Monitoring of Host Suitability and Defense-Related Genes in Wheat to Bipolaris sorokiniana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Material
2.2. DNA Extraction
2.3. DNA Sequencing and Phylogeny
2.4. Pathogenicity Tests of Isolates and Host Susceptabilty of Wheat Cultivars
2.5. Primer Design
2.6. Detection of Bipolaris sorokiniana Infection in Wheat Tissues
2.7. Expression Analysis of Defense-Related Genes in Wheat to the Pathogen
3. Results
3.1. The Identification of Pathogen Isolates
3.2. Aggressiveness of the Pathogen Isolates and Disease Reaction of Wheat
3.3. Designing Species-Specific Primers
3.4. Detection of Bipolaris sorokiniana Infection in Wheat Tissues
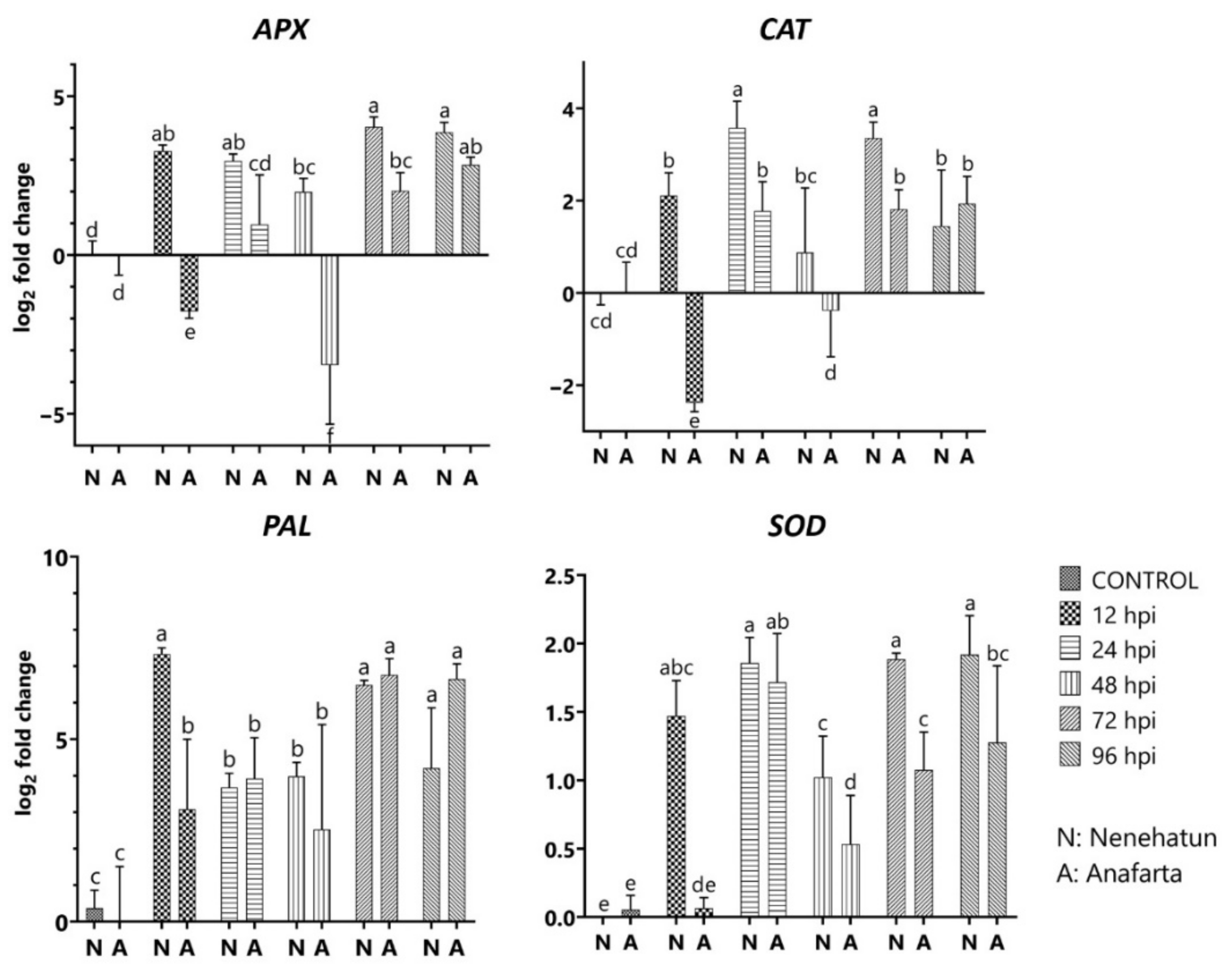
3.5. Differential Expression of Defense-Related Genes in Wheat Cultivars
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, D.P. (Ed.) Management of Wheat and Barley Diseases; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Al-Sadi, A.M. Bipolaris sorokiniana-induced black point, common root rot, and spot blotch diseases of wheat: A review. Front. Cell. Infect. Microbiol. 2021, 11, 118. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kumar, N.; Prajapati, S.; Maurya, S. Review on spot blotch of wheat: An emerging threat to wheat basket in changing climate. J. Pharmacogn. Phytochem. 2020, 9, 1985–1997. [Google Scholar]
- Tunali, B.; Nicol, J.M.; Hodson, D.; Uçkun, Z.; Büyük, O.; Erdurmuş, D.; Bağci, S.A. Root and crown rot fungi associated with spring, facultative, and winter wheat in Turkey. Plant Dis. 2008, 92, 1299–1306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Özer, G.; Paulitz, T.C.; Imren, M.; Alkan, M.; Muminjanov, H.; Dababat, A.A. Identity and pathogenicity of fungi associated with crown and root rot of dryland winter wheat in Azerbaijan. Plant Dis. 2020, 104, 2149–2157. [Google Scholar] [CrossRef]
- Gupta, P.K.; Chand, R.; Vasistha, N.K.; Pandey, S.P.; Kumar, U.; Mishra, V.K.; Joshi, A.K. Spot blotch disease of wheat: The current status of research on genetics and breeding. Plant Pathol. 2018, 67, 508–531. [Google Scholar] [CrossRef]
- Ghazvini, H.; Tekauz, A. Molecular diversity in the barley pathogen Bipolaris sorokiniana (Cochliobolus sativus). Australas. Plant Pathol. 2012, 41, 283–293. [Google Scholar] [CrossRef]
- Al-Sadi, A.M. Variation in resistance to spot blotch and the aggressiveness of Bipolaris sorokiniana on barley and wheat cultivars. J. Plant Pathol. 2016, 98, 97–103. [Google Scholar] [CrossRef]
- James, W.C. Assessment of plant diseases and losses. Annu. Rev. Phytopathol. 1974, 12, 27–48. [Google Scholar] [CrossRef]
- Adlakha, K.L.; Wilcoxson, R.D.; Raychaudhuri, S.P. Resistance of wheat to leaf spot caused by Bipolaris sorokiniana. Plant Dis. 1984, 68, 320–321. [Google Scholar] [CrossRef] [Green Version]
