically in SMD [39].
3. Results
3.1. Evolution for Decreased Sucrose Sensitivity
The previously constructed strain IMK1010 is devoid of all hexose transporters, disaccharide transporters and disaccharide hydrolases and is therefore unable to grow on any sugar substrate [35]. MAL11, encoding a proton-symporter with affinity for maltose, sucrose and glucose, was introduced in IMK1010 via the episomal expression vector pUDE432. Subsequently, growth of the resulting strain (IMZ786) was investigated on synthetic medium (SM) with only glucose, maltose or sucrose as the carbon source, and on SM with mixtures of glucose with either sucrose or maltose. Although introduction of MAL11 was expected to enable the uptake of glucose, sucrose and maltose, in IMZ786 it only complemented the growth on medium with glucose (SMD, Figure 1). We hypothesized that the inability to grow on disaccharides was caused by the absence of disaccharide hydrolysis activity in IMZ786, which is essential for their further metabolism [40] (Supplementary Data S2). Moreover, IMZ786 was found to be sensitive to the presence of either sucrose or maltose, as could be observed from its inability to grow on SMD with the addition of 20 g L−1 of sucrose or maltose (Figure 1). We attributed this detrimental effect to the proton symport mechanism of the constitutively expressed Mal11, which enabled proton-motive force-driven intracellular accumulation of the disaccharides that, in this specific strain background, could not be relieved by their degradation or export. To investigate whether this inability to grow on SMD in the presence of sucrose or maltose was not only due to competitive inhibition of the transporter, cultures of IMK1010 (sugar0) and IMZ786 (sugar0 MAL11), which were pre-grown in SM with 2% (v/v) ethanol as the carbon source, were transferred to SM with 2% (v/v) ethanol and 20 g L−1 sucrose. While the optical density of IMK1010 in the presence of sucrose increased from 0.11 to 17.9 in 45.5 h, that of IMZ786 only increased from 0.07 to 0.12 within the same time span. These results indicate that growth on carbon sources not transported by Mal11, such as ethanol, which passively diffuses across the plasma membrane, is strongly impaired and likely caused by Mal11-mediated intracellular accumulation of sucrose.
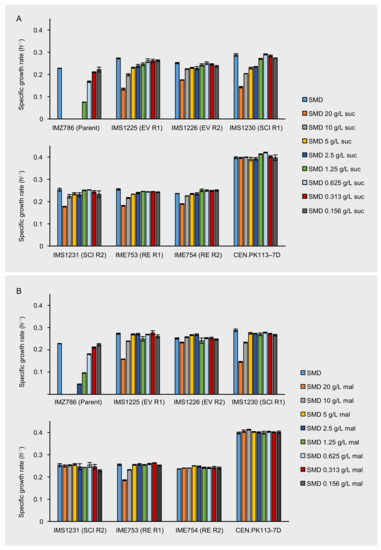
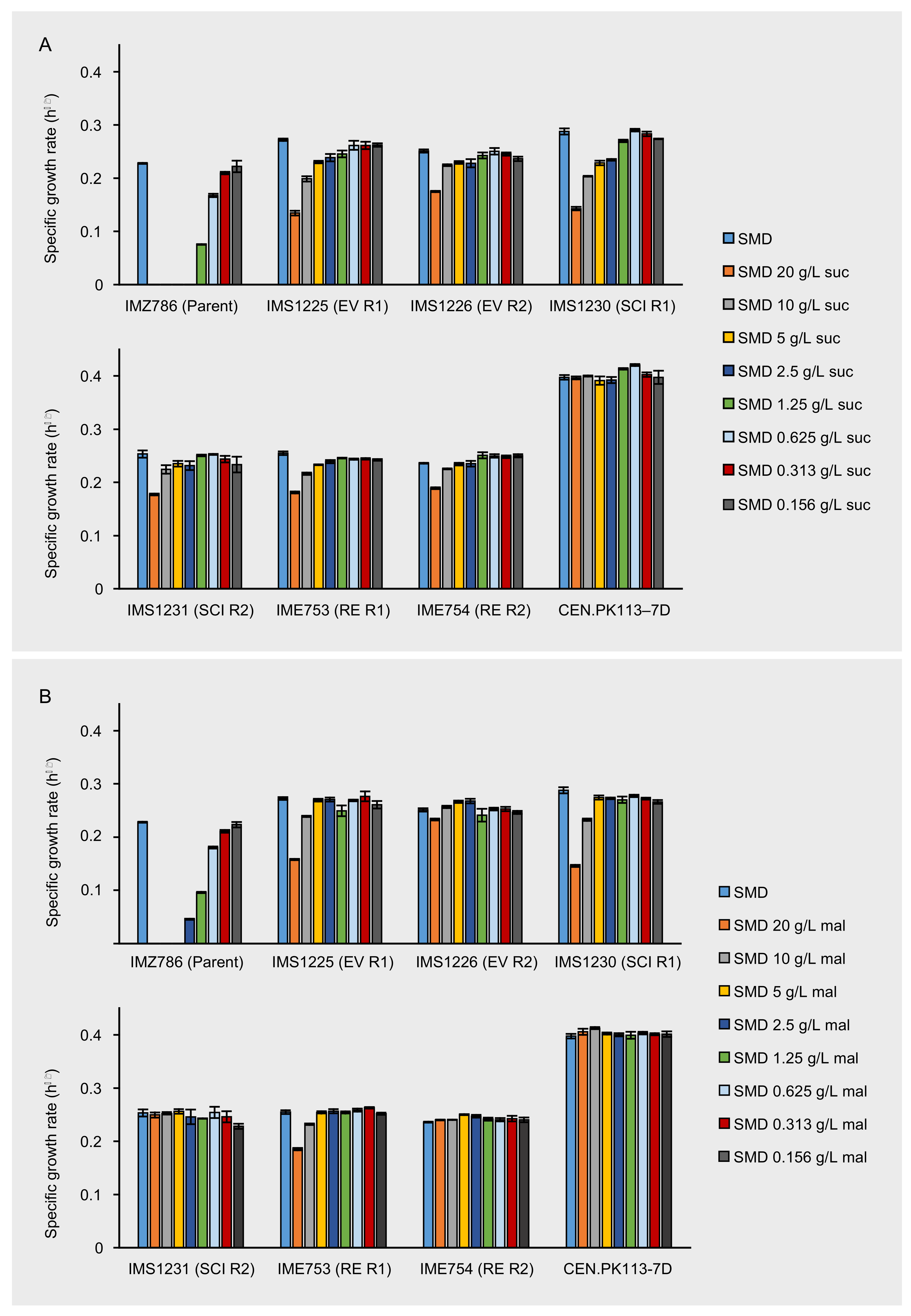
Figure 1.
