Morphological, Molecular and Metabolic Characterization of the Pigmented Fungus Subramaniula asteroids
Abstract
:1. Introduction
2. Materials and Methods
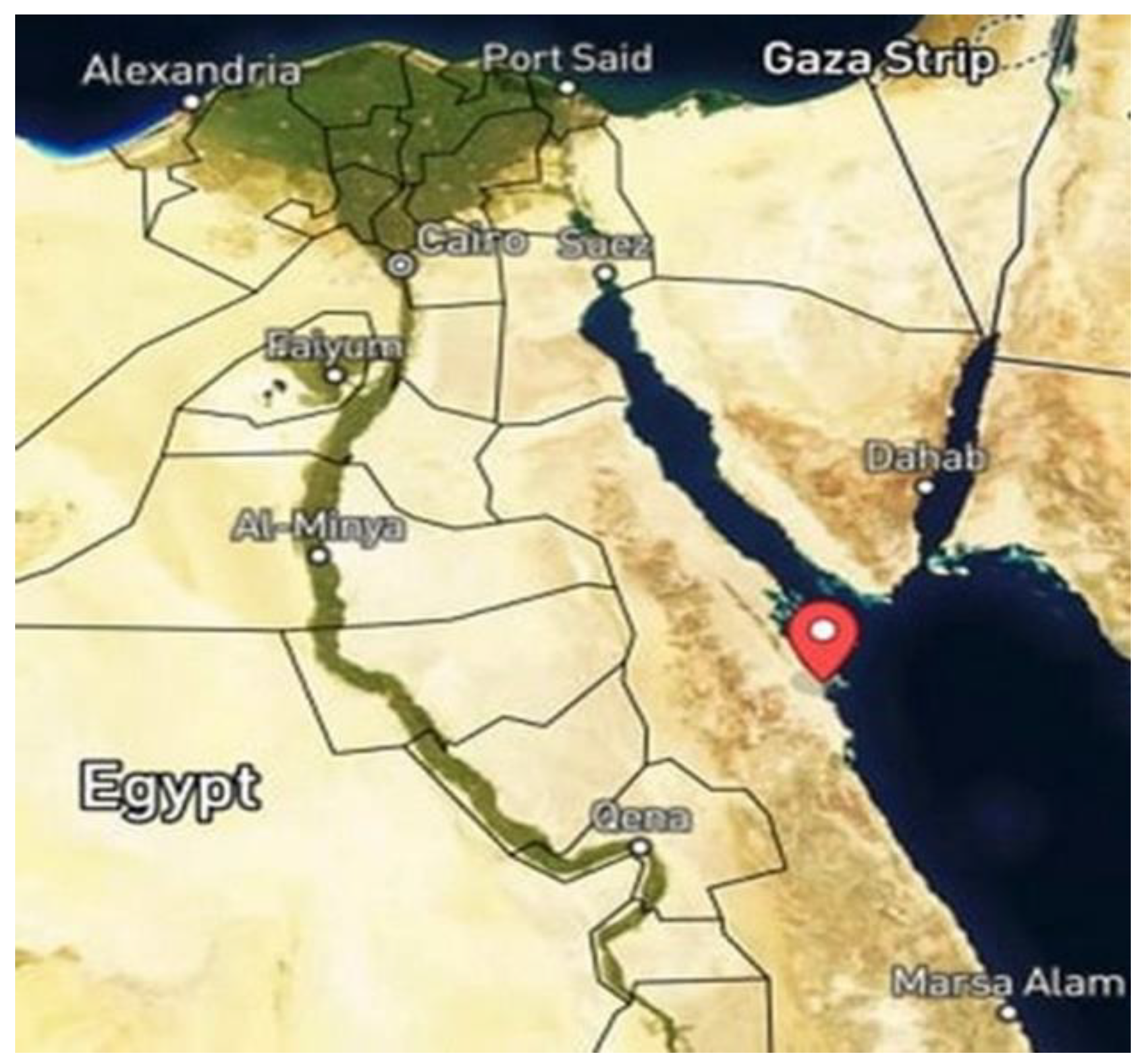
2.1. Collection of the Studied Soil Samples
2.2. Fungal Isolation and Morphological Characterization
2.3. Submerged Cultivation of the Fungal Isolate
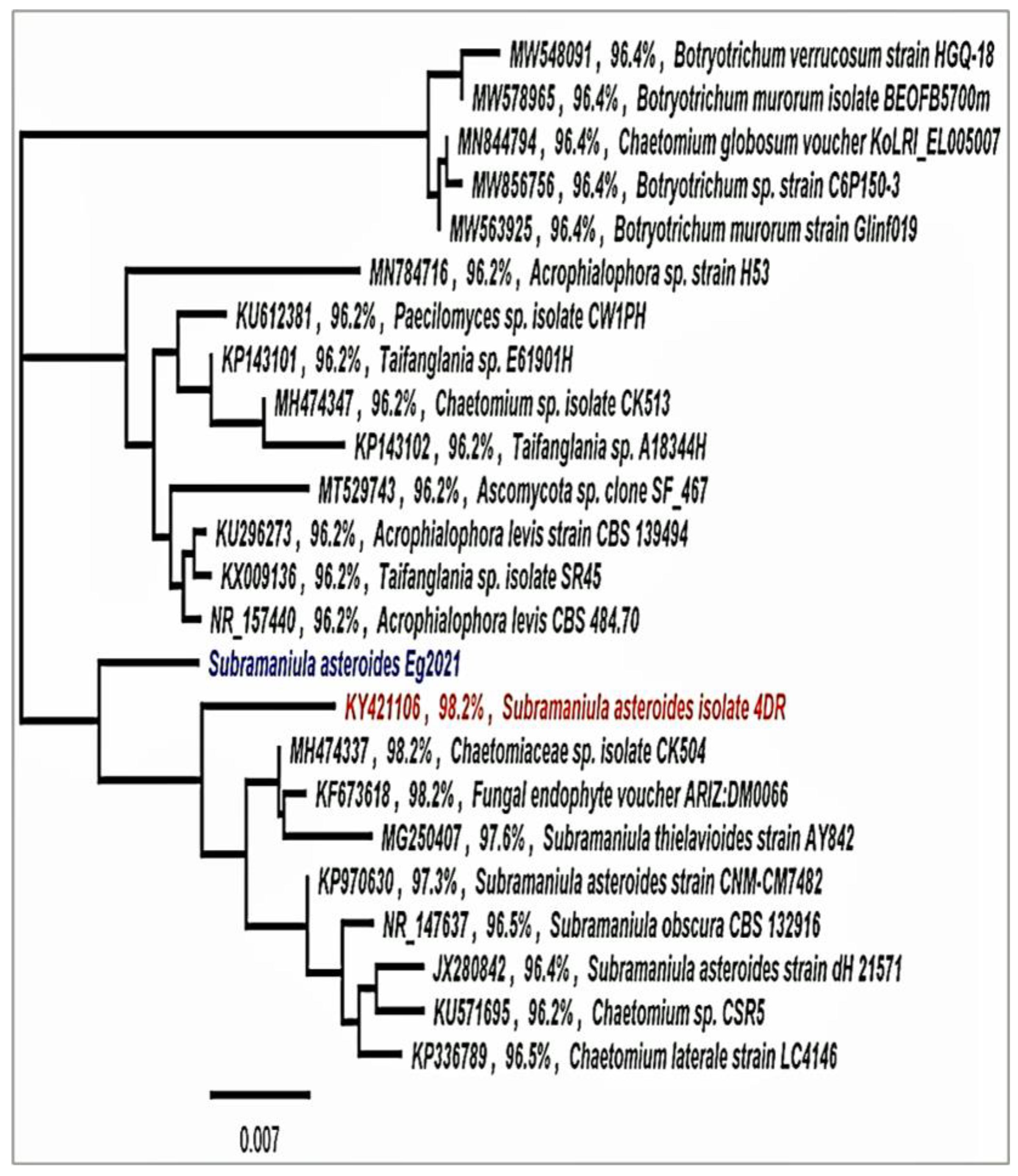
2.4. Molecular Identification
2.4.1. Extraction of Fungal DNA and Polymerase Chain Reaction (PCR) Amplification
2.4.2. Sequencing and Analysis
2.4.3. Polar and Non-Polar Fungal Metabolites Extraction
2.4.4. NMR Data Collection and Metabolite Identification
2.4.5. GC/MS Data and Metabolite Identification
3. Results
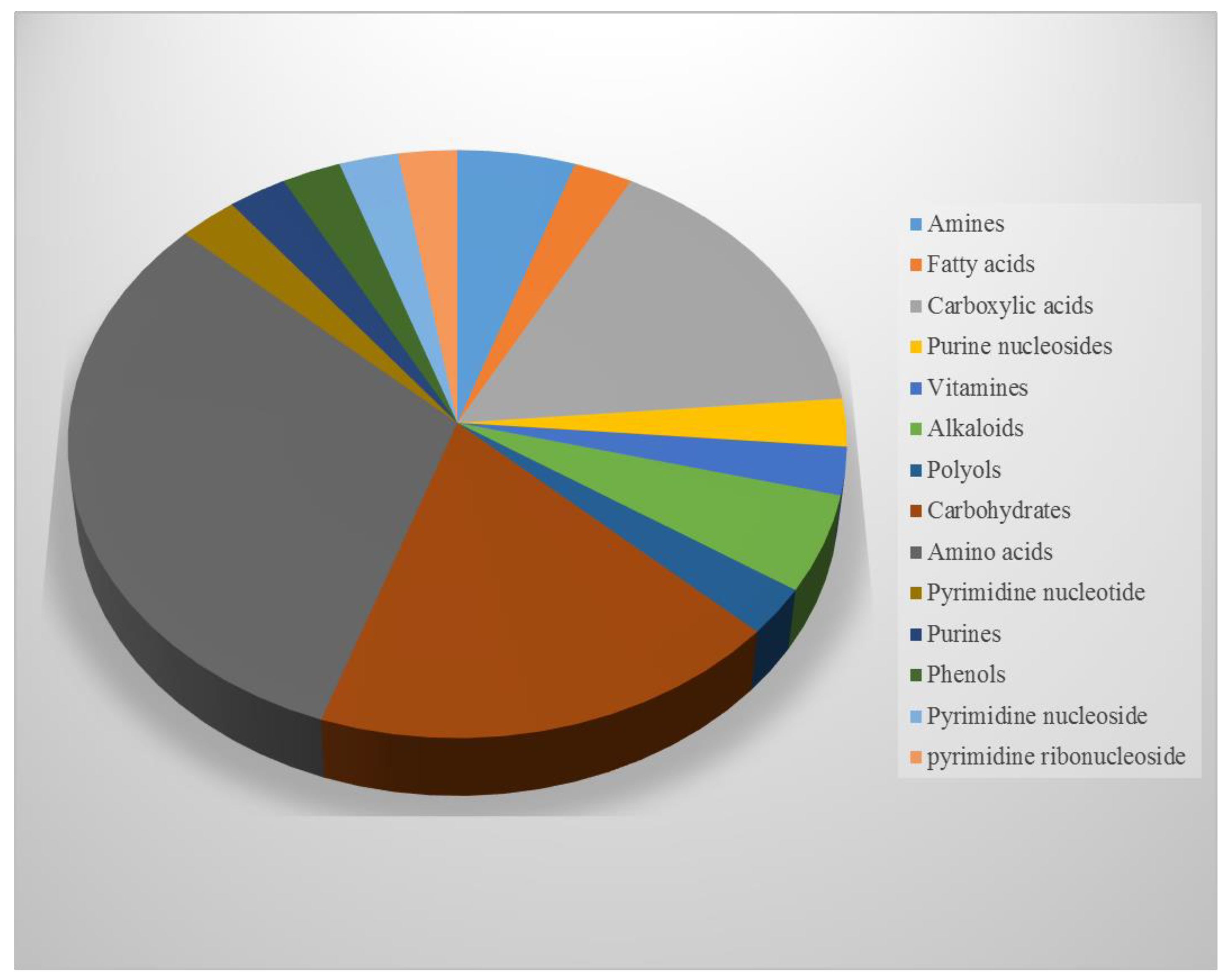
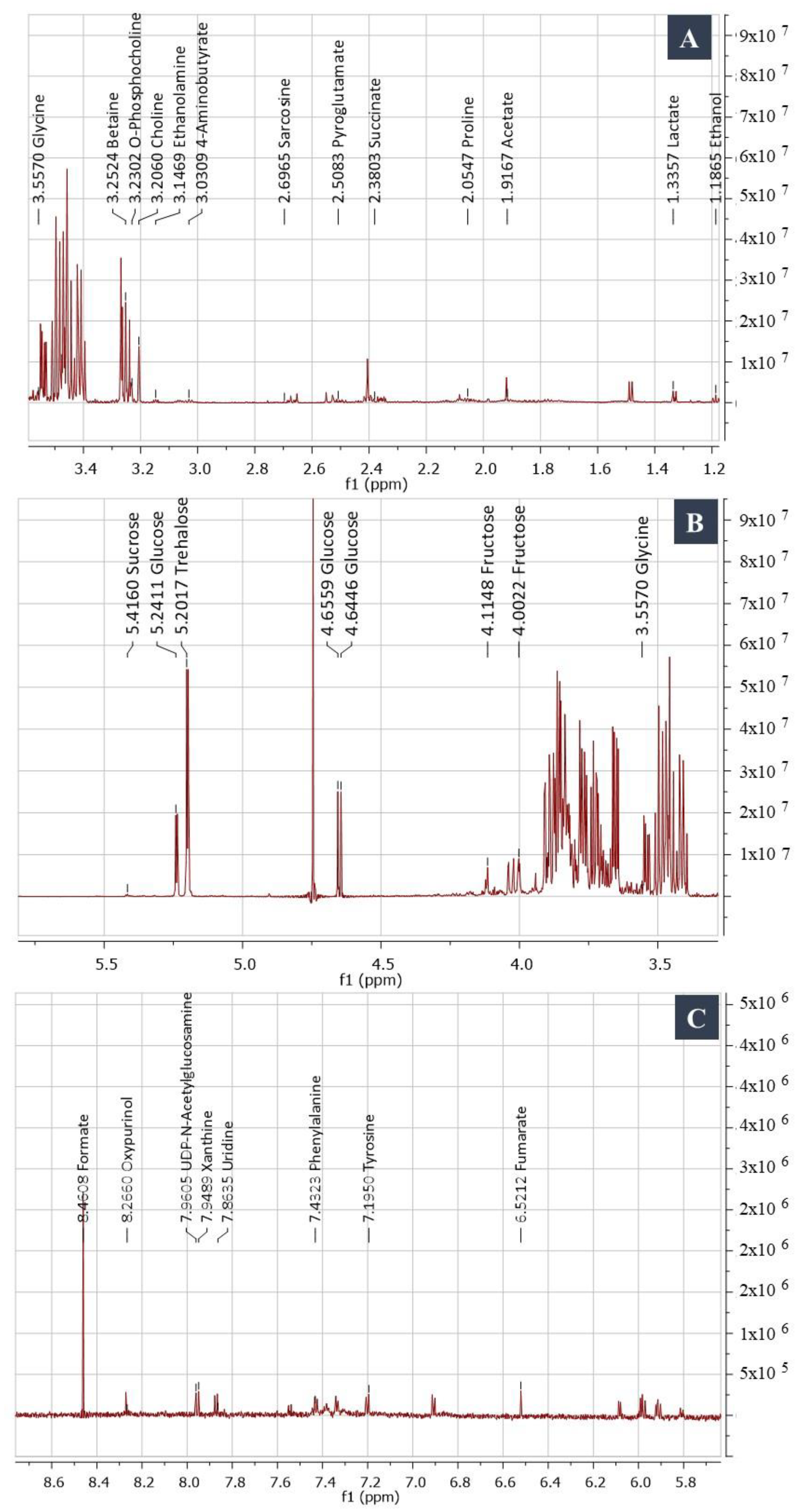
NMR Spectroscopy and GC/MS-Based Metabolic Profiling
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahmed, S.A.; Khan, Z.; Wang, X.; Moussa, T.A.; Al-Zahrani, H.S.; Almaghrabi, O.A.; Sutton, D.A.; Ahmad, S.; Groenewald, J.Z.; Alastruey-Izquierdo, A.; et al. Chaetomium-like Fungi Causing Opportunistic Infections in Humans: A Possible Role for Extremotolerance. Fungal Divers. 2015, 76, 11–26. [Google Scholar] [CrossRef]
- Santos, D.W.C.L.; Padovan, A.C.B.; Melo, A.S.A.; Gonçalves, S.S.; Azevedo, V.R.; Ogawa, M.M.; Veras, T.; Freitas, S.; Colombo, A.L. Molecular Identification of Melanised Non-Sporulating Moulds: A Useful Tool for Studying the Epidemiology of Phaeohyphomycosis. Mycopathologia 2013, 175, 445–454. [Google Scholar] [CrossRef] [PubMed]
- de Hoog, G.S.; van Diepeningen, A.D.; Mahgoub, E.S.; van de Sande, W.W.J. New Species of Madurella, Causative Agents of Black-Grain Mycetoma. J. Clin. Microbiol. 2012, 50, 988–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hubka, V.; Mencl, K.; Skorepova, M. Phaeohyphomycosis and Onychomycosis Due to Chaetomium Spp., Including the Fi Rst Report of Chaetomium Brasiliense Infection. Med. Mycol. 2011, 49, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ghavam, M.; Afzali, A.; Manca, M.L. Chemotype of Damask Rose with Oleic Acid (9 Octadecenoic Acid) and Its Antimicrobial Effectiveness. Sci. Rep. 2021, 11, 8027. [Google Scholar] [CrossRef]