- Fetch, T.G., Jr.; Steffenson, B.J. Rating scales for assessing infection responses of barley infected with Cochliobolus sativus. Plant Dis. 1999, 83, 213–217. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Ramasamy, R.P. Current and prospective methods for plant disease detection. Biosensors 2015, 5, 537–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsedaley, B. A review on disease detection, pathogen identification and population genetics in fungi. J. Biol. Agric. Healthc. 2015, 5, 6–20. [Google Scholar]
- Brunner, K.; Mach, R.L. Quantitative detection of fungi by molecular methods: A case study on Fusarium. In Molecular Identification of Fungi; Gherbawy, Y., Voigt, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 179–193. [Google Scholar] [CrossRef]
- Manamgoda, D.S.; Cai, L.; McKenzie, E.H.; Crous, P.W.; Madrid, H.; Chukeatirote, E.; Shivas, R.G.; Tan, Y.P.; Hyde, K.D. A phylogenetic and taxonomic re-evaluation of the Bipolaris-Cochliobolus-Curvularia complex. Fungal Divers. 2012, 56, 131–144. [Google Scholar] [CrossRef]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal identification using molecular tools: A primer for the natural products research community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef] [PubMed]
- Matusinsky, P.; Frei, P.; Mikolasova, R.; Svacinova, I.; Tvaruzek, L.; Spitzer, T. Species–specific detection of Bipolaris sorokiniana from wheat and barley tissues. Crop Prot. 2010, 29, 1325–1330. [Google Scholar] [CrossRef]
- Aggarwal, R.; Gupta, S.; Banerjee, S.; Singh, V.B. Development of a SCAR marker for detection of Bipolaris sorokiniana causing spot blotch of wheat. Can. J. Microbiol. 2011, 57, 934–942. [Google Scholar] [CrossRef] [PubMed]
- Fajolu, O.L.; Wadl, P.A.; Vu, A.L.; Gwinn, K.D.; Scheffler, B.E.; Trigiano, R.N.; Ownley, B.H. Development and characterization of simple sequence repeats for Bipolaris sorokiniana and cross transferability to related species. Mycologia 2013, 105, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Orina Aleksandra, S.; Orina, A.S.; Gavrilova, O.P.; Gagkaeva, T.Y. Adaptation of the quantitative PCR method for the detection of the main representatives of cereal grain mycobiota. Microbiol. Indep. Res. J. 2018, 5, 78–83. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Van Strien, E.A. The families of pathogenesis–related proteins, their activities, and comparative analysis of PR1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- Thakur, M.; Sohal, B.S. Role of elicitors in inducing resistance in plants against pathogen infection: A review. Int. Sch. Res. Notices 2013, 2013, 762412. [Google Scholar] [CrossRef] [Green Version]
- Dubey, N.; Singh, K. Role of NBS–LRR proteins in plant defense. In Molecular Aspects of Plant–Pathogen Interaction; Springer: Singapore, 2018; pp. 115–138. [Google Scholar] [CrossRef]
- Van Loon, L.C. Induced resistance in plants and the role of pathogenesis-related proteins. Eur. J. Plant Pathol. 1997, 103, 753–765. [Google Scholar] [CrossRef]
- Sels, J.; Mathys, J.; De Coninck, B.M.; Cammue, B.P.; De Bolle, M.F. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Grover, A. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212, 29–37. [Google Scholar] [CrossRef]