Maximum specific growth rates of IMZ786 (sugar0 MAL11) and strains that originate thereof, which were the evolved populations (EV) after laboratory evolution in glucose-limited continuous cultures with increasing addition of sucrose in two independent reactors (R1 and R2), the corresponding single-colony isolates (SCI) and the corresponding reverse engineered strains (RE). (A) Maximum specific growth rates on SMD within the presence of different concentrations of sucrose (suc). (B) Maximum specific growth rates on SMD with in the presence of different concentrations of maltose (mal). Data represent average and mean deviation of three replicate experiments. To validate the experimental procedure and data analysis, the growth rates of laboratory strain CEN.PK113-7D were also determined in all conditions.
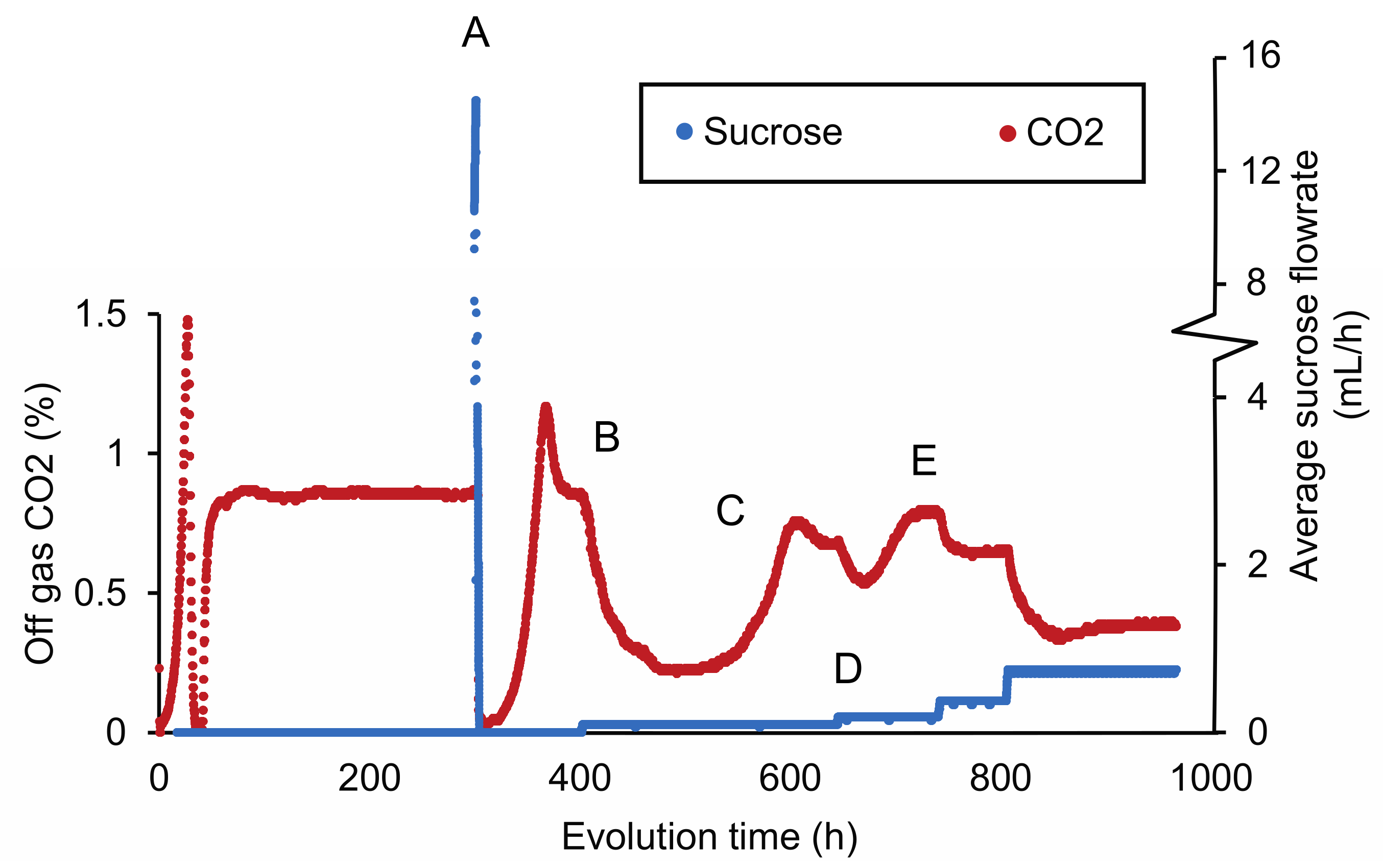
The sensitivity of IMZ786 towards sucrose when grown on glucose inspired us to investigate whether this property could enable a laboratory evolution strategy to decrease the substrate specificity of Mal11 for sucrose, while retaining its glucose transport activity. To this end, IMZ786 was first grown in two independent anaerobic glucose-limited continuous cultures at a dilution rate of 0.07 h−1, and online measurements of the CO2 concentrations in the reactor off-gas were used as a means to monitor metabolic activity and growth (Figure 2). After a stable off-gas CO2 concentration of ~0.83% was observed, we then investigated whether the presence of sucrose was also toxic to IMZ786 under glucose-limited conditions by separately adding a concentrated sucrose solution (500 g L−1) to the culture in one-second pulses, which resulted an average flowrate of ~16 mL h−1. Indeed, a near instant decrease of the CO2 production towards 0.08% was observed (within 1 h), indicating that growth and metabolic activity had ceased due to the addition of sucrose (Figure 2A). To prevent culture washout, the addition of sucrose was stopped until a stable CO2 concentration of ~0.85% was observed again. Then, the frequency at which the pulses of the sucrose solution were added was adjusted to obtain an average flowrate of ~0.09 mL h−1, which again led to a gradual decrease in CO2 production (Figure 2B), indicating significant sucrose toxicity in the culture. After approximately 480 h, the off-gas CO2 concentration first stabilized at 0.22%, and then increased to reach a new stable value at ~0.7% (Figure 2C). We hypothesized that this decrease, followed by an increase in CO2 production, was the result of washout of the original strain IMZ786, followed by the emergence and enrichment of less sensitive mutants. To increase the selective pressure on sucrose insensitivity, the frequency of the sucrose pulses was then manually increased (Figure 2D), after which the CO2 concentration was allowed to stabilize again (Figure 2E). This cycle was repeated until a stable CO2 output (0.53% for the first replicate, 0.68% for the second) was observed at an average flowrate of 1.6 mL h−1 (Supplementary Figure S1, Supplementary Data S1), which, under the assumptions of steady-state, ideal mixing and that sucrose is not consumed, corresponded to approximately 11.4 g L−1 of sucrose in the reactor. After a total of approximately 350 generations, evolved populations were stocked from each reactor, resulting in IMS1225 and IMS1226, from which single cell lines were isolated (IMS1230 and IMS1231, respectively).
