Cloning and Molecular Characterization of CmOxdc3 Coding for Oxalate Decarboxylase in the Mycoparasite Coniothyrium minitans
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Strains, Cultural Media and Cultural Conditions
2.2. Gene Cloning and Analysis
2.3. DNA Extraction and Southern Blotting
2.4. RNA Extraction and Quantification of Gene Expression
2.5. Gene Expression Analysis of CmOxdc3
2.6. Gene Knockout and Complementation
2.7. Determination of Sensitivity to OA
2.8. Assaying OA Degradation and Mycoparasitic Activities
2.9. Confocal Microscopy
2.10. Statistical Analysis
3. Results
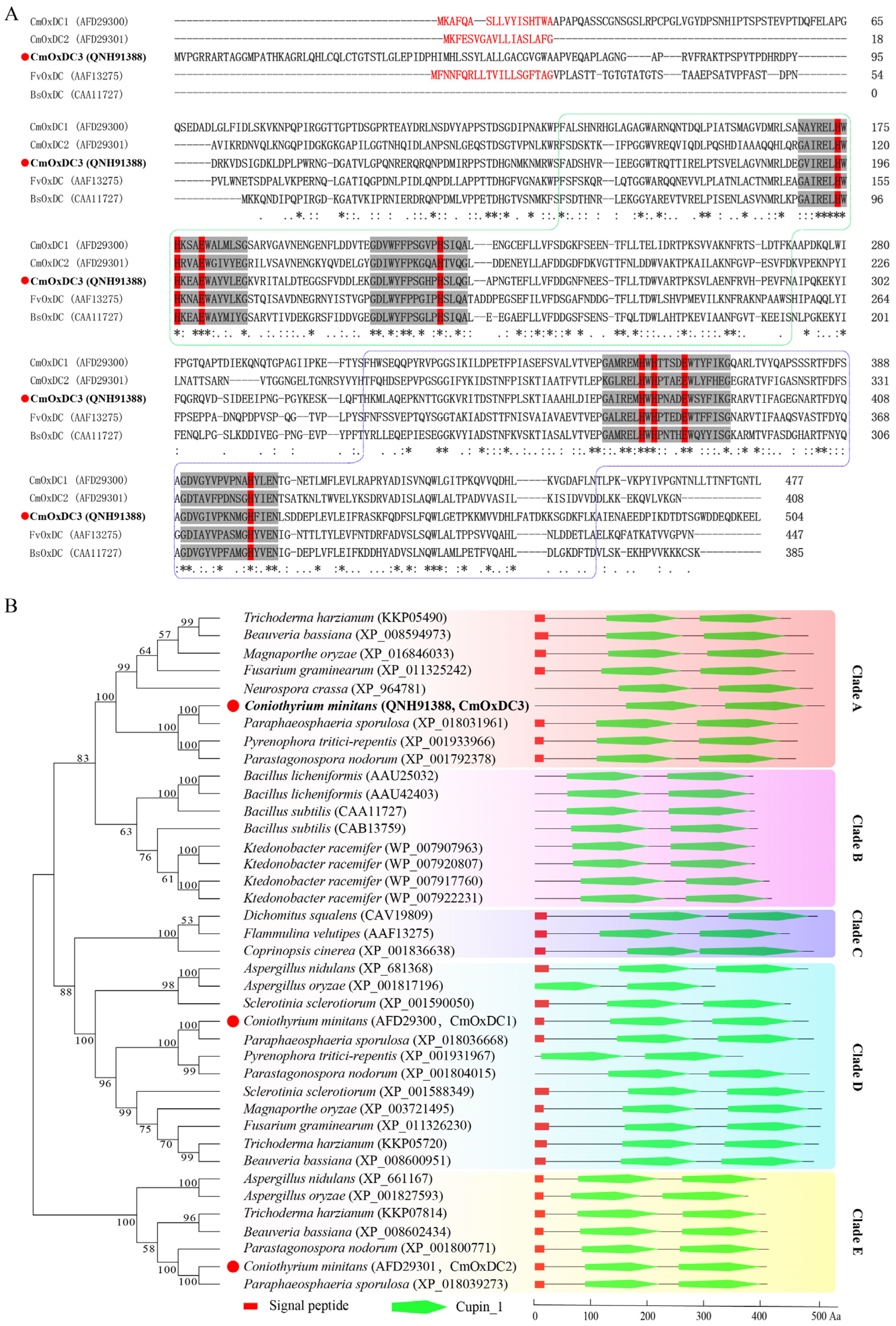
3.1. Identification and Characteristics of CmOxdc3
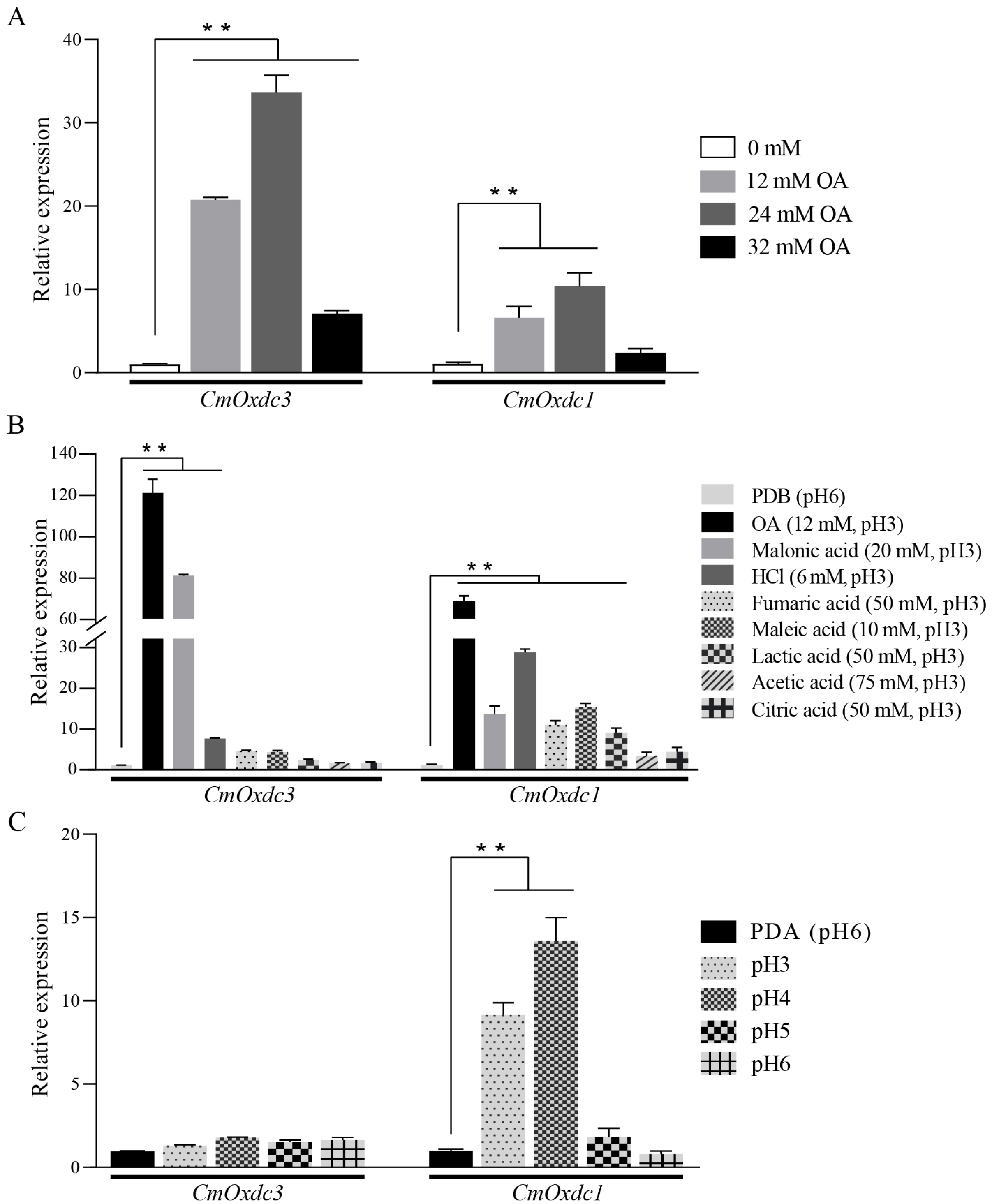
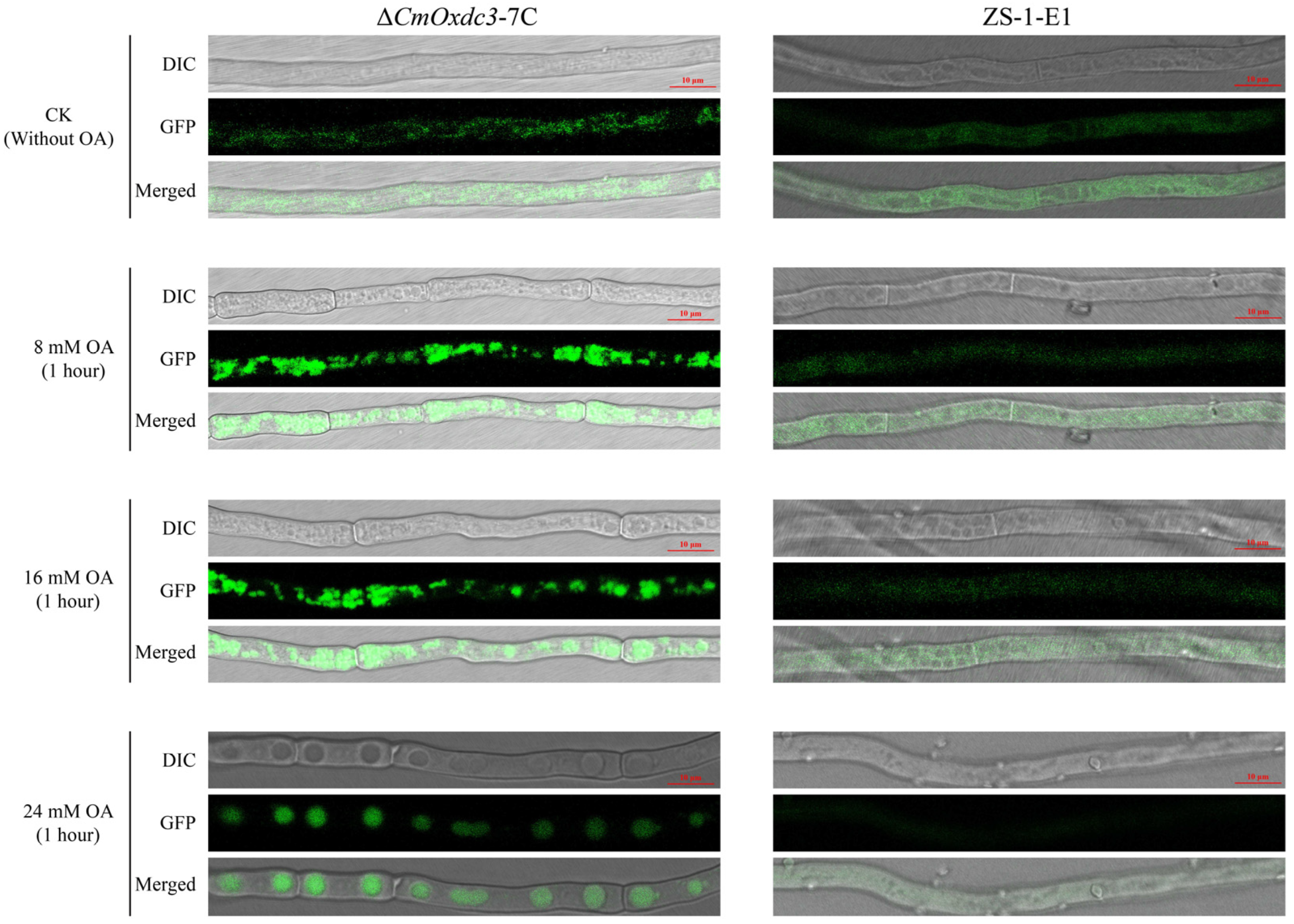
3.2. Expression Pattern and Subcellular Localization of CmOxdc3
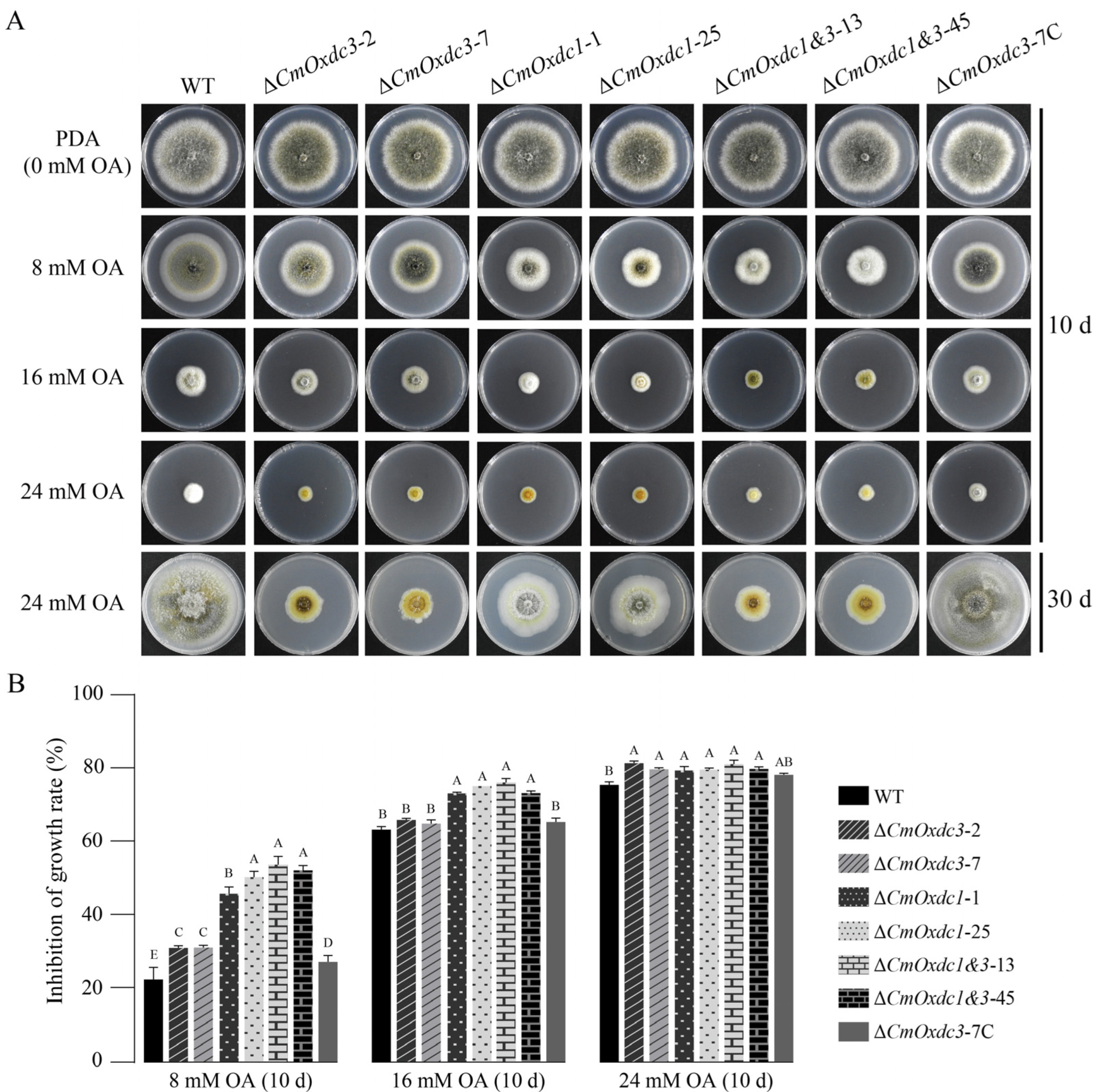
3.3. Effect of Disruption of CmOxdc3 on Sensitivity to OA
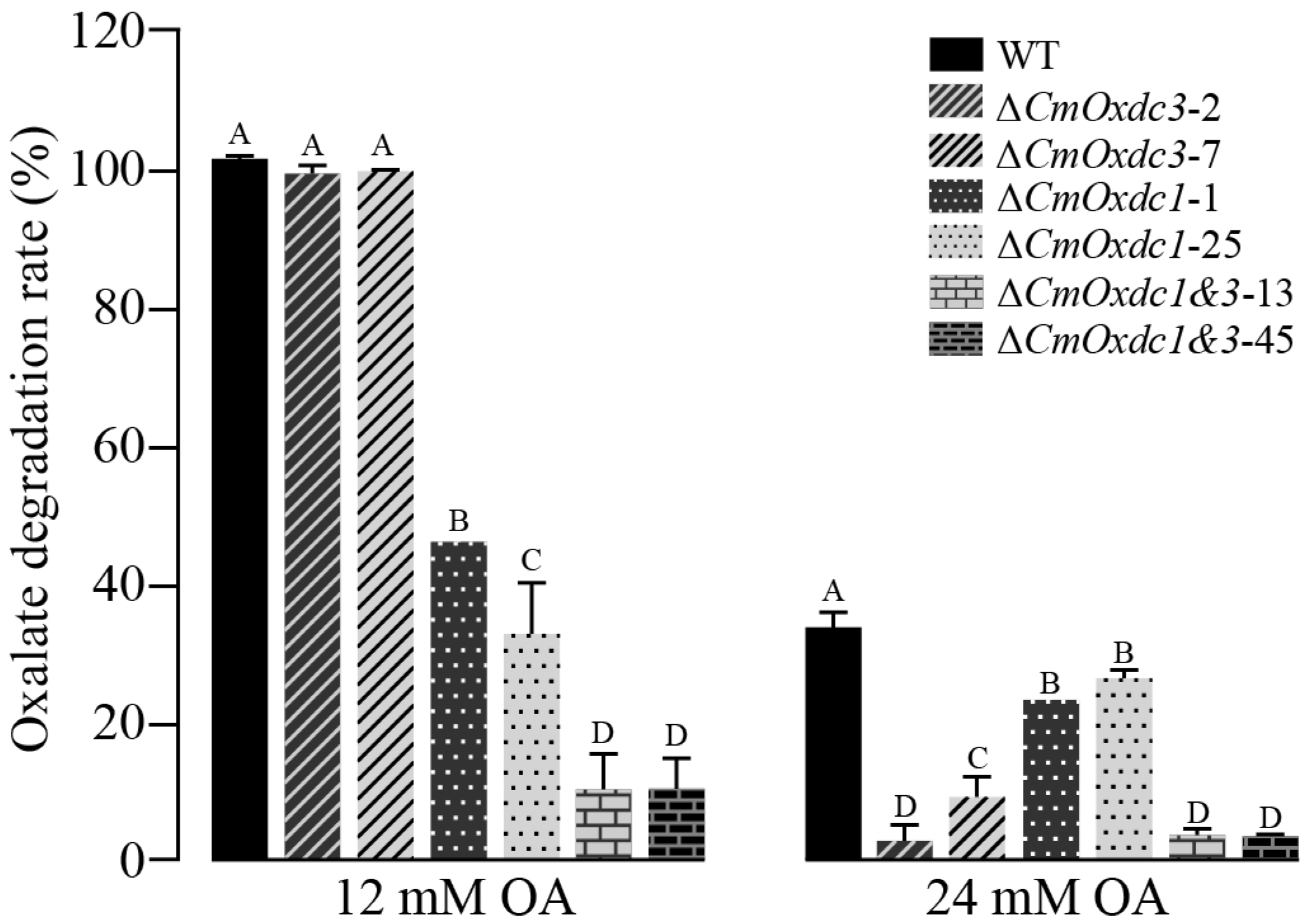
3.4. Effect of Disruption of CmOxdc3 on OA Degradation
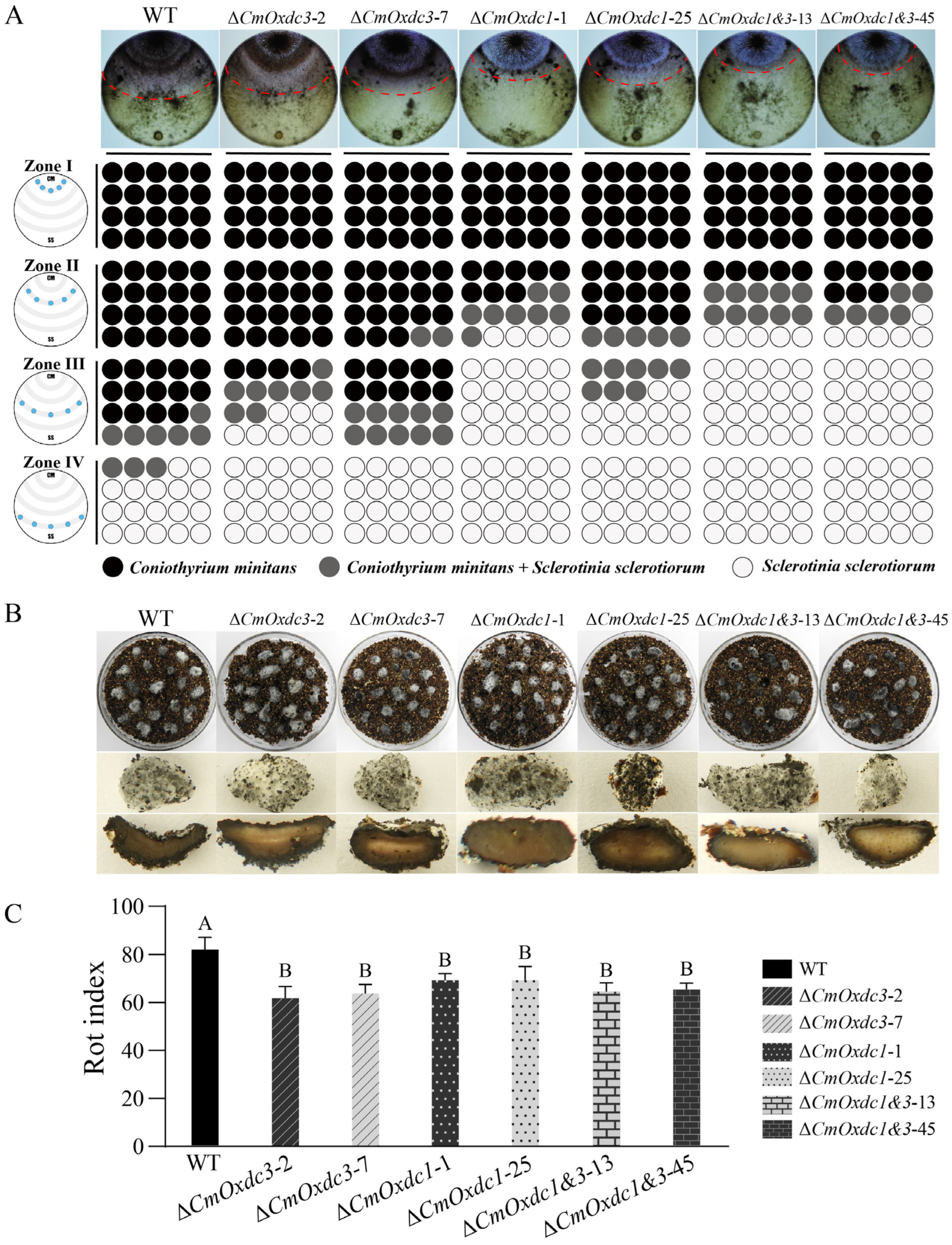
3.5. Effect of Disruption of CmOxdc3 on Mycoparasitism on the Host Hyphae
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munir, E.; Yoon, J.J.; Tokimatsu, T.; Hattori, T.; Shimada, M. A physiological role for oxalic acid biosynthesis wood-rotting basidiomycete Fomitopsis palustris. Proc. Natl. Acad. Sci. USA 2001, 98, 11126–11130. [Google Scholar] [CrossRef] [PubMed]
- Peck, A.B.; Canales, B.K.; Nguyen, C. Oxalate-degrading microorganisms or oxalate-degrading enzymes: Which is the future therapy for enzymatic dissolution of calcium-oxalate uroliths in recurrent stone disease? Urolithiasis 2016, 44, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Petroski, W.; Minich, D.M. Is there such a thing as “Anti-nutrients”? A narrative review of perceived problematic plant compounds. Nutrients 2020, 12, 2929. [Google Scholar] [CrossRef] [PubMed]