- Balasubramanian, V.; Vashisht, D.; Cletus, J.; Sakthivel, N. Plant β-1, 3-glucanases: Their biological functions and transgenic expression against phytopathogenic fungi. Biotechnol. Lett. 2012, 34, 1983–1990. [Google Scholar] [CrossRef] [PubMed]
- Rong, W.; Luo, M.; Shan, T.; Wei, X.; Du, L.; Xu, H.; Zhang, Z. A wheat cinnamyl alcohol dehydrogenase TaCAD12 contributes to host resistance to the sharp eyespot disease. Front. Plant Sci. 2016, 7, 1723. [Google Scholar] [CrossRef] [Green Version]
- Lincoln, J.E.; Sanchez, J.P.; Zumstein, K.; Gilchrist, D.G. Plant and animal PR1 family members inhibit programmed cell death and suppress bacterial pathogens in plant tissues. Mol. Plant Pathol. 2018, 19, 2111–2123. [Google Scholar] [CrossRef] [Green Version]
- Mackintosh, C.A.; Lewis, J.; Radmer, L.E.; Shin, S.; Heinen, S.J.; Smith, L.A.; Muehlbauer, G.J. Overexpression of defense response genes in transgenic wheat enhances resistance to Fusarium head blight. Plant Cell Rep. 2007, 26, 479–488. [Google Scholar] [CrossRef] [Green Version]
- Eissa, H.F.; Hassanien, S.E.; Ramadan, A.M.; El–Shamy, M.M.; Saleh, O.M.; Shokry, A.M.; Bahieldin, A. Developing transgenic wheat to encounter rusts and powdery mildew by overexpressing barley chi26 gene for fungal resistance. Plant Methods 2017, 13, 41. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, H.; Wei, X.; Zhang, J.; Wang, H.; Liu, D. Expression analysis and functional characterization of a pathogen–induced thaumatin–like gene in wheat conferring enhanced resistance to Puccinia triticina. J. Plant Interact. 2017, 12, 332–339. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Yuan, S.; Wu, W.; Yang, Y.; Cui, Z.; Wang, H.; Liu, D. TaTLP1 interacts with TaPR1 to contribute to wheat defense responses to leaf rust fungus. PLoS Genet. 2020, 16, e1008713. [Google Scholar] [CrossRef]
- Spanic, V.; Vuletic, M.V.; Abicic, I.; Marcek, T. Early response of wheat antioxidant system with special reference to Fusarium head blight stress. Plant Physiol. Biochem. 2017, 115, 34–43. [Google Scholar] [CrossRef]
- Hao, Q.; Wang, W.; Han, X.; Wu, J.; Lyu, B.; Chen, F.; Fu, D. Isochorismate-based salicylic acid biosynthesis confers basal resistance to Fusarium graminearum in barley. Mol. Plant Pathol. 2018, 19, 1995–2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, C.S.; Hannan, A.; Monsur, A.; Sagor, G.H.M. Screening and biochemical characterization of wheat cultivars resistance to Magnaporthe oryzae pv. triticum (MoT). J. Plant Stress Physiol. 2020, 6, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Sivanesan, A. Graminicolous Species of Bipolaris, Curvularia, Drechslera, Exserohilum and Their Teleomorphs; Mycological Papers No. 158; C.A.B. International: Wallingford, UK, 1987; 261p. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar] [CrossRef]