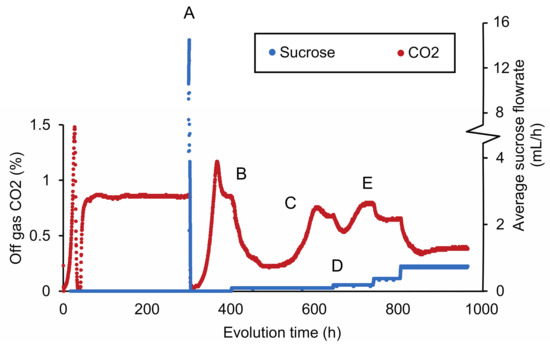
Figure 2.
CO2 concentrations in the reactor off-gas and average flowrate of a 500 g L−1 sucrose solution during adaptive laboratory evolution of IMZ786 (sugar0 MAL11) in anaerobic glucose-limited chemostats at a dilution rate of 0.07 h−1 with pulse-wise addition of sucrose. The off-gas CO2 concentrations were used as a read-out to monitor growth and metabolic activity. One representative of the two independent evolution experiments is shown. (A) After a stable CO2 concentration of ~0.83% in the reactor off-gas was observed, sucrose was administered separately to the cultures at an average flowrate of 16 mL h−1. At this flowrate, the CO2 concentration decreased to 0.08% within 1 h. To subsequently prevent washout of cells and allow for evolution, the addition of sucrose was temporarily stopped. (B) After ~400 h, the average sucrose flowrate was set to 0.09 mL h−1. This resulted in a decrease of the off-gas CO2 concentration to a value of 0.22%. (C) The decrease in the off-gas CO2 concentration was, at the 480 h time point, followed by a strong increase, until it stabilized at ~0.64%. (D) Subsequently, the average flowrate of sucrose was increased to 0.19 mL h−1 and the culture was again left to evolve until a stable CO2 output was reached (E). An overview of the full experiment, which lasted ~3400 h, is presented in Supplementary Figure S1.
3.2. Evolved Strains Are Less Sensitive to Sucrose and Maltose
To investigate whether the bioreactor-derived strains were less sensitive to the presence of the disaccharides sucrose and maltose, the maximum specific growth rates of the evolved populations IMS1230 and IMS1231 were determined in SMD medium with various concentrations of either sucrose or maltose and compared to those of the unevolved parental strain IMZ786. Once again, the toxic effect that sucrose has on IMZ786 was apparent by the absence of growth on SMD in the presence of ≥2.5 g L−1 of sucrose, and only at a concentration of 0.16 g L−1 of sucrose no effect on the growth rate could be observed compared to growth on SMD without sucrose (Figure 1A).
On the contrary, the evolved strains IMS1225 and IMS1226 exhibited growth in all tested conditions, and in each condition, their growth rates were higher than that of the unevolved parental strain. For IMS1225, no effect on the growth rate could be observed between the SMD control and cultures with sucrose concentrations up to 0.625 g L−1, and for IMS1226 up to 1.25 g L−1 (Figure 1A). The presence of maltose had a similar effect on the growth rates of the three strains, although the toxicity effect was slightly less apparent than for sucrose (Figure 1B). For instance, the parental strain IMZ786 also exhibited growth in the presence of up to 2.5 g L−1 of maltose and compared to the growth rates of IMS1225 and IMS1226 on SMD, no effects were observed upon addition of up to 5 g L−1 and 10 g L−1 of maltose, respectively. Strikingly, the addition of 20 g L−1 of maltose only led to a 7% decrease of the growth rate of IMS1226. These results indicate that the evolution not only affected the sensitivity towards sucrose, but also its isomer maltose. Moreover, the growth rates of both strains on SMD without sucrose or maltose were 19.4% (IMS1225) and 10.1% (IMS1226) higher than that of the parental strain. In all conditions, growth rates of IMS1230 were highly similar to those of IMS1225, whereas the growth rates of IMS1231 were highly similar to those of IMS1226, indicating that the isolated single colonies are phenotypical representatives of the evolved population.