- Noonan, S.C.; Savage, G.P. Oxalate content of foods and its effect on humans. Asia Pac. J. Clin. Nutr. 1999, 8, 64–74. [Google Scholar] [PubMed]
- Kim, K.S.; Min, J.Y.; Dickman, M.B. Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant-Microbe Interact. 2008, 21, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Dutton, M.V.; Evans, C.S. Oxalate production by fungi: Its role in pathogenicity and ecology in the soil environment. Can. J. Microbiol. 1996, 42, 881–895. [Google Scholar] [CrossRef]
- Aguiar, A.; de Souza-Cruz, P.B.; Ferraz, A. Oxalic acid, Fe3+-reduction activity and oxidative enzymes detected in culture extracts recovered from Pinus taeda wood chips biotreated by Ceriporiopsis subvermispora. Enzym. Microb. Technol. 2006, 38, 873–878. [Google Scholar] [CrossRef]
- Cotton, P.; Kasza, Z.; Bruel, C.; Rascle, C.; Fèvre, M. Ambient pH controls the expression of endopolygalacturonase genes in the necrotrophic fungus Sclerotinia sclerotiorum. FEMS Microbiol. Lett. 2003, 227, 163–169. [Google Scholar] [CrossRef]
- Williams, B.; Kabbage, M.; Kim, H.J.; Britt, R.; Dickman, M.B. Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 2011, 7, e1002107. [Google Scholar] [CrossRef]
- Kabbage, M.; Williams, B.; Dickman, M.B. Cell death control: The interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 2013, 9, e1003287. [Google Scholar] [CrossRef]