- Berbee, M.L.; Pirseyedi, M.; Hubbard, S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 1999, 91, 964–977. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real–time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Desmond, O.J.; Edgar, C.I.; Manners, J.M.; Maclean, D.J.; Schenk, P.M.; Kazan, K. Methyl jasmonate induced gene expression in wheat delays symptom development by the crown rot pathogen Fusarium pseudograminearum. Physiol. Mol. Plant Pathol. 2005, 67, 171–179. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, C.; Cheng, Y.; Wang, X.; Li, F.; Han, Q.; Kang, Z. Histological and molecular studies of the non–host interaction between wheat and Uromyces fabae. Planta 2011, 234, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Özer, G.; İmren, M.; Özdemir, F.; Morgounov, A.; Dababat, A.A. First report of common root rot on triticale caused by Bipolaris sorokiniana in Kazakhstan. Plant Dis. 2020, 104, 2735. [Google Scholar] [CrossRef]
- Özer, G.; Imren, M.; Alkan, M.; Paulitz, T.C.; Bayraktar, H.; Palacıoğlu, G.; Dababat, A.A. Molecular and pathogenic characterization of Cochliobolus anamorphs associated with common root rot of wheat in Azerbaijan. Phytopathol. Mediterr. 2020, 59, 147–158. [Google Scholar] [CrossRef]
- Kang, R.; Hu, Y.; Wang, L.; Xie, S.; Li, Y.; Yuan, H.; Li, H. Pathogenicity variation and DNA polymorphism of Bipolaris sorokiniana infecting winter wheat in the Huanghuai floodplain of China. Plant Pathol. 2021, 70, 87–99. [Google Scholar] [CrossRef]
- Sultana, S.; Adhikary, S.K.; Islam, M.M.; Rahman, S.M.M. Evaluation of pathogenic variability based on leaf blotch disease development components of Bipolaris sorokiniana in Triticum aestivum and agroclimatic origin. Plant Pathol. J. 2018, 34, 93. [Google Scholar] [CrossRef]
- Duveiller, E.; Garcia Altamirano, I. Pathogenicity of Bipolaris sorokiniana isolates from wheat roots, leaves and grains in Mexico. Plant Pathol. 2000, 49, 235–242. [Google Scholar] [CrossRef]
- Jaiswal, S.K.; Prasad, L.C.; Sharma, S.; Kumar, S.; Prasad, R.; Pandey, S.P.; Joshi, A.K. Identification of molecular marker and aggressiveness for different groups of Bipolaris sorokiniana isolates causing spot blotch disease in wheat (Triticum aestivum L.). Curr. Microbiol. 2007, 55, 135–141. [Google Scholar] [CrossRef]
- Liatukas, Ž.; Ruzgas, V. Resistance of European winter wheat cultivars to spot blotch at juvenile growth stages. Phytopathol. Mediterr. 2011, 50, 350–358. [Google Scholar] [CrossRef]
- Bockus, W.W.; Bowden, R.L.; Hunger, R.M.; Morril, W.L.; Murray, T.D.; Smiley, R.W. Diseases caused by fungi and fungus–like organisms. In Compendium Wheat Diseaseand Pests; Bockus, W.W., Bowden, R.L., Hunger, R.M., Morrill, W.L., Murray, T.D., Smiley, R.W., Eds.; APS Press: St. Paul, MN, USA, 2010; pp. 15–86. [Google Scholar] [CrossRef]
- Özer, G.; Bayraktar, H. Development of conventional and real-time PCR assays to detect Alternaria burnsii in cumin seed. Gesunde Pflanz. 2019, 71, 205–212. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, L.; Meng, C.; Zhang, D.; Xu, N.; Yu, J. Establishment and application of a multiplex PCR assay for detection of Rhizoctonia cerealis, Bipolaris sorokiniana, and Fusarium spp. in winter wheat. J. Plant Pathol. 2020, 102, 19–27. [Google Scholar] [CrossRef]