3.3. Mutations Occurred in MAL11 during Evolution
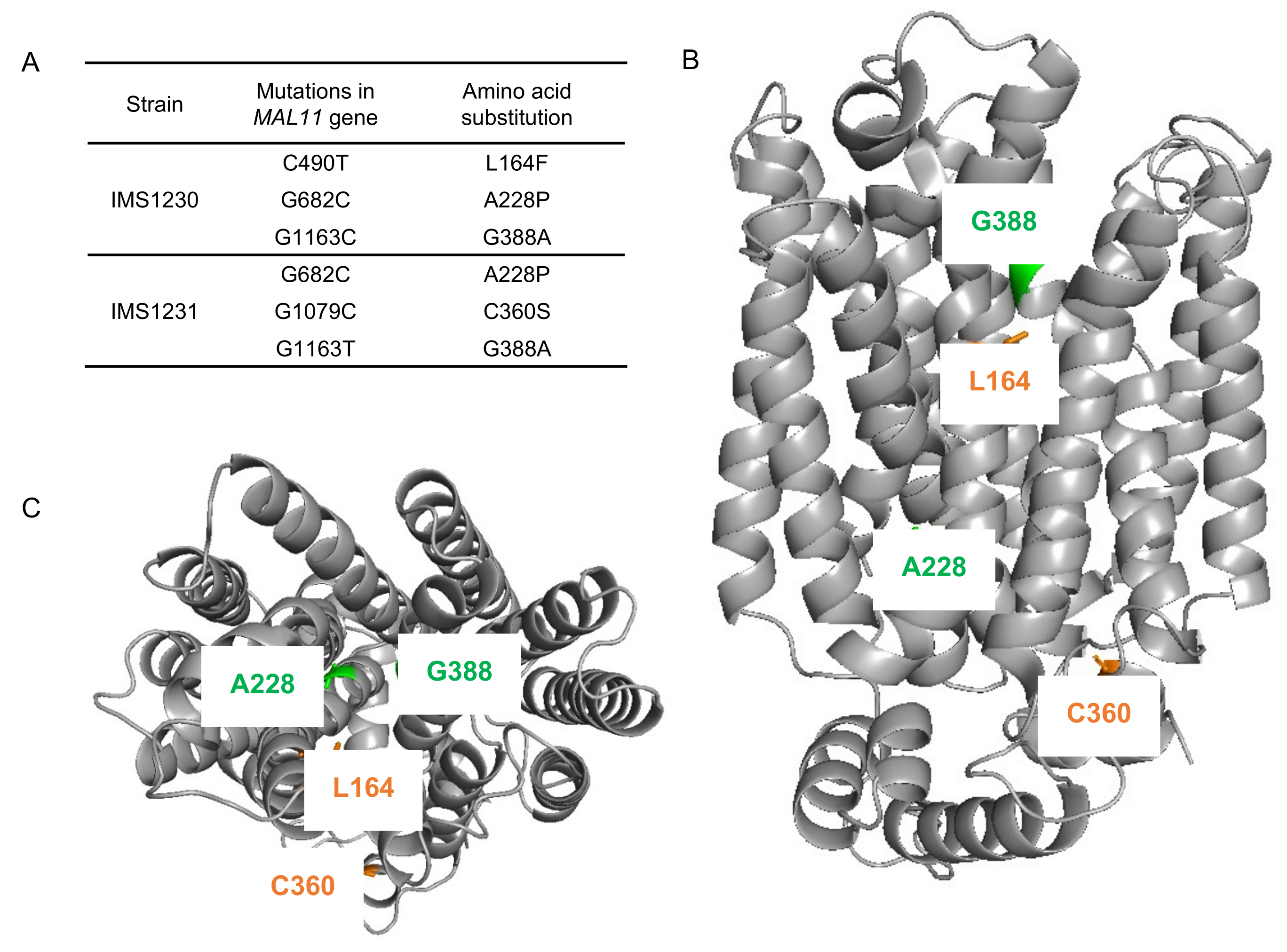
To investigate whether mutations occurred in the transporter gene MAL11 during the laboratory evolution experiment, the plasmids from single-colony isolates IMS1230 and IMS1231 were isolated and the MAL11 open reading frame was sequenced. Strikingly, three non-synonymous mutations had occurred in each independently-evolved strain: C490T, G682C and G1163C in IMS1230 and G682C, G1079C and G1163T in IMS1231 (Figure 3A). Although G1163 was mutated to a different base in each strain, the resulting amino acid substitution in the protein was identical, indicating that there were two common amino acid changes (Ala-228-Pro and Gly-388-Ala) in Mal11 that evolved independently in the two cultures.
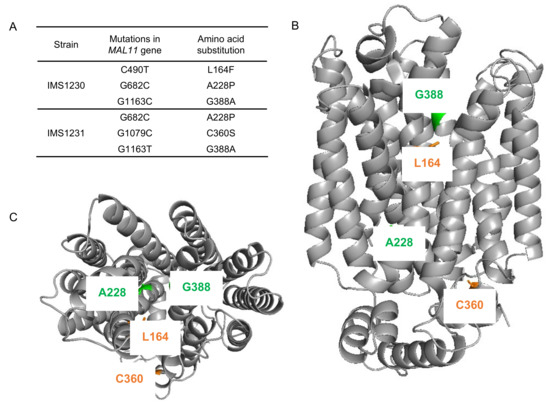
Figure 3.
Mutations identified in MAL11 after evolution of IMZ786 (sugar0 MAL11) in two independent, glucose-limited continuous cultures with increasing addition of sucrose. (A) In single-colony isolates from two independent reactors, three mutations were identified in MAL11. (B) (side view), (C) (top view). Structural model of Mal11, highlighting the location of the amino acid changes resulting from the mutations that occurred during evolution. Mutations that were identical in both strains are indicated in green, and mutations that were unique in each strain are indicated in orange. All structure figures were prepared with PyMol.
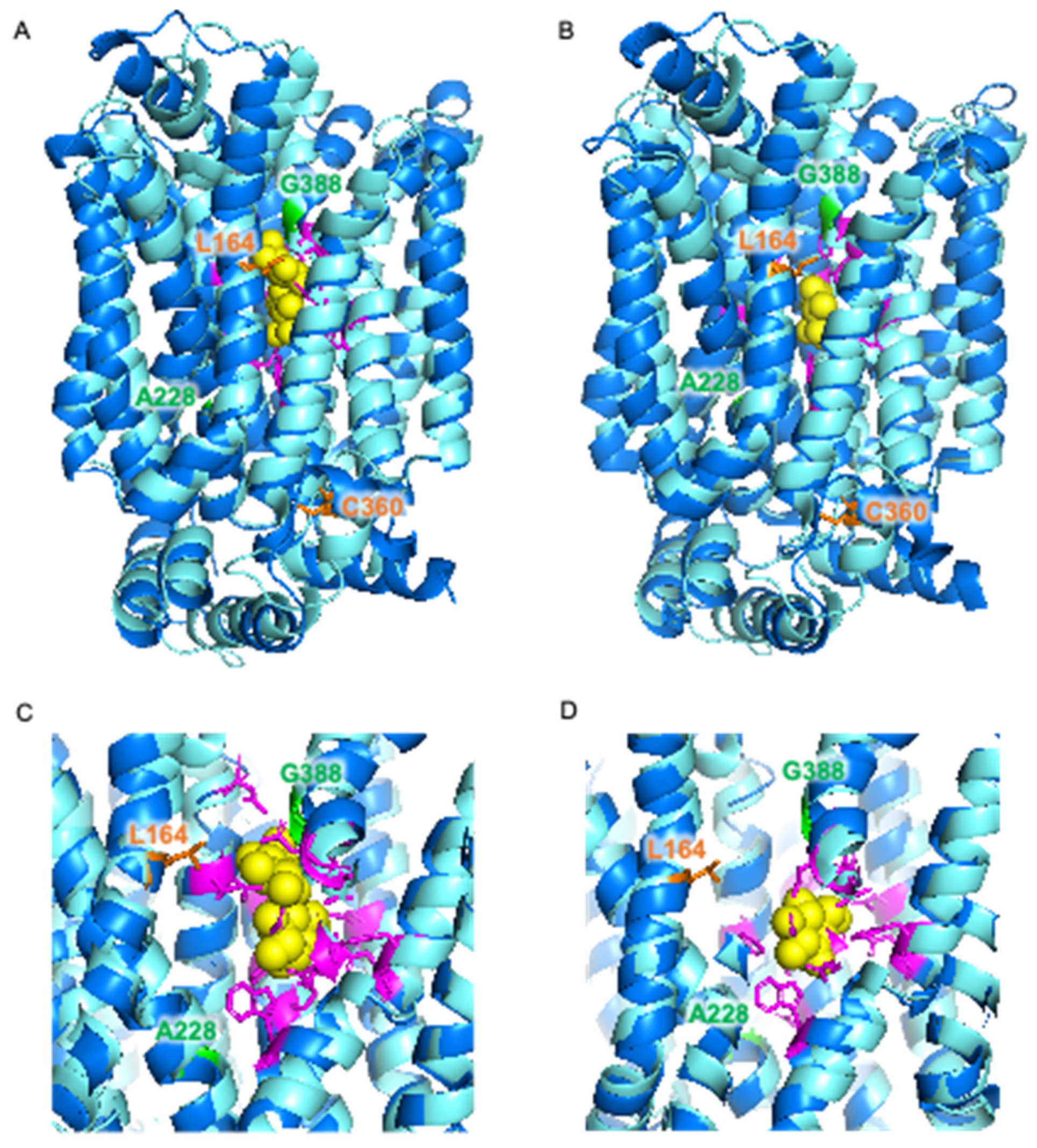
The locations of these mutations were investigated using a previously constructed homology model of Mal11 [41] (Figure 3B,C). The mutations that where shared in the two evolved strains were both found to be located in the central cavity of the protein, as well as the Leu-164-Phe mutation that was identified in IMS1230, albeit at different heights in the protein structure. To investigate the proximity of these mutations to the sugar binding sites, the Mal11 structure was compared to that of the homologous human GLUT3 sugar transporter, for which crystal structures have been determined for the glucose- and maltose-bound proteins [42]. These crystal structures showed that binding of one of the glucose units comprising maltose in GLUT3 completely overlaps with binding of a glucose molecule. Superimposition of the Mal11 homology model with the two GLUT3 structures suggested that the three mutations in the central cavity of Mal11 might be relatively close to the amino acids that form the sugar binding pocket (Figure 4). Strikingly, L164F and G388A appeared to be in near proximity to the glucose monomer of maltose that does not overlap with glucose.