- Liang, X.F.; Moomaw, E.W.; Rollins, J.A. Fungal oxalate decarboxylase activity contributes to Sclerotinia sclerotiorum early infection by affecting both compound appressoria development and function. Mol. Plant Pathol. 2015, 16, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Escutia, M.R.; Bowater, L.; Edwards, A.; Bottrill, A.R.; Burrell, M.R.; Polanco, R.; Vicuña, R.; Bornemann, S. Cloning and sequencing of two Ceriporiopsis subvermispora bicupin oxalate oxidase allelic isoforms: Implications for the reaction specificity of oxalate oxidases and decarboxylases. Appl. Environ. Microb. 2005, 71, 3608–3616. [Google Scholar] [CrossRef]
- Xu, L.S.; Li, G.Q.; Jiang, D.H.; Chen, W.D. Sclerotinia sclerotiorum: An evaluation of virulence theories. Annu. Rev. Phytopathol. 2018, 56, 311–338. [Google Scholar] [CrossRef] [PubMed]
- Tanner, A.; Bornemann, S. Bacillus subtilis YvrK is an acid-induced oxalate decarboxylase. J. Bacteriol. 2000, 182, 5271–5273. [Google Scholar] [CrossRef] [PubMed]
- Mäkelä, M.R.; Hildén, K.; Lundell, T.K. Oxalate decarboxylase: Biotechnological update and prevalence of the enzyme in filamentous fungi. Appl. Microbiol. Biotechnol. 2010, 87, 801–814. [Google Scholar] [CrossRef]
- Zeng, L.M.; Zhang, J.; Han, Y.C.; Yang, L.; Wu, M.D.; Jiang, D.H.; Chen, W.D.; Li, G.Q. Degradation of oxalic acid by the mycoparasite Coniothyrium minitans plays an important role in interacting with Sclerotinia sclerotiorum. Environ. Microbiol. 2014, 16, 2591–2610. [Google Scholar] [CrossRef]
- Kamthan, A.; Kamthan, M.; Kumar, A.; Sharma, P.; Ansari, S.; Thakur, S.S.; Chaudhuri, A.; Datta, A. A calmodulin like EF hand protein positively regulates oxalate decarboxylase expression by interacting with E-box elements of the promoter. Sci. Rep. 2015, 5, 14578. [Google Scholar] [CrossRef]