- Bayraktar, H.; Özer, G.; Aydoğan, A.; Palacıoğlu, G. Determination of Ascochyta blight disease in chickpea using real–time PCR. J. Plant Dis. Prot. 2016, 123, 109–117. [Google Scholar] [CrossRef]
- Daniëls, B.; Dreesen, R.; Davey, M.W.; Keulemans, J. Real–time PCR as a promising tool to monitor growth of Venturia spp. in scab–susceptible and–resistant apple leaves. Eur. J. Plant Pathol. 2012, 134, 821–833. [Google Scholar] [CrossRef]
- Leiminger, J.; Bäßler, E.; Knappe, C.; Bahnweg, G.; Hausladen, H. Quantification of disease progression of Alternaria spp. on potato using real-time PCR. Eur. J. Plant Pathol. 2015, 141, 295–309. [Google Scholar] [CrossRef]
- Zhu, F.; Xu, M.; Wang, S.; Jia, S.; Zhang, P.; Lin, H.; Xi, D. Prokaryotic expression of pathogenesis related protein 1 gene from Nicotiana benthamiana: Antifungal activity and preparation of its polyclonal antibody. Biotechnol. Lett. 2012, 34, 919–924. [Google Scholar] [CrossRef] [PubMed]
- Molina, A.; Görlach, J.; Volrath, S.; Ryals, J. Wheat genes encoding two types of PR1 proteins are pathogen inducible, but do not respond to activators of systemic acquired resistance. Mol. Plant-Microbe Interact. 1999, 12, 53–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, Y.C.; Outram, M.A.; Breen, S.; Wang, C.; Dagvadorj, B.; Winterberg, B.; Solomon, P.S. PR1-mediated defense via C-terminal peptide release is targeted by a fungal pathogen effector. New Phytol. 2021, 229, 3467–3480. [Google Scholar] [CrossRef]
- Soltanloo, H.; Khorzoghi, E.G.; Ramezanpour, S.; Arabi, M.K.; Pahlavani, M.H. The expression profile of ‘Chi-1, Glu-2, Glu-3 and PR1. 2’ genes in scab–resistant and susceptible wheat cultivars during enfection by ‘Fusarium graminearum’. Plant Omics 2010, 3, 162–166. [Google Scholar]
- Muhae-Ud-Din, G.; Chen, D.; Liu, T.; Chen, W.; Gao, L. Characterization of the wheat cultivars against Tilletia controversa Kühn, causal agent of wheat dwarf bunt. Sci. Rep. 2020, 10, 9029. [Google Scholar] [CrossRef]
- Wu, S.W.; Wang, H.W.; Yang, Z.D.; Kong, L.R. Expression comparisons of pathogenesis–related (PR) genes in wheat in response to infection/infestation by Fusarium, Yellow dwarf virus (YDV) aphid–transmitted and hessian fly. J. Integr. Agric. 2014, 13, 926–936. [Google Scholar] [CrossRef]
- Li, W.L.; Faris, J.D.; Muthukrishnan, S.; Liu, D.J.; Chen, P.D.; Gill, B.S. Isolation and characterization of novel cDNA clones of acidic chitinases and β-1, 3-glucanases from wheat spikes infected by Fusarium graminearum. Theor. Appl. Genet. 2001, 102, 353–362. [Google Scholar] [CrossRef]
- Casassola, A.; Brammer, S.P.; Chaves, M.S.; Martinelli, J.A.; Stefanato, F.; Boyd, L.A. Changes in gene expression profiles as they relate to the adult plant leaf rust resistance in the wheat cv. Toropi. Physiol. Mol. Plant Pathol. 2015, 89, 49–54. [Google Scholar] [CrossRef]