- Garcia-Perez, I.; Posma, J.M.; Serrano-Contreras, J.I.; Boulangé, C.L.; Chan, Q.; Frost, G.; Stamler, J.; Elliott, P.; Lindon, J.C.; Holmes, E.; et al. Identifying Unknown Metabolites Using NMR-Based Metabolic Profiling Techniques. Nat. Protoc. 2020, 15, 2538–2567. [Google Scholar] [CrossRef]
- Kang, D.; Kim, J.; Choi, J.N.; Liu, K.H.; Lee, C.H. Chemotaxonomy of Trichoderma Spp. Using Mass Spectrometry-Based Metabolite Profiling. J. Microbiol. Biotechnol. 2011, 21, 5–13. [Google Scholar] [CrossRef] [Green Version]
- Fancy, S.A.; Rumpel, K. GC-MS-Based Metabolomics. In Biomarker Methods in Drug Discovery and Development; Wang, F., Ed.; Humana Press: Totowa, NJ, USA, 2008. [Google Scholar]
- Ja’afaru, M.I. Screening of Fungi Isolated from Environmental Samples for Xylanase and Cellulase Production. ISRN Microbiol. 2013, 2013, 283423. [Google Scholar] [CrossRef] [Green Version]
- Janli, D.; Purwanto, M.G.M.; Artadana, I.B.; Askitosari, T.D. Extraction and Toxicity Assay of Mycotoxin from Entomopathogenic Fungi Isolate of Kusuma Agrowisata Orchard Batu, Jawa Timur, Indonesia. KnE Life Sci. 2017, 3, 63. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Hae Choi, Y.; Casas Tapias, E.; Kyong Kim, H.; Lefeber, A.W.; Erkelens, C.; ThJ Verhoeven, J.; Brzin, J.; Zel, J.; Verpoorte, R. Metabolic Discrimination of Catharanthus Roseus Leaves Infected by Phytoplasma Using 1 H-NMR Spectroscopy and Multivariate Data Analysis 1. Plant Physiol. 2004, 135, 2398–2410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Southam, A.D.; Hines, A.; Viant, M.R. High-Throughput Tissue Extraction Protocol for NMR- and MS-Based Metabolomics. Anal. Biochem. 2008, 372, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Tuft, S.J.; Tullo, A.B. Fungal Keratitis in the United Kingdom 2003–2005. Eye 2009, 23, 1308–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gower, E.W.; Keay, L.J.; Oechsler, R.A.; Iovieno, A.; Alfonso, E.C.; Jones, D.B.; Colby, K.; Tuli, S.S.; Patel, S.R.; Lee, S.M.; et al. Trends in Fungal Keratitis in the United States, 2001 to 2007. Ophthalmology 2010, 117, 2263–2267. [Google Scholar] [CrossRef]
- Farooq, A.V.; Shukla, D. Herpes Simplex Epithelial and Stromal Keratitis: An Epidemiologic Update. Surv. Ophthalmol. 2012, 57, 448–462. [Google Scholar] [CrossRef] [Green Version]
- Fanselow, N.; Sirajuddin, N.; Yin, X.T.; Huang, A.J.W.; Stuart, P.M. Acanthamoeba Keratitis, Pathology, Diagnosis and Treatment. Pathogens 2021, 10, 323. [Google Scholar] [CrossRef]