- Hou, N.; Wang, Q.R.; Sun, Y.; Li, X.Y.; Song, Q.Y.; Jiang, X.X.; Li, B.X.; Zhao, X.Y.; Zang, H.L.; Li, D.P.; et al. A novel biodemulsifier of Bacillus mojavensis XH1 – Oxalate decarboxylase with the potential for demulsification of oilfield emulsion. J. Hazard. Mater. 2021, 407, 124737. [Google Scholar] [CrossRef]
- Gupta, S.; Kanwar, S.S. Molecular characterization and in silico analysis of oxalate decarboxylase of Pseudomonas sp. OxDC12. J. Biomol. Struct. Dyn. 2022, 1–15. [Google Scholar] [CrossRef]
- Wu, X.Q.; Lyu, Y.P.; Ren, H.; Zhou, F.Y.; Zhang, X.J.; Zhao, X.Y.; Zhang, G.Z.; Yang, H.T. Degradation of oxalic acid by Trichoderma afroharzianum and its correlation with cell wall degrading enzymes in antagonizing Botrytis cinerea. J. Appl. Microbiol. 2022, 133, 2680–2693. [Google Scholar] [CrossRef]
- Azam, M.; Kesarwani, M.; Chakraborty, S.; Natarajan, K.; Datta, A. Cloning and characterization of the 5’-flanking region of the oxalate decarboxylase gene from Flammulina velutipes. Biochem. J. 2002, 367, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Micales, J.A. Localization and induction of oxalate decarboxylase in the brown-rot wood decay fungus Postia placenta. Int. Biodeter. Biodegr. 1997, 39, 125–132. [Google Scholar] [CrossRef]
- Dutton, M.V.; Kathiara, M.; Gallagher, I.M.; Evans, C.S. Purification and characterization of oxalate decarboxylase from Coriolus versicolor. FEMS Microbiol. Lett. 1994, 116, 321–326. [Google Scholar] [CrossRef]
- Kathiara, M.; Wood, D.A.; Evans, C.S. Detection and partial characterization of oxalate decarboxylase from Agaricus bisporus. Mycol. Res. 2000, 104, 345–350. [Google Scholar] [CrossRef]
- Mäkelä, M.; Galkin, K.; Hatakka, A.; Lundell, T. Production of organic acids and oxalate decarboxylase in lignin-degrading white rot fungi. Enzyme Microb. Technol. 2002, 30, 542–549. [Google Scholar] [CrossRef]