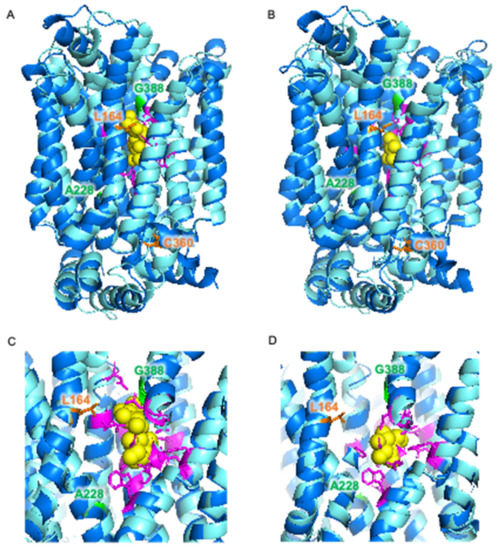
Figure 4.
Superimposition of homology model of S. cerevisiae Mal11 (turquoise) [41] and crystal structures of human GLUT3 (blue) [42] bound to maltose (PDB: 4ZWB) (A,C) and bound to glucose (PDB: 4ZW9) (B,D). Glucose and maltose are represented by yellow spheres. Amino acid residues involved in sugar binding in GLUT3 are shown in magenta, and amino acid residues in Mal11 that were substituted during evolution of IMZ786 are shown in orange and green.
3.4. Mutations in MAL11 Are Responsible for Decreased Sucrose and Maltose Sensitivity
To investigate whether the mutations that were identified in MAL11 were responsible for the decreased sensitivity of the evolved strains towards sucrose and maltose, the plasmids containing the mutated variants of MAL11 were isolated from IMS1230 and IMS1231 and reintroduced in the unevolved IMK1010 (sugar0) strain background. When the resulting strains (IME753 and IME754) were grown on SMD in the presence of different concentrations of sucrose or maltose (Figure 1), they performed similarly to their corresponding evolved strain from which the respective MAL11 allele originated. These observations indicate that the mutations in MAL11 are solely responsible for the evolved phenotype and no other genomic mutations contributed to the decreased sensitivity of evolved strains towards sucrose and maltose.
4. Discussion
In this study, we successfully applied an evolutionary engineering strategy to decrease the sensitivity of a MAL11-expressing strain lacking maltose or sucrose hydrolases towards sucrose while maintaining its ability to consume glucose. Concurrently, the sensitivity towards maltose was also decreased, which suggests that the mechanism behind the decreased sensitivity towards both disaccharides is highly similar. Expression of the evolved MAL11 alleles in an unevolved strain background and characterization of the resulting strains confirmed that the mutations in MAL11, which result in amino acid changes in the Mal11 protein, underlie the evolved phenotype. We hypothesize that the improved growth performance in the presence of these sugars could be due to (one of) the following alterations: (1) a decreased binding affinity of Mal11 for disaccharides; (2) an overall lower disaccharide transport capacity of the protein, resulting in less toxic accumulation of disaccharides in growing cultures; (3) glucose-specialized uptake, due to glucose inhibition of disaccharide uptake, similar to what is observed for Hxt transporters in mixtures of glucose and pentose sugars in S. cerevisiae [7]; (4) loss of proton coupling to sugar uptake, resulting in a facilitated diffusion mechanism. In the latter case, intracellular accumulation of disaccharides is limited to their extracellular concentrations. Future work to investigate which mechanism is responsible for the evolved phenotype could involve the determination of kinetic parameters of the Mal11 variants (Km, Ki and Vmax) via transport assays with radiolabeled sucrose, maltose and glucose [41,43,44]. The accumulation ratio (ratio between intra- and extracellular sugar concentrations) combined with proton-uptake assays could also provide insight into the transport mechanisms of the evolved Mal11 proteins. Additionally, the effect of expressing these transporters on strain physiology, and more specifically, anaerobic biomass yield on sugar, can also be used to elucidate their transport mechanism [23,36,45].
Two identical amino acid substitutions (A228P and G388A) occurred in both independent evolution lines, which suggests that these two amino acid residues play an important role in determining the substrate specificity of Mal11 towards glucose and disaccharides and that their substitution resulted in the most substantial fitness benefit under the conditions of the evolution experiment. In previous work, it was found that in Escherichia coli XylE and the human GLUT transporters, which are homologous to Mal11, the position that corresponds to Gly-388 in Mal11 is occupied by a tyrosine residue [46]. In those transporters, this tyrosine residue is involved in closing off the sugar-binding site from the aqueous extracellular space [47,48]. Since in Mal11, a tyrosine residue is located one helix-turn above this position, the location of this hydrophobic residue may sterically dictate the size of the sugar that can be transported by the protein [46]. We speculate that the substitution of Gly-388 by alanine, which has a larger hydrophobic side chain, results in a functional similarity to the tyrosine residues occupying this position in the homologous monosaccharide transporters. A similar mechanism could be envisioned as a result of the L164F mutation, which was also predicted to be in close proximity to the ‘additional’ glucose unit of Mal11-bound maltose and does not overlap with glucose binding (Figure 4). Here, substitution of the leucine side chain by the bulky aromatic side chain of phenylalanine could sterically hinder the binding of disaccharides. The roles of the other amino acid residues in Mal11 that were mutated during evolution have not been predicted previously, which further demonstrates the power of evolutionary engineering to pinpoint amino acid residues with important roles in the transport mechanism. Testing the effects of the individual and different combinations of the mutations in Mal11 identified in this study on sucrose, maltose and glucose transport capacity could provide valuable insights into the importance of each mutation for the evolved phenotype and potential synergistic effects of mutation combinations.