- Zhang, H.; Qiu, Y.; Yuan, C.; Chen, X.; Huang, L. Fine–tuning of PR genes in wheat responding to different Puccinia rust species. J. Plant Physiol. Pathol. 2018, 6, 2. [Google Scholar] [CrossRef]
- Campbell, J.; Zhang, H.; Giroux, M.J.; Feiz, L.; Jin, Y.; Wang, M.; Huang, L. A mutagenesis–derived broad–spectrum disease resistance locus in wheat. Theor. Appl. Genet. 2012, 125, 391–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhang, Y.; Zhang, W.; Zhang, J.; Wang, H.; Liu, D. Expression profiles of pathogenesis–related gene, TaLr35PR1, as it relate to Lr35–mediated adult plant leaf rust resistance. Plant Mol. Biol. Rep. 2016, 34, 1127–1135. [Google Scholar] [CrossRef]
- Iqbal, M.J.; Yaegashi, S.; Ahsan, R.; Shopinski, K.L.; Lightfoot, D.A. Root response to Fusarium solani f. sp. glycines: Temporal accumulation of transcripts in partially resistant and susceptible soybean. Theor. Appl. Genet. 2005, 110, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, Q.; Yi, L.; Song, X.; Li, L.; Deng, G.; Long, H. PAL-mediated SA biosynthesis pathway contributes to nematode resistance in wheat. Plant J. 2021, 107, 698–712. [Google Scholar] [CrossRef]
- Cass, C.L.; Peraldi, A.; Dowd, P.F.; Mottiar, Y.; Santoro, N.; Karlen, S.D.; Sedbrook, J.C. Effects of Phenylalanine Ammonia Lyase (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J. Exp. Bot. 2015, 66, 4317–4335. [Google Scholar] [CrossRef] [Green Version]
- Christensen, A.B.; Thordal–Christensen, H.; Zimmermann, G.; Gjetting, T.; Lyngkjær, M.F.; Dudler, R.; Schweizer, P. The germinlike protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley. Mol. Plant-Microbe Interact. 2004, 17, 109–117. [Google Scholar] [CrossRef] [Green Version]
- Debona, D.; Rodrigues, F.Á.; Rios, J.A.; Nascimento, K.J.T. Biochemical changes in the leaves of wheat plants infected by Pyricularia oryzae. Phytopathology 2012, 102, 1121–1129. [Google Scholar] [CrossRef] [Green Version]





| Wheat Cultivar | Source | Wheat Type | Disease Severity * |
|---|---|---|---|
| Kırik | East Anatolian Agricultural Research Institute | Winter | 7.97 a |
| Lancer | East Anatolian Agricultural Research Institute | Winter | 7.50 a |
| Doğu-88 | East Anatolian Agricultural Research Institute | Winter | 6.27 b |
| Nenehatun | East Anatolian Agricultural Research Institute | Winter | 6.13 b |
| Damla | Trakya Agricultural Research Institute | Winter | 6.03 b |
| Palandöken-97 | East Anatolian Agricultural Research Institute | Winter | 5.43 bc |
| Karasu-90 | East Anatolian Agricultural Research Institute | Winter | 5.30 bd |
| Ceyhan-99 | Eastern Mediterranean Agricultural Research Institute | Spring | 5.20 be |
| Ayyıldız | East Anatolian Agricultural Research Institute | Winter | 4.87 cf |
| Es-26 | Transitional Zone Agricultural Research Institute | Winter | 4.60 cg |
| Sarıbaşak | Eastern Mediterranean Agricultural Research Institute | Spring | 4.60 cg |