- Sato, S.; Liu, F.; Koc, H.; Tien, M. Expression analysis of extracellular proteins from Phanerochaete chrysosporium grown on different liquid and solid substrates. Microbiology 2007, 153, 3023–3033. [Google Scholar] [CrossRef]
- Ding, L.N.; Li, T.; Guo, X.J.; Li, M.; Liu, X.Y.; Cao, J.; Tan, X.L. Sclerotinia stem rot resistance in rapeseed: Recent progress and future prospects. J. Agric. Food Chem. 2021, 69, 2965–2978. [Google Scholar] [CrossRef]
- De Vrije, T.; Antoine, N.; Buitelaar, R.M.; Bruckner, S.; Dissevelt, M.; Durand, A.; Gerlagh, M.; Jones, E.E.; Lüth, P.; Oostra, J.; et al. The fungal biocontrol agent Coniothyrium minitans: Production by solid-state fermentation, application and marketing. Appl. Microbiol. Biotechnol. 2001, 56, 58–68. [Google Scholar] [CrossRef]
- Zeng, W.T.; Wang, D.C.; Kirk, W.; Hao, J.J. Use of Coniothyrium minitans and other microorganisms for reducing Sclerotinia sclerotiorum. Biol. Control 2012, 60, 225–232. [Google Scholar] [CrossRef]
- Elsheshtawi, M.; Elkhaky, M.T.; Sayed, S.R.; Bahkali, A.H.; Mohammed, A.A.; Gambhir, D.; Mansour, A.S.; Elgorban, A.M. Integrated control of white rot disease on beans caused by Sclerotinia sclerotiorum using Contans® and reduced fungicides application. Saudi J. Biol. Sci. 2017, 24, 405–409. [Google Scholar] [CrossRef]
- Sun, X.P.; Zhao, Y.; Jia, J.C.; Xie, J.T.; Cheng, J.S.; Liu, H.Q.; Jiang, D.H.; Fu, Y.P. Uninterrupted expression of CmSIT1 in a sclerotial parasite Coniothyrium minitans leads to reduced growth and enhanced antifungal ability. Front. Microbiol. 2017, 8, 2208. [Google Scholar] [CrossRef] [PubMed]
- Nicot, P.C.; Avril, F.; Duffaud, M.; Leyronas, C.; Troulet, C.; Villeneuve, F.; Bardin, M. Differential susceptibility to the mycoparasite Paraphaeosphaeria minitans among Sclerotinia sclerotiorum isolates. Trop. Plant Pathol. 2019, 44, 82–93. [Google Scholar] [CrossRef]
