Phylogenetic Comparison of Swainsonine Biosynthetic Gene Clusters among Fungi
Abstract
:1. Introduction
2. Materials and Methods
3. Results
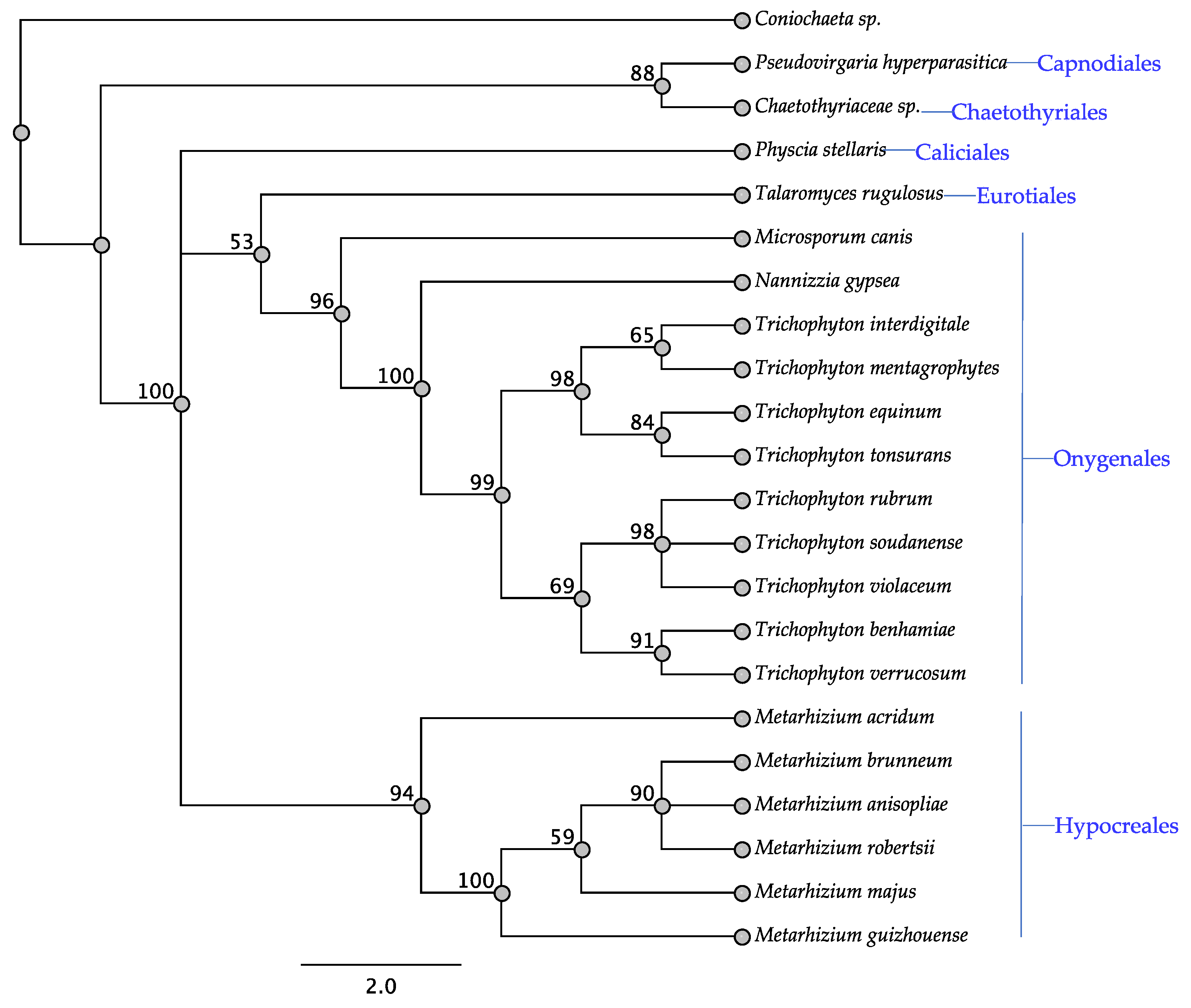
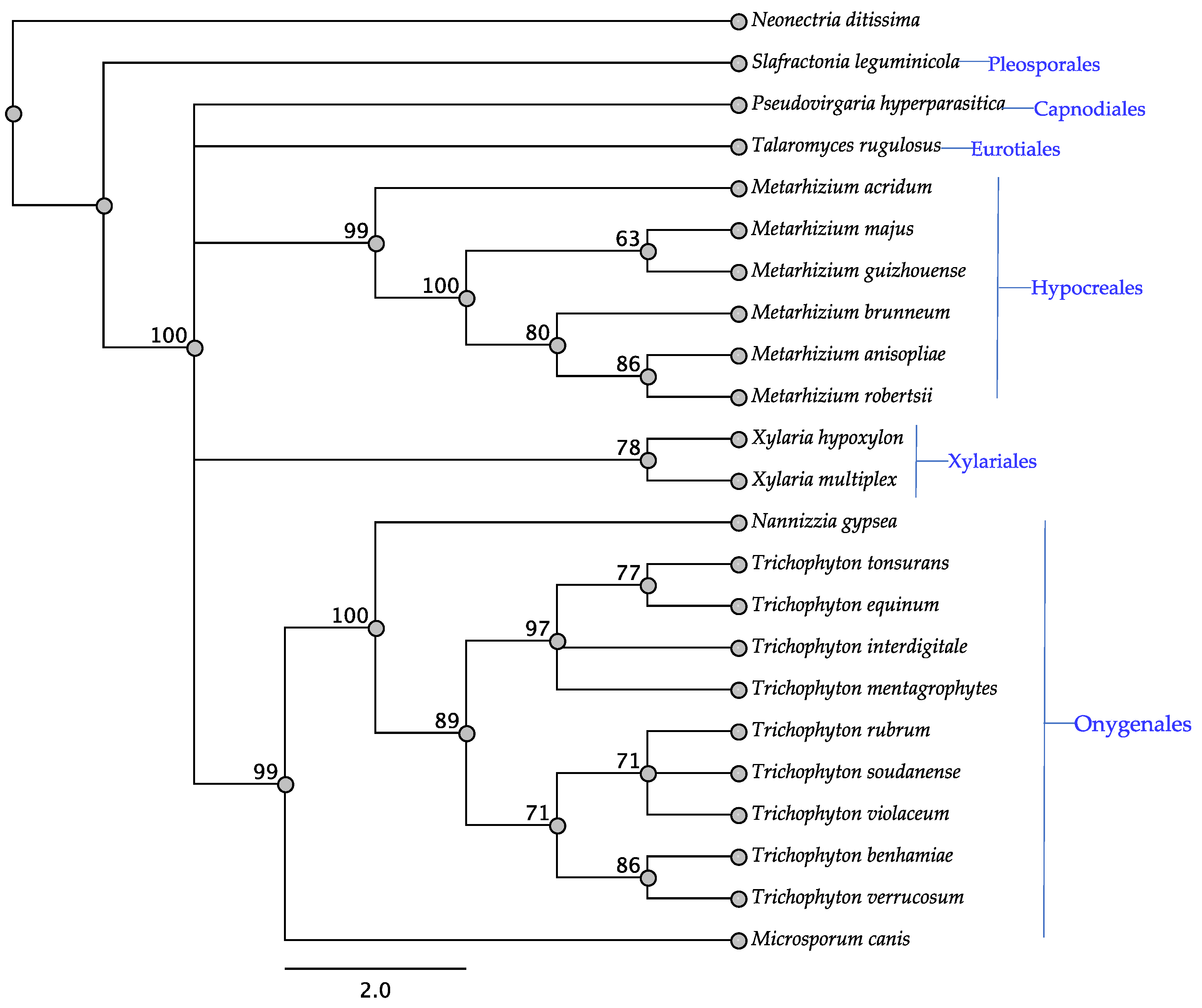
3.1. Swn Genes
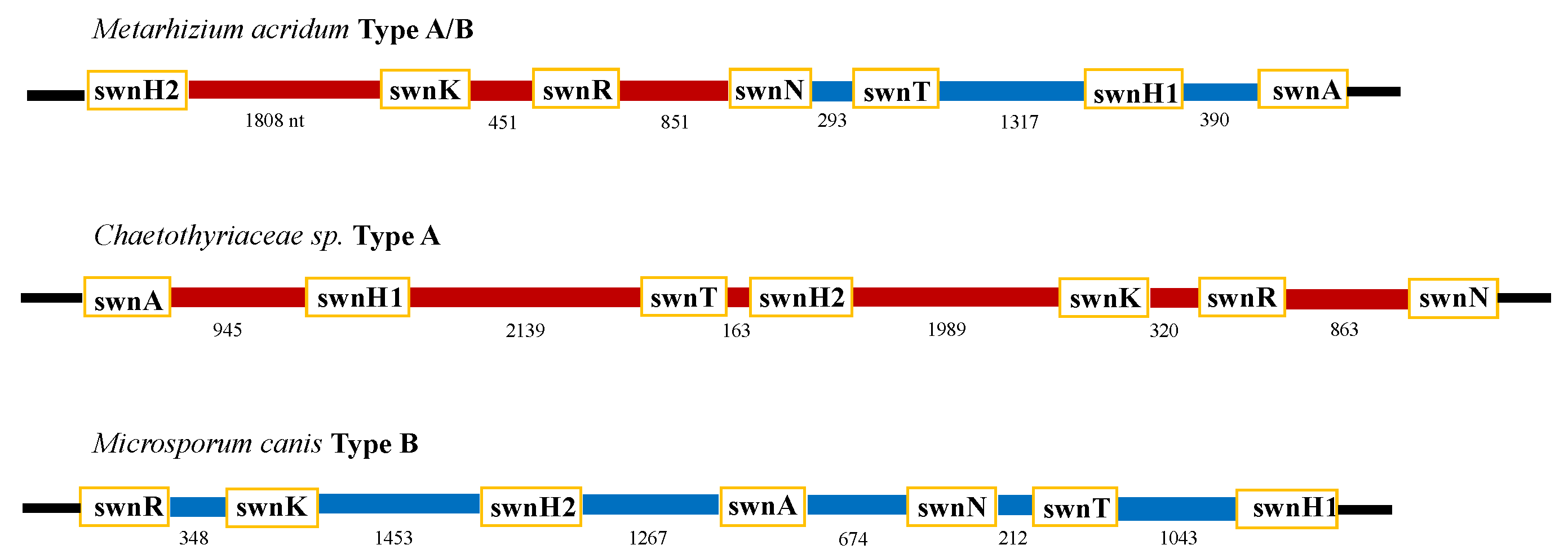