- Han, H.; Gao, Y.; Chai, M.; Zhang, X.; Liu, S.; Huang, Y.; Jin, Q.; Grzybowski, A.; Ji, J.; Yao, K. Biofilm Microenvironment Activated Supramolecular Nanoparticles for Enhanced Photodynamic Therapy of Bacterial Keratitis. J. Control Release 2020, 327, 676–687. [Google Scholar] [CrossRef]
- Donovan, C.; Arenas, E.; Ayyala, R.S.; Margo, C.E.; Espana, E.M. Fungal Keratitis: Mechanisms of Infection and Management Strategies. Surv. Ophthalmol. 2021, 67, 758–769. [Google Scholar] [CrossRef]
- Xie, L.; Zhong, W.; Shi, W.; Sun, S. Spectrum of Fungal Keratitis in North China. Ophthalmology 2006, 113, 1943–1948. [Google Scholar] [CrossRef]
- Xie, L.; Zhai, H.; Zhao, J.; Sun, S.; Shi, W.; Dong, X. Antifungal Susceptibility for Common Pathogens of Fungal Keratitis in Shandong Province, China. Am. J. Ophthalmol. 2008, 146, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Bourcier, T.; Sauer, A.; Dory, A.; Denis, J.; Sabou, M. Fungal Keratitis. J. Fr. Ophtalmol. 2017, 40, e307–e313. [Google Scholar] [CrossRef] [PubMed]
- Cultrera, R.; Torelli, R.; Sarnicola, C.; Segala, D.; Mengoli, A.; Chiaretto, G.; Perri, P.; Sanguinetti, M. Identification and Molecular Characterization of Subramaniula Asteroides Causing Human Fungal Keratitis: A Case Report. BMC Infect. Dis. 2021, 21, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V. Metabolic Engineering of Filamentous Fungi. In Metabolic Engineering: Concepts and Applications, Volume 13a; Wiley: New York, NY, USA, 2021. [Google Scholar]
- George, T.K.; Devadasan, D.; Jisha, M.S. Chemotaxonomic Profiling of Penicillium Setosum Using High-Resolution Mass Spectrometry (LC-Q-ToF-MS). Heliyon 2019, 5, e02484. [Google Scholar] [CrossRef] [Green Version]
- Beale, D.J.; Pinu, F.R.; Kouremenos, K.A.; Poojary, M.M.; Narayana, V.K.; Boughton, B.A.; Kanojia, K.; Dayalan, S.; Jones, O.A.H.; Dias, D.A. Review of Recent Developments in GC-MS Approaches to Metabolomics-Based Research. Metabolomics 2018, 14, 152. [Google Scholar] [CrossRef]
- Tripathi, T.; Dhobi, M.; Kalaiselvan, V. Comparative Metabolic Profiling, Isolation of Alkylated Phenols and Antioxidant Activity of Roots of Plumbago Species Using GC-MS and NMR Based Metabolomics Study. Nat. Prod. Res. 2022; online ahead of print. [Google Scholar]