To the best of our knowledge, this is the first demonstration of an evolutionary engineering strategy where proton-coupled substrate uptake is used as a counter-selective pressure to change the substrate specificity of a transporter. One of the mutations that we identified corresponded to an amino acid residue whose role could be predicted based on known crystal structures. This strategy could therefore also be valuable to identify important amino acid residues in other, less well studied transport proteins for which a crystal structure is not available yet. When selecting targets for such an evolutionary strategy, we consider three criteria to be especially important.
First, the transporter of interest should remain essential for growth under the selective conditions. Otherwise, cells are likely to ‘escape’ the selective pressure by evolving loss-of-function mutations in the targeted transporter. For example, when the aim of an evolution experiment is to evolve the amino acid permease Put4, which actively transports alanine, glycine and proline [49], into a proline-specific transporter, Put4-mediated proline transport can be made essential for growth by using a strain background in which all other related transporter genes have been deleted, as well as PRO3. The PRO3 deletion results in a proline-auxotrophic strain that relies on uptake of proline from the medium [50], while transport of alanine and glycine, for which S. cerevisiae is prototrophic, can be lost.
Second, the counter-selectable ‘substrate’ should not be metabolized once taken up by cells, to allow for toxic accumulation. In this study, the absence of disaccharide hydrolases was sufficient to prevent intracellular conversion of sucrose or maltose. Another relatively straightforward application could be similar to the aforementioned transporters with affinity for both pentose and hexose sugars, where deletion of hexokinase-encoding genes is sufficient to prevent the conversion of glucose, while the introduction of a novel metabolic route allows for pentose conversion [27,28,29,51]. Such a strain design could therefore also be used for the evolution of proton-coupled hexose/pentose transporters, such as Gxs1 from Candida intermedia [52]. However, this criterium might be less straightforward with other substrates, especially when considering that the ability to grow on the essential substrate should be maintained. For example, when aiming to decrease the glucose affinity of MAL11, a difficulty arises by the fact that utilization of sucrose and maltose is also dependent on the hexokinase reaction. In such cases, the deletion of hexokinase genes cannot be used to disable glucose conversion and instead the use of non-metabolizable glucose analogs (for instance, 2-deoxy-glucose [53]) in cultures could provide the desired counter-selective pressure on glucose transport by its intracellular accumulation.
Third, the driving force of the transport process mediated by the targeted transporter should be sufficient to achieve (toxic) levels of accumulation of the counter-selectable substrate. This can be especially challenging if the proton-coupled transport process does not result in net transfer of charge due to transport of an anion, as is for example the case for proton-coupled monocarboxylic-acid transporters such as Jen1 [54]. Proton symport of the negatively-charged lactate, acetate, pyruvate and propionate anions via this transporter is electroneutral, therefore only the chemical component of the proton-motive force contributes to its driving force. To promote toxic accumulation of lactate without the contribution of the electrical component of the proton-motive force, the evolution could be performed at a low extracellular pH to maximize the chemical potential difference of protons across the plasma membrane [55]. An evolution experiment aimed at losing the ability of Jen1 to transport lactate could then be performed in a strain devoid of all other lactic acid importers [56,57] and lactate dehydrogenase activity [58].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8040358/s1, Figure S1: CO2 concentrations in the reactor off-gas and average flowrate of a 500 g L−1 sucrose solution during adaptive laboratory evolution of IMZ786 (sugar0 MAL11) in anaerobic glucose-limited chemostats with pulse-wise addition of sucrose.; Data S1: Online measurements during evolution of IMZ786; Data S2: Growth profiler data for determination of growth rates of strains constructed in this study.
Author Contributions
Conceptualization, R.M.; methodology, S.C.d.V.; formal analysis, S.C.d.V.; investigation, S.C.d.V.; resources, R.M.; data curation, S.C.d.V.; writing—original draft preparation, S.C.d.V.; writing—review and editing, R.M.; visualization, S.C.d.V.; supervision, R.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to thank Erik de Hulster for performing growth experiments on SM with ethanol and sucrose.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, L.-Q.; Cheung, L.S.; Feng, L.; Tanner, W.; Frommer, W.B. Transport of sugars. Annu. Rev. Biochem. 2015, 84, 865–894. [Google Scholar] [CrossRef]
- Saier, M.H.J. Transport protein evolution deduced from analysis of sequence, topology and structure. Curr. Opin. Struct. Biol. 2016, 38, 9–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leandro, M.J.; Sychrová, H.; Prista, C.; Loureiro-Dias, M.C. The osmotolerant fructophilic yeast Zygosaccharomyces rouxii employs two plasmamembrane fructose uptake systems belonging to a new family of yeast sugar transporters. Microbiology 2011, 157, 601–608. [Google Scholar] [CrossRef] [Green Version]
- Han, E.-K.; Cotty, F.; Sottas, C.; Jiang, H.; Michels, C.A. Characterization of AGT1 encoding a general α-glucoside transporter from Saccharomyces. Mol. Microbiol. 1995, 17, 1093–1107. [Google Scholar] [CrossRef] [PubMed]
- Wieczorke, R.; Krampe, S.; Weierstall, T.; Kreidel, K.; Hollenberg, C.P.; Boles, E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999, 464, 123–128. [Google Scholar] [CrossRef] [Green Version]
- Stambuk, B.U.; da Silva, M.A.; Panek, A.D.; de Araujo, P.S. Active α-glucoside transport in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 1999, 170, 105–110. [Google Scholar] [CrossRef]