- Yang, R.; Han, Y.C.; Li, G.Q.; Jiang, D.H.; Huang, H.C. Suppression of Sclerotinia sclerotiorum by antifungal substances produced by the mycoparasite Coniothyrium minitans. Eur. J. Plant Pathol. 2007, 119, 411–420. [Google Scholar] [CrossRef]
- Yang, R.; Han, Y.C.; Li, G.Q.; Jiang, D.H.; Huang, H.C. Effects of ambient pH and nutritional factors on antifungal activity of the mycoparasite Coniothyrium minitans. Biol. Control 2008, 44, 116–127. [Google Scholar] [CrossRef]
- Lou, Y.; Han, Y.C.; Yang, L.; Wu, M.D.; Zhang, J.; Cheng, J.S.; Wang, M.Y.; Jiang, D.H.; Chen, W.D.; Li, G.Q. CmpacC regulates mycoparasitism, oxalate degradation and antifungal activity in the mycoparasitic fungus Coniothyrium minitans. Environ. Microbiol. 2015, 17, 4711–4729. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Li, G.Q.; Han, Y.C.; Jiang, D.H.; Huang, H.C. Degradation of oxalic acid by Coniothyrium minitans and its effects on production and activity of β-1, 3-glucanase of this mycoparasite. Biol. Control 2007, 43, 1–11. [Google Scholar] [CrossRef]
- Ren, L.; Li, G.Q.; Jiang, D.H. Characterization of some culture factors affecting oxalate degradation by the mycoparasite Coniothyrium minitans. J. Appl. Microbiol. 2010, 108, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.S.; Jiang, D.H.; Yi, X.H.; Fu, Y.P.; Li, G.Q.; Whipps, J.M. Production, survival and efficacy of Coniothyrium minitans conidia produced in shaken liquid culture. FEMS Microbiol. Lett. 2003, 227, 127–131. [Google Scholar] [CrossRef]