- Zhang, B.; Powers, R.; O’day, E.M. Metabolites Article Evaluation of Non-Uniform Sampling 2D 1 H-13 C HSQC Spectra for Semi-Quantitative Metabolomics. Metabolites 2020, 10, 203. [Google Scholar] [CrossRef]
- Tronchin, G.; Pihet, M.; Bezerra, L.M.L.; Bouchara, J.-P. Adherence Mechanisms in Human Pathogenic Fungi. Med Mycol. 2008, 46, 749–772. [Google Scholar] [CrossRef] [Green Version]
- Thammahong, A.; Puttikamonkul, S.; Perfect, J.R.; Brennan, R.G.; Cramer, R.A. Central Role of the Trehalose Biosynthesis Pathway in the Pathogenesis of Human Fungal Infections: Opportunities and Challenges for Therapeutic Development. Microbiol. Mol. Biol. Rev. 2017, 81, e00053-16. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Jiang, X.; Wu, W.; Wang, M.; Hamid, M.I.; Xiang, M.; Liu, X. Genomic, Transcriptomic, and Proteomic Analysis Provide Insights Into the Cold Adaptation Mechanism of the Obligate Psychrophilic Fungus Mrakia Psychrophila. G3 Genes Genomes Genet. 2016, 6, 3603–3613. [Google Scholar] [CrossRef] [Green Version]
- Zaragoza, O.; Blazquez, M.A.; Gancedo, C. Disruption of the Candida Albicans TPS1 Gene Encoding Trehalose-6-Phosphate Synthase Impairs Formation of Hyphae and Decreases Infectivity. J. Bacteriol. 1998, 180, 3809–3815. [Google Scholar] [CrossRef]
- Alvarez-Peral, F.J.; Zaragoza, O.; Pedren, Y.; Argu, J.-C. Protective Role of Trehalose during Severe Oxidative Stress Caused by Hydrogen Peroxide and the Adaptive Oxidative Stress Response in Candida Albicans. Microbiology 2002, 148, 2599–2606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petzold, E.W.; Himmelreich, U.; Mylonakis, E.; Rude, T.; Toffaletti, D.; Cox, G.M.; Miller, J.L.; Perfect, J.R. Characterization and Regulation of the Trehalose Synthesis Pathway and Its Importance in the Pathogenicity of Cryptococcus Neoformans. Infect. Immun. 2006, 74, 5877–5887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garbe, E.; Vylkova, S. Role of Amino Acid Metabolism in the Virulence of Human Pathogenic Fungi. Curr. Clin. Microbiol. Rep. 2019, 6, 108–119. [Google Scholar] [CrossRef] [Green Version]
- Gerald, F.; Silao, S.; Ryman, K.; Wallströ, A.; Id, B.; Udekwuid, K.; Ljungdahlid, O. Mitochondrial Proline Catabolism Activates Ras1/CAMP/PKA-Induced Filamentation in Candida Albicans. PLoS Genet. 2019, 15, e1007976. [Google Scholar] [CrossRef] [Green Version]