- Nijland, J.G.; Driessen, A.J.M. Engineering of pentose transport in Saccharomyces cerevisiae for biotechnological applications. Front. Bioeng. Biotechnol. 2020, 7, 464. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, K.M.V.; Mendes, V.; Carraro, C.B.; Taveira, I.C.; Oshiquiri, L.H.; Gupta, V.K.; Silva, R.N. Sugar transporters from industrial fungi: Key to improving second-generation ethanol production. Renew. Sustain. Energy Rev. 2020, 131, 109991. [Google Scholar] [CrossRef]
- Agrimi, G.; Steiger, M.G. Metabolite transport and its impact on metabolic engineering approaches. FEMS Microbiol. Lett. 2021, 368, fnaa211. [Google Scholar] [CrossRef]
- Soares-Silva, I.; Ribas, D.; Sousa-Silva, M.; Azevedo-Silva, J.; Rendulić, T.; Casal, M. Membrane transporters in the bioproduction of organic acids: State of the art and future perspectives for industrial applications. FEMS Microbiol. Lett. 2020, 367, fnaa118. [Google Scholar] [CrossRef]
- von Heijne, G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 2006, 7, 909–918. [Google Scholar] [CrossRef]
- Moraes, I.; Evans, G.; Sanchez-Weatherby, J.; Newstead, S.; Stewart, P.D.S. Membrane protein structure determination—The next generation. Biochim. Biophys. Acta 2014, 1838, 78–87. [Google Scholar] [CrossRef] [Green Version]
- Grisshammer, R.; Tate, C.G. Overexpression of integral membrane proteins for structural studies. Quar. Rev. Biophys. 2009, 28, 315–422. [Google Scholar] [CrossRef] [PubMed]
- Arinaminpathy, Y.; Khurana, E.; Engelman, D.M.; Gerstein, M.B. Computational analysis of membrane proteins: The largest class of drug targets. Drug Discov. Today 2009, 14, 1130–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Packer, M.S.; Liu, D.R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015, 16, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Yuen, C.M.; Liu, D.R. Dissecting protein structure and function using directed evolution. Nat. Methods 2007, 4, 995–997. [Google Scholar] [CrossRef]
- Dragosits, M.; Mattanovich, D. Adaptive laboratory evolution—Principles and applications for biotechnology. Microb. Cell Factories 2018, 12, 17–64. [Google Scholar] [CrossRef] [Green Version]
- Mans, R.; Daran, J.-M.G.; Pronk, J.T. Under pressure: Evolutionary engineering of yeast strains for improved performance in fuels and chemicals production. Curr. Opin. Biotechnol. 2018, 50, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, T.E.; Salazar, M.J.; Weng, L.L.; Palsson, B.O.; Feist, A.M. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 2019, 56, 1–16. [Google Scholar] [CrossRef]
- Lim, H.G.; Eng, T.; Banerjee, D.; Alarcon, G.; Lau, A.K.; Park, M.; Simmons, B.A.; Palsson, B.O.; Singer, S.W.; Mukhopadhyay, A.; et al. Generation of Pseudomonas putida KT2440 strains with efficient utilization of xylose and galactose via adaptive laboratory evolution. ACS Sustain. Chem. Eng. 2021, 9, 11512–11523. [Google Scholar] [CrossRef]
- Marques, W.L.; van der Woude, L.N.; Luttik, M.A.H.; van den Broek, M.; Nijenhuis, J.M.; Pronk, J.T.; van Maris, A.J.A.; Mans, R.; Gombert, A.K. Laboratory evolution and physiological analysis of Saccharomyces cerevisiae strains dependent on sucrose uptake via the Phaseolus vulgaris Suf1 transporter. Yeast 2018, 35, 639–652. [Google Scholar] [CrossRef]
- Henderson, R.K.; de Valk, S.C.; Poolman, B.; Mans, R. Energy coupling of membrane transport and efficiency of sucrose dissimilation in yeast. Metab. Eng. 2021, 65, 243–254. [Google Scholar] [CrossRef]
- Apel, A.R.; Ouellet, M.; Szmidt-Middleton, H.; Keasling, J.D.; Mukhopadhyay, A. Evolved hexose transporter enhances xylose uptake and glucose/xylose co-utilization in Saccharomyces cerevisiae. Sci. Rep. 2016, 6, 19512. [Google Scholar] [CrossRef]
- Brickwedde, A.; van den Broek, M.; Geertman, J.-M.; Magalhães, F.; Kuijpers, N.; Gibson, B.; Pronk, J.T.; Daran, J.-M.G. Evolutionary engineering in chemostat cultures for improved maltotriose fermentation kinetics in Saccharomyces pastorianus lager brewing yeast. Front. Microbiol. 2017, 8, 1690. [Google Scholar] [CrossRef] [PubMed]
- Jansen, M.L.A.; Daran-Lapujade, P.; de Winde, J.H.; Piper, M.D.W.; Pronk, J.T. Prolonged maltose-limited cultivation of Saccharomyces cerevisiae selects for cells with improved maltose affinity and hypersensitivity. Appl. Environ. Microbiol. 2004, 70, 1956–1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farwick, A.; Bruder, S.; Schadeweg, V.; Oreb, M.; Boles, E. Engineering of yeast hexose transporters to transport D-xylose without inhibition by D-glucose. Proc. Natl. Acad. Sci. USA 2014, 111, 5159–5164. [Google Scholar] [CrossRef] [Green Version]
- Nijland, J.G.; Shin, H.Y.; de Jong, R.M.; de Waal, P.P.; Klaassen, P.; Driessen, A.J.M. Engineering of an endogenous hexose transporter into a specific D-xylose transporter facilitates glucose-xylose co-consumption in Saccharomyces cerevisiae. Biotechnol. Biofuels 2014, 7, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Schmitz, O.; Alper, H.S. Enabling glucose/xylose co-transport in yeast through the directed evolution of a sugar transporter. Appl. Microbiol. Biotechnol. 2016, 100, 10215–10223. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, C.P.; Juroszek, J.; Beavan, M.J.; Ruby, F.M.S.; de Morais, S.M.F.; Rose, A.H. Ethanol dissipates the proton-motive force across the plasma-membrane of Saccharomyces cerevisiae. J. Gen. Microbiol. 1986, 132, 369–377. [Google Scholar] [CrossRef]
- Postma, E.P.; Verduyn, C.; Kuiper, K.; Scheffers, W.A.; Van Dijken, J.P. Substrate-accelerated death of Saccharornyces cerevisiae CBS 8066 under maltose stress. Yeast 1990, 6, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Entian, K.-D.; Kötter, P. Yeast genetic strain and plasmid collections. Methods Microbiol. 2007, 36, 629–666. [Google Scholar] [CrossRef]
- Gietz, R.D.; Woods, R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002, 350, 87–96. [Google Scholar] [CrossRef]
- van den Broek, M.; Bolat, I.; Nijkamp, J.F.; Ramos, E.; Luttik, M.A.H.; Koopman, F.; Geertman, J.-M.; de Ridder, D.; Pronk, J.T.; Daran, J.-M. Chromosomal copy number variation in Saccharomyces pastorianus is evidence for extensive genome dynamics in industrial lager brewing strains. Appl. Environ. Microbiol. 2015, 81, 6253–6267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Valk, S.C.; Bouwmeester, S.E.; de Hulster, E.; Mans, R. Engineering proton-coupled hexose uptake in Saccharomyces cerevisiae for improved ethanol yield. Biotechnol. Biofuels Bioproducts, under review.