- Zhao, H.Z.; Zhou, T.; Xie, J.T.; Cheng, J.S.; Chen, T.; Jiang, D.H.; Fu, Y.P. Mycoparasitism illuminated by genome and transcriptome sequencing of Coniothyrium minitans, an important biocontrol fungus of the plant pathogen Sclerotinia sclerotiorum. Microb. Genom. 2020, 6, e000345. [Google Scholar] [CrossRef]
- Patel, D.; Shittu, T.A.; Baroncelli, R.; Muthumeenakahi, S.; Osborne, T.H.; Janganan, T.K.; Sreenivasaprasad, S. Genome sequence of the biocontrol agent Coniothyrium minitans Conio (IMI 134523). Mol. Plant-Microbe Interact. 2021, 34, 222–225. [Google Scholar] [CrossRef]
- Anand, R.; Dorrestein, P.C.; Kinsland, C.; Begley, T.P.; Ealick, S.E. Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 Å resolution. Biochemistry 2002, 41, 7659–7669. [Google Scholar] [CrossRef] [PubMed]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.D.; Hyde, K.D.; Liew, E.C.Y. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol. 2000, 147, 617–630. [Google Scholar] [CrossRef]
- Xu, Y.P.; Wang, Y.C.; Zhao, H.Z.; Wu, M.D.; Zhang, J.; Chen, W.D.; Li, G.Q.; Yang, L. Genome-wide identification and expression analysis of the bZIP transcription factors in the mycoparasite Coniothyrium minitans. Microorganisms 2020, 8, 1045. [Google Scholar] [CrossRef] [PubMed]
- Kershaw, M.J.; Talbot, N.J. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc. Natl. Acad. Sci. USA 2009, 106, 15967–15972. [Google Scholar] [CrossRef]
- Gao, T.Y.; Hao, F.M.; Yang, D.; Bie, Z.L.; Li, G.Q. Oxalic acid produced by Aspergillus niger Y-1 is effective for suppression of bacterial fruit blotch of watermelon seedlings. Biol. Control 2017, 112, 28–33. [Google Scholar] [CrossRef]
- Förster, C.; Kane, P.M. Cytosolic Ca2+ homeostasis is a constitutive function of the V-ATPase in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 38245–38253. [Google Scholar] [CrossRef]