- Augustin, A.J.; Boker, T.; Blumenröder, S.H.; Lutz, J.; Spitznas, M. Free Radical Scavenging and Antioxidant Activity of Allopurinol and Oxypurinol in Experimental Lens-Induced Uveitis. Investig. Ophthalmol. Vis. Sci. 1994, 35, 3897–3904. [Google Scholar]
- Baldus, S.; Köster, R.; Chumley, P.; Heitzer, T.; Rudolph, V.; Ostad, M.A.; Warnholtz, A.; Staude, H.J.; Thuneke, F.; Koss, K.; et al. Oxypurinol Improves Coronary and Peripheral Endothelial Function in Patients with Coronary Artery Disease. Free Radic. Biol. Med. 2005, 39, 1184–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, N.; Kumar Shreshtha, A.; Thakur, M.S.; Patra, S. Xanthine Scaffold: Scope and Potential in Drug Development. Heliyon 2018, 4, 829. [Google Scholar] [CrossRef] [Green Version]
- Youssef, N.H.; Qari, S.H.; Behiry, S.I.; Dessoky, E.S.; El-Hallous, E.I.; Elshaer, M.M.; Kordy, A.; Maresca, V.; Abdelkhalek, A.; Heflish, A.A.; et al. Antimycotoxigenic Activity of Beetroot Extracts against Alternaria Alternata Mycotoxins on Potato Crop. Appl. Sci. 2021, 11, 4239. [Google Scholar] [CrossRef]
- Chen, S.; Liu, J.; Gong, H.; Yang, D. Identification and Antibacterial Activity of Secondary Metabolites from Taxus Endophytic Fungus. Chin. J. Biotechnol. 2009, 25, 368–374. [Google Scholar]
- Aparna, V.; Dileep, K.V.; Mandal, P.K.; Karthe, P.; Sadasivan, C.; Haridas, M. Anti-Inflammatory Property of n-Hexadecanoic Acid: Structural Evidence and Kinetic Assessment. Chem. Biol. Drug Des. 2012, 80, 434–439. [Google Scholar] [CrossRef]
- Ravi, L.; Krishnan, K. Cytotoxic Potential of N-Hexadecanoic Acid Extracted from Kigelia Pinnata Leaves. Asian J. Cell Biol. 2016, 12, 20–27. [Google Scholar] [CrossRef] [Green Version]
- Sales-Campos, H.; Reis de Souza, P.; Crema Peghini, B.; Santana da Silva, J.; Ribeiro Cardoso, C. An Overview of the Modulatory Effects of Oleic Acid in Health and Disease. Mini-Rev. Med. Chem. 2013, 13, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Hameed, I.H.; Hussein, H.J.; Kareem, M.A.; Hamad, N.S. Identification of Five Newly Described Bioactive Chemical Compounds in Methanolic Extract of Mentha Viridis by Using Gas Chromatography-Mass Spectrometry (GC-MS). J. Pharmacogn. Phytother. 2015, 7, 107–125. [Google Scholar] [CrossRef]