| Soyer-02 | Transitional Zone Agricultural Research Institute | Winter | 4.47 ch |
| Müfitbey | Transitional Zone Agricultural Research Institute | Winter | 4.47 ch |
| Bezostaya-1 | Transitional Zone Agricultural Research Institute | Winter | 4.40 cı |
| Seri-2013 | Eastern Mediterranean Agricultural Research Institute | Spring | 4.40 cı |
| Nevzatbey | Black Sea Agricultural Research Institute | Winter | 4.40 cı |
| Adana-99 | Eastern Mediterranean Agricultural Research Institute | Spring | 4.37 cı |
| Köprü | Trakya Agricultural Research Institute | Winter | 4.33 cı |
| Ekinoks | Eastern Mediterranean Agricultural Research Institute | Spring | 4.27 dj |
| İzgi-2001 | Transitional Zone Agricultural Research Institute | Winter | 4.26 dj |
| Yüksel | Trakya Agricultural Research Institute | Winter | 4.13 ek |
| Bereket | Trakya Agricultural Research Institute | Winter | 4.13 ek |
| Gökkan | Eastern Mediterranean Agricultural Research Institute | Spring | 4.03 fk |
| Yunus | Transitional Zone Agricultural Research Institute | Winter | 3.93 fl |
| Alturna | East Anatolian Agricultural Research Institute | Winter | 3.90 fm |
| Gerek-79 | Transitional Zone Agricultural Research Institute | Winter | 3.90 fm |
| Aldane | Trakya Agricultural Research Institute | Winter | 3.87 fm |
| Sönmez-2001 | Transitional Zone Agricultural Research Institute | Winter | 3.87 fm |
| Candaş | Eastern Mediterranean Agricultural Research Institute | Spring | 3.83 fn |
| Alparslan | East Anatolian Agricultural Research Institute | Winter | 3.83 fn |
| Harmankaya-99 | Transitional Zone Agricultural Research Institute | Winter | 3.77 fn |
| Sultan-95 | Transitional Zone Agricultural Research Institute | Winter | 3.77 fn |
| Sakin | Black Sea Agricultural Research Institute | Winter | 3.73 fn |
| Kirve | Black Sea Agricultural Research Institute | Spring | 3.60 go |
| Nacibey | Transitional Zone Agricultural Research Institute | Winter | 3.60 go |
| Özcan | Black Sea Agricultural Research Institute | Winter | 3.53 go |
| Canik-2003 | Black Sea Agricultural Research Institute | Winter | 3.50 go |
| Osmaniyem | Eastern Mediterranean Agricultural Research Institute | Spring | 3.47 go |
| Çetinel-2000 | Transitional Zone Agricultural Research Institute | Winter | 3.37 hp |
| Daphan | East Anatolian Agricultural Research Institute | Winter | 3.30 ıq |
| Tekirdağ | Trakya Agricultural Research Institute | Winter | 3.30 ıq |
| Saban | Trakya Agricultural Research Institute | Winter | 3.30 ıq |
| Pehlivan | Trakya Agricultural Research Institute | Winter | 3.13 jo |
| Altındane | Black Sea Agricultural Research Institute | Spring | 3.03 kq |
| Mesut | Transitional Zone Agricultural Research Institute | Winter | 2.87 lq |
| Alpu-2001 | Transitional Zone Agricultural Research Institute | Winter | 2.87 lq |
| Selimiye | Trakya Agricultural Research Institute | Winter | 2.80 lq |
| Altay | Transitional Zone Agricultural Research Institute | Winter | 2.77 mr |
| Yıldırım | East Anatolian Agricultural Research Institute | Winter | 2.77 mr |