- Huang, C.; Chang, A. pH-dependent cargo sorting from the Golgi. J. Biol. Chem. 2011, 286, 10058–10065. [Google Scholar] [CrossRef]
- Yang, X.X.; Cui, H.; Cheng, J.S.; Xie, J.T.; Jiang, D.H.; Hsiang, T.; Fu, Y.P. A HOPS protein, CmVps39, is required for vacuolar morphology, autophagy, growth, conidiogenesis and mycoparasitic functions of Coniothyrium minitans. Environ. Microbiol. 2016, 18, 3785–3797. [Google Scholar] [CrossRef]
- Tian, L.; Li, J.J.; Huang, C.M.; Zhang, D.D.; Xu, Y.; Yang, X.Y.; Song, J.; Wang, D.; Qiu, N.W.; Short, D.P.G.; et al. Cu/Zn superoxide dismutase (VdSOD1) mediates reactive oxygen species detoxification and modulates virulence in Verticillium dahliae. Mol. Plant Pathol. 2021, 22, 1092–1108. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Wu, M.; Zhang, J.; Li, G.; Yang, L. Cloning and Molecular Characterization of CmOxdc3 Coding for Oxalate Decarboxylase in the Mycoparasite Coniothyrium minitans. J. Fungi 2022, 8, 1304. https://doi.org/10.3390/jof8121304
Xu Y, Wu M, Zhang J, Li G, Yang L. Cloning and Molecular Characterization of CmOxdc3 Coding for Oxalate Decarboxylase in the Mycoparasite Coniothyrium minitans. Journal of Fungi. 2022; 8(12):1304. https://doi.org/10.3390/jof8121304
Chicago/Turabian StyleXu, Yuping, Mingde Wu, Jing Zhang, Guoqing Li, and Long Yang. 2022. "Cloning and Molecular Characterization of CmOxdc3 Coding for Oxalate Decarboxylase in the Mycoparasite Coniothyrium minitans" Journal of Fungi 8, no. 12: 1304. https://doi.org/10.3390/jof8121304
APA StyleXu, Y., Wu, M., Zhang, J., Li, G., & Yang, L. (2022). Cloning and Molecular Characterization of CmOxdc3 Coding for Oxalate Decarboxylase in the Mycoparasite Coniothyrium minitans. Journal of Fungi, 8(12), 1304. https://doi.org/10.3390/jof8121304