| Compound Name | Chemical Formula | Weight (Da) | Assigned 13C Chemical Shift from 1H-13C HSQC (2D NMR) | Assigned 1H Chemical Shift from 1H (1D NMR) | Coupling Constant | |
|---|---|---|---|---|---|---|
| 1 | 3-Hydroxyisovalerate | C5H10O3 | 118.13 | 30.6 (CH3) 52.0 (CH2) | 1.26 (CH3, s) 2.35 (CH2, s) | - - |
| 2 | 4-Aminobutyrate | C4H9NO2 | 103.11 | 26.4 (CH2) 37.0 (CH2) 42.0 (CH2) | 1.90 (CH2, m) 2.28 (CH2, t) 2.99 (CH2, t) | - 7.35 7.40 |
| 3 | Acetate | C2H4O2 | 60.05 | 26.2 (CH3) | 1.92 (CH3, s) | - |
| 4 | Alanine | C3H7NO2 | 89.09 | 53.5 (Cα) 18.9 (Cβ) | 3.78 (Hα, q) 1.46 (Hβ, d) | 7.2, 7.26 |
| 5 | Arginine | C6H14N4O2 | 174.20 | 26.6 (Cγ) 30.3 (Cβ) 57.1 (Cα) | 1.64 (Hγ, m) 1.92 (Hβ, m) 3.75 (Hα, t) | - - 6.2 |
| 6 | Betaine | C5H11NO2 | 117.14 | 69.2 (CH2) 56.2 (CH3) | 3.90 (Hα, s) 3.26 (Hβ, s) | - - |
| 7 | Choline | C5H14NO | 104.17 | 58.6 (Cα) 56.8 (Cγ) 70.4 (Cβ) | 4.06 (Hα, m) 3.21 (Hγ, s) 3.52 (Hβ, m) | - - - |
| 8 | Ethanol | C2H6O | 46.06 | 119.5 (CH3) 60.3 (CH2) | 1.17 (CH3, t) 3.63 (CH2, q) | 6.7 6.7 |
| 9 | Ethanolamine | C2H7NO | 61.08 | 60.5 (CH2) - | 3.8 (CH2, t) 3.13 (CH2, t) | 5.21 |
| 10 | Formate | CH2O2 | 46.02 | - | 8.44 (CH, s) | - |
| 11 | Fructose | C6H12O6 | 180.15 | 78.2 (3CH) 66.1 (6CH2) 72.0 (5CH) 66.6 (1CH) | 4.12 (3CH, dd) 4.02 (6CH2, dd) 4.00 (5CH, m) 3.56 (1CH, m) | 12.72, 1.04 7.7, 5.4 |
| 12 | Fumarate | C4H4O4 | 116.07 | 138.0 (CH) | 6.49 (CH, s) | - |
| 13 | Gallate | C7H6O5 | 170.11 | 111.8 (CH) | 7.00 (CH, s) | - |
| 14 | Glucose | C6H12O6 | 180.15 | 98.9 (1βCH) 94.8 (1αCH) 74.4 (2αCH) 72.6 (4CH) 63.7 (6CH) | 4.66 (1βCH, d) 5.24 (1αCH, d) 3.53(2αCH, m) 3.41 (4CH, m) 3.74 (5βCH, m) | 7.88 3.68 - - - |
| 15 | Glucose-6-P | C6H13O9P | 260.13 | 94.9 (1αCH) 98.9 (1βCH) 65.4 (5βCH) | 5.2 (1αCH, d) 4.6 (1βCH, d) 4.0 (5βCH, m) | 3.75 8.0 - |
| 16 | Glutamate | C5H9NO4 | 147.12 | 29.7 (Cβ) 36.2 (Cγ) 57.3 (Cα) | 2.04 (Hβ, m) 2.35 (Hγ, m) 3.74 (Hα, dd) | - - 7.1, 4.7 |
| 17 | Glycine | C2H5NO2 | 75.06 | 44.3 (Cα) | 3.56 (Hα, s) | - |
| 18 | Isoleucine | C6H13NO2 | 131.17 | 26.8 (Cγ) 17.5 (Cγ) | 1.29 (Hγ, m) 1.00 (Hγ, d) 0.91 (Hδ, t) | - 7.0 7.1 |
| 19 | Lactate | C3H6O3 | 90.07 | 22.1 (CH3) | 1.33 (CH3, d) | 6.88 |
| 20 | Leucine | C6H13NO2 | 131.17 | 24.7 (Cδ) | 0.98 (Hδ, t) | 6.1 |