| Altınbaşak | Eastern Mediterranean Agricultural Research Institute | Spring | 2.70 mr |
| Abide | Trakya Agricultural Research Institute | Winter | 2.53 or |
| Gelibolu | Trakya Agricultural Research Institute | Winter | 2.27 pr |
| Yakamoz | Eastern Mediterranean Agricultural Research Institute | Spring | 2.17 qr |
| Koç-2015 | Bati Akdeniz Agricultural Research Institute | Spring | 1.63 r |
| Anafarta | Trakya Agricultural Research Institute | Winter | 1.63 r |
| Target Genes | Primer Names | Sequence (5′–3′) | References |
|---|---|---|---|
| ITS | BsITSF | TTCTGGGAGACTCGCCTTA | |
| BsITSR | GTCTTGATGGATTACCGTCCTT | ||
| GAPDH | BS_F01 | CCATTCACGCATATTAAAGCTG | in this study |
| BS_R01 | CTCTGGTGAAAGGTTCTGGATT | ||
| SSU | BsSSUF | GCGAAGGCAAACCTCTATGTA | |
| BsSSUR | CGTCCCTCAACGTCAGTTATAG | ||
| β-tubulin | β-tubulin_F | GCCATGTTCAGGAGGAAGG | [45] |
| β-tubulin_R | CTCGGTGAACTCCATCTCGT | ||
| PR1 | TaPR1_F | GAGAATGCAGACGCCCAAGC | |
| TaPR1_R | CTGGAGCTTGCAGTCGTTGATC | ||
| PR2 | TaPR2_F | AGGATGTTGCTTCCATGTTTGCCG | |
| TaPR2_R | AAGTAGATGCGCATGCCGTTGATG | ||
| PR3 | TaPR3_F | TACTGCTTCAAGGACCAGATAGAC | |
| TaPR3_R | CACCAGGTTCGGGTTGTTTA | ||
| PR5 | TaPR5_F | CAAGCAGTGGTATCAACGCAGAG | |
| TaPR5_R | GTGAAGCCACAGTTGTTCTTGATGTT | ||
| PR10 | TaPR10_F | TTAAACCAGCACGAGAAACATCAG | |
| TaPR10_R | ATCCTCCCTCGATTATTCTCACG | ||
| Phenylalanine ammonia-lyase | TaPAL_F | CGTCAAGAGCTGTGTGAAGATGG | [46] |
| TaPAL_R | GGTAGTTGGAGCTGCAAGGGTC | ||
| Catalase | TaCAT_F | TGCCTGTGTTTTTTATCCGAGA | |
| TaCAT_R | CTGCTGATTAAGGTGTAGGTGTTGA | ||
| Superoxide dismutase | TaSOD_F | CGATAGCCAGATTCCTTTGACT | |
| TaSOD_R | GAAACCAGCGACCTACAACG | ||
| Ascorbate-peroxidase | TaAPX_F | GGTTTGAGTGACCAGGACATTG | |
| TaAPX_R | GCATCCTCATCCGCAGCAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alkan, M.; Bayraktar, H.; İmren, M.; Özdemir, F.; Lahlali, R.; Mokrini, F.; Paulitz, T.; Dababat, A.A.; Özer, G. Monitoring of Host Suitability and Defense-Related Genes in Wheat to Bipolaris sorokiniana. J. Fungi 2022, 8, 149. https://doi.org/10.3390/jof8020149
Alkan M, Bayraktar H, İmren M, Özdemir F, Lahlali R, Mokrini F, Paulitz T, Dababat AA, Özer G. Monitoring of Host Suitability and Defense-Related Genes in Wheat to Bipolaris sorokiniana. Journal of Fungi. 2022; 8(2):149. https://doi.org/10.3390/jof8020149
Chicago/Turabian StyleAlkan, Mehtap, Harun Bayraktar, Mustafa İmren, Fatih Özdemir, Rachid Lahlali, Fouad Mokrini, Timothy Paulitz, Abdelfattah A. Dababat, and Göksel Özer. 2022. "Monitoring of Host Suitability and Defense-Related Genes in Wheat to Bipolaris sorokiniana" Journal of Fungi 8, no. 2: 149. https://doi.org/10.3390/jof8020149
APA StyleAlkan, M., Bayraktar, H., İmren, M., Özdemir, F., Lahlali, R., Mokrini, F., Paulitz, T., Dababat, A. A., & Özer, G. (2022). Monitoring of Host Suitability and Defense-Related Genes in Wheat to Bipolaris sorokiniana. Journal of Fungi, 8(2), 149. https://doi.org/10.3390/jof8020149