- Marques, W.L.; Mans, R.; Henderson, R.K.; Marella, E.R.; Horst, J.T.; Hulster, E.; Poolman, B.; Daran, J.M.; Pronk, J.T.; Gombert, A.K.; et al. Combined engineering of disaccharide transport and phosphorolysis for enhanced ATP yield from sucrose fermentation in Saccharomyces cerevisiae. Metab. Eng. 2018, 45, 121–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verduyn, C.; Postma, E.; Scheffers, W.A.; van Dijken, J.P. Effect of benzoic acid on metabolic fluxes in yeasts: A continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 1992, 8, 501–517. [Google Scholar] [CrossRef] [Green Version]
- Verduyn, C.; Postma, E.; Scheffers, W.A.; van Dijken, J.P. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J. Gen. Microbiol. 1990, 136, 395–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nijkamp, J.F.; van den Broek, M.; Datema, E.; de Kok, S.; Bosman, L.; Luttik, M.A.; Daran-Lapujade, P.; Vongsangnak, W.; Nielsen, J.; Heijne, W.H.M.; et al. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb. Cell Factories 2012, 11, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques, W.L.; Mans, R.; Marella, E.R.; Cordeiro, R.L.; Van den Broek, M.; Daran, J.-M.G.; Pronk, J.T.; Gombert, A.K.; van Maris, A.J.A. Elimination of sucrose transport and hydrolysis in Saccharomyces cerevisiae: A platform strain for engineering sucrose metabolism. FEMS Yeast Res. 2017, 17, fox006. [Google Scholar] [CrossRef]
- Henderson, R.K.; Poolman, B. Proton-solute coupling mechanism of the maltose transporter from Saccharomyces cerevisiae. Sci. Rep. 2017, 7, 14375. [Google Scholar] [CrossRef]
- Deng, D.; Sun, P.; Yan, C.; Ke, M.; Jiang, X.; Xiong, L.; Ren, W.; Hirata, K.; Yamamoto, M.; Fan, S.; et al. Molecular basis of ligand recognition and transport by glucose transporters. Nature 2015, 526, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Postma, E.; Scheffers, W.A.; Van Dijken, J.P. Adaptation of the Kinetics of Glucose Transport to Environmental Conditions in the Yeast Candida utilis CBS 621: A Continuous-culture Study. J. Gen. Microbiol. 1988, 134, 1109–1116. [Google Scholar] [CrossRef] [Green Version]
- Santos, E.; Rodriguez, L.; Elorza, M.V.; Sentandreu, R. Uptake of sucrose by Saccharomyces cerevisiae. Arch. Biochem. Biophys. 1982, 216, 652–660. [Google Scholar] [CrossRef]
- Weusthuis, R.A.; Adams, H.; Scheffers, W.A.; van Dijken, J.P. Energetics and kinetics of maltose transport in Saccharomyces cerevisiae: A continuous culture study. Appl. Environ. Microbiol. 1993, 59, 3102–3109. [Google Scholar] [CrossRef] [Green Version]
- Henderson, R.K.; Ayudya, L.N.; Poolman, B. Characterization of Maltose-Binding Residues in the Central Cavity of Mal11. Mech. Proton-Coupled Transp. By Maltose Permease Saccharomyces cerevisiae. Ph.D. Thesis, Groningen University, Groningen, The Netherlands, 2019. [Google Scholar]
- Wisedchaisri, G.; Park, M.-S.; Iadanza, M.G.; Zheng, H.; Gonen, T. Proton-coupled sugar transport in the prototypical major facilitator superfamily protein XylE. Nat. Commun. 2014, 5, 4621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, N. Structural biology of the Major Facilitator Superfamily transporters. Annu. Rev. Biophys. 2015, 44, 257–283. [Google Scholar] [CrossRef]
- Regenberg, B.; Düring-Olsen, L.; Kielland-Brandt, M.C.; Holmberg, S. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet. 1999, 36, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Brandriss, M.C. Isolation and Preliminary Characterization of Saccharomyces cerevisiae Proline Auxotrophs. J. Bacteriol. 1979, 138, 816–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhoeven, M.D.; Bracher, J.M.; Nijland, J.G.; Bouwknegt, J.; Daran, J.-M.G.; Driessen, A.J.M.; van Maris, A.J.A.; Pronk, J.T. Laboratory evolution of a glucose-phosphorylation deficient, arabinose-fermenting S. cerevisiae strain reveals mutations in GAL2 that enable glucose-insensitive l-arabinose uptake. FEMS Yeast Res. 2018, 18, foy062. [Google Scholar] [CrossRef] [PubMed]
- Leandro, M.J.; Gonçalves, P.; Spencer-Martins, I. Two glucose/xylose transporter genes from the yeast Candida intermedia: First molecular characterization of a yeast xylose–H+ symporter. Biochem. J. 2006, 395, 543–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heredia, C.F.; de la Fuenta, G.; Sols, A. Metabolic studies with 2-deoxyhexoses. I. Mechanisms of inhibition of growth and fermentation in baker's yeast. Biochim. Biophys. Acta 1964, 86, 216–223. [Google Scholar] [CrossRef]
- Casal, M.; Paiva, S.; Andrade, R.P.; Gancedo, C.; Leão, C. The lactate-proton symport of Saccharomyces cerevisiae is encoded by JEN1. J. Bacteriol. 1999, 181, 2620–2623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cássio, F.; Leão, C.; van Uden, N. Transport of Lactate and Other Short-Chain Monocarboxylates in the Yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1987, 53, 509–513. [Google Scholar] [CrossRef] [Green Version]
- Baldi, N.; de Valk, S.C.; Sousa-Silva, M.; Casal, M.; Soares-Silva, I.; Mans, R. Evolutionary engineering reveals amino acid substitutions in Ato2 and Ato3 that allow improved growth of Saccharomyces cerevisiae on lactic acid. FEMS Yeast Res. 2021, 21, foab033. [Google Scholar] [CrossRef] [PubMed]
- Mans, R.; Hassing, J.-E.; Wijsman, M.; Giezekamp, A.; Pronk, J.T.; Daran, J.-M.G.; Van Maris, A.J.A. A CRISPR/Cas9-based exploration into the elusive mechanism for lactate export in Saccharomyces cerevisiae. FEMS Yeast Res. 2017, 17, fox085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guiard, B. Structure, expression and regulation of a nuclear gene encoding a mitochondrial protein: The yeast L(+)-lactate cytochrome c oxidoreductase (cytochrome b2). EMBO J. 1985, 4, 3265–3272. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).