| 22 | Methylsuccinate | C5H8O4 | 132.11 | 19.7 (CH3) 44.9 (CH2) | 1.01 (CH3, d) 2.11 (CH2, m) | 6.7 |
| 23 | O-Phosphocholine | C5H15NO4P | 184.15 | 56.5 (CH3) 86.9 (CH2) 60.6 (CH2) | 3.20 (CH3, s) 3.58 (CH2, m) 4.15 (CH2, m) | - - - |
| 24 | Oxypurinol | C5H4N4O2 | 152.11 | - | 8.20 (CH, s) | - |
| 25 | Phenylalanine | C9H11NO2 | 165.18 | 39.2 (Cβ) 58.6 (Cα) 132.1(Cδ) 130.4(Cζ) 131.8(Cε) | 3.10 (Hβ, dd) 3.39 (Hα, q) 7.31 (Hδ, m) 7.36 (Hζ, m) 7.41(Hε, m) | 15.4,6.3 |
| 26 | Proline | C5H9NO2 | 115.13 | 64.2 (Cα) 49.0 (Cδ) 31.9 (Cβ) 26.7 (Cγ) | 4.13 (Hα, m) 3.41 (Hδ, m) 2.36 (Hβ, m) - | - - |
| 27 | Pyroglutamate | C5H7NO3 | 129.11 | 27.9 (Cβ) 60.9 (Cα) | 2.40 (Hγ,m) 4.16 (Hα, dd) | 9.0, 5.9 |
| 29 | Sarcosine | C3H7NO2 | 89.09 | 35.5 (CH3) 53.4 (CH2) | 2.73 (CH3, s) 3.61 (CH2, s) | - - |
| 31 | Succinate | C4H6O4 | 118.08 | 37.1 (CH2) | 2.41 (CH2, s) | - |
| 32 | Sucrose | C12H22O11 | 342.29 | 95.1 (1CH) 79.8 (3′CH) 76.8 (4′CH) 73.9 (2CH) 72.1 (3CH) 65.3(6CH2) 64.2 (1′CH2) 72.1 (3CH) | 5.42 (1CH, d) 4.22 (3′CH, d) 4.06 (4′CH, t) 3.56 (2CH, m) 3.48 (3CH, m) 3.83 (6CH2, m) 3.69 (1′CH2, s) 3.48 (3CH, m) | 3.89 8.8 38.6 |
| 34 | Trehalose | C12H22O11 | 342.30 | 95.8 (1CH) 73.6 (2CH) | 5.20 (1CH, d) 3.66 (2CH, dd) 3.46 (CH, t) | 3.73 10, 3.97 9.54 |
| 35 | Tyrosine | C9H11NO3 | 181.18 | 38.2 (Cβ) 118.9(Cε) 133.9(Cδ) | 3.10 (Hβ, dd) 6.88 (Hε, m) 7.18 (Hδ, m) | 7.30, 14.30 |
| 36 | UMP | C9H13N2O9P | 324.18 | 105.2 (CH) 90.9 (CH) | 5.97 (CH, m) 5.98 (CH, m) | - - |
| 37 | Uridine | C9H12N2O6 | 244.20 | 72.0 (CH) 92.0 (CH) | 4.21 (CH, dd) 5.90 (CH, d) | 9.0, 4.3 9.0 |
| 38 | Valine | C5H11NO2 | 117.14 | 63.0 (Cα) 20.8 (Cγ) 19.5 (Cγ) | 3.60 (Hα, d) 0.98 (Hγ, d) 1.03 (Hγ, d) | 4.54 7.00 7.00 |
| 39 | Xanthine | C5H4N4O2 | 152.11 | 140.3 (CH) | 7.90 (CH, s) | - |
| Compound Name | Chemical Formula | Weight (Da) | RT (min) | Area % | |
|---|---|---|---|---|---|
| 1 | Tetrahydro-2-(12-pentadecynyloxy)-2H-pyran | C20H36O2 | 308 | 7.20 | 1.56 |
| 2 | 1-Dodecanamine, N,N-dimethyl- | C14H31N | 213 | 11.95 | 27.96 |
| 3 | 2,4-Di-tert-butylphenol | C14H22O | 206 | 12.22 | 1.30 |
| 4 | 1-Tetradecanamine, N,N-dimethyl- | C16H35N | 241 | 15.75 | 10.10 |
| 5 | 2H-Pyran-3-ol, tetrahydro-2,2,6-trimethyl-6-(4-meth yl-3-cyclohexen-1-yl)-, [3S-[3à,6à(R*)]] | C16H26O2 | 250 | 16.55 | 1.30 |
| 6 | Phen-1,4-diol, 2,3-dimethyl-5-trifluoromethyl- | C9H9F3O2 | 206 | 18.64 | 0.92 |
| 7 | 7,9-Di-tert-butyl-1-oxaspiro(4,5)dec a-6,9-diene-2,8-dione | C17H24O3 | 276 | 19.51 | 0.93 |
| 8 | Cyclopropanebutanoic acid, 2-[[2-[[2-[(2-pentylcyclopropyl)meth yl] cyclopropyl]methyl]cyclopropyl] methyl]-, methyl ester | C25H42O2 | 374 | 19.61 | 0.91 |
| 9 | n-Hexadecanoic acid (palmitic acid) | C16H32O2 | 256 | 20.28 | 4.13 |
| 10 | Hexadecanoic acid, ethyl ester | C18H36O2 | 284 | 20.72 | 7.05 |
| 11 | 9-Octadecenoic acid (Z)-, methyl ester | C19H36O2 | 296 | 22.37 | 1.41 |
| 12 | 1-(N-Benzyl-N-methylamino)-4-methoxybutan-2-one | C13H19NO2 | 221 | 22.55 | 4.68 |
| 13 | Oleic acid | C18H34O2 | 282 | 23.03 | 4.85 |
| 14 | Ethyl (9E,12E)-octadeca-9,12-dienoate | C20H36O2 | 308 | 23.30 | 8.31 |
| 15 | Ethyl Oleate | C20H38O2 | 310 | 23.39 | 9.27 |
| 16 | octadecenoic acid ethyl ester | C20H40O2 | 312 | 23.79 | 1.94 |
| 17 | (1RS,2RS,1’SR)-2-[(Dimethylamino) phenylmethyl] cycloctanol | C17H27NO | 261 | 25.60 | 1.25 |
| 18 | Squalene | C30H50 | 410 | 32.05 | 6.42 |
| 19 | 9(11)-Dehydroergosterol tosylate | C35H48O3S | 548 | 33.57 | 2.09 |
| 20 | Glycidyl oleate | C21H38O3 | 338 | 35.96 | 1.49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Sayed, H.; Osman, M.E.; Abdelsalam, A.; Boroujerdi, A.; Sonbol, H.; Elsaba, Y.M. Morphological, Molecular and Metabolic Characterization of the Pigmented Fungus Subramaniula asteroids. J. Fungi 2022, 8, 1149. https://doi.org/10.3390/jof8111149
El-Sayed H, Osman ME, Abdelsalam A, Boroujerdi A, Sonbol H, Elsaba YM. Morphological, Molecular and Metabolic Characterization of the Pigmented Fungus Subramaniula asteroids. Journal of Fungi. 2022; 8(11):1149. https://doi.org/10.3390/jof8111149
Chicago/Turabian StyleEl-Sayed, Heba, Mohamed E. Osman, Asmaa Abdelsalam, Arezue Boroujerdi, Hana Sonbol, and Yasmin M. Elsaba. 2022. "Morphological, Molecular and Metabolic Characterization of the Pigmented Fungus Subramaniula asteroids" Journal of Fungi 8, no. 11: 1149. https://doi.org/10.3390/jof8111149
APA StyleEl-Sayed, H., Osman, M. E., Abdelsalam, A., Boroujerdi, A., Sonbol, H., & Elsaba, Y. M. (2022). Morphological, Molecular and Metabolic Characterization of the Pigmented Fungus Subramaniula asteroids. Journal of Fungi, 8(11), 1149. https://doi.org/10.3390/jof8111149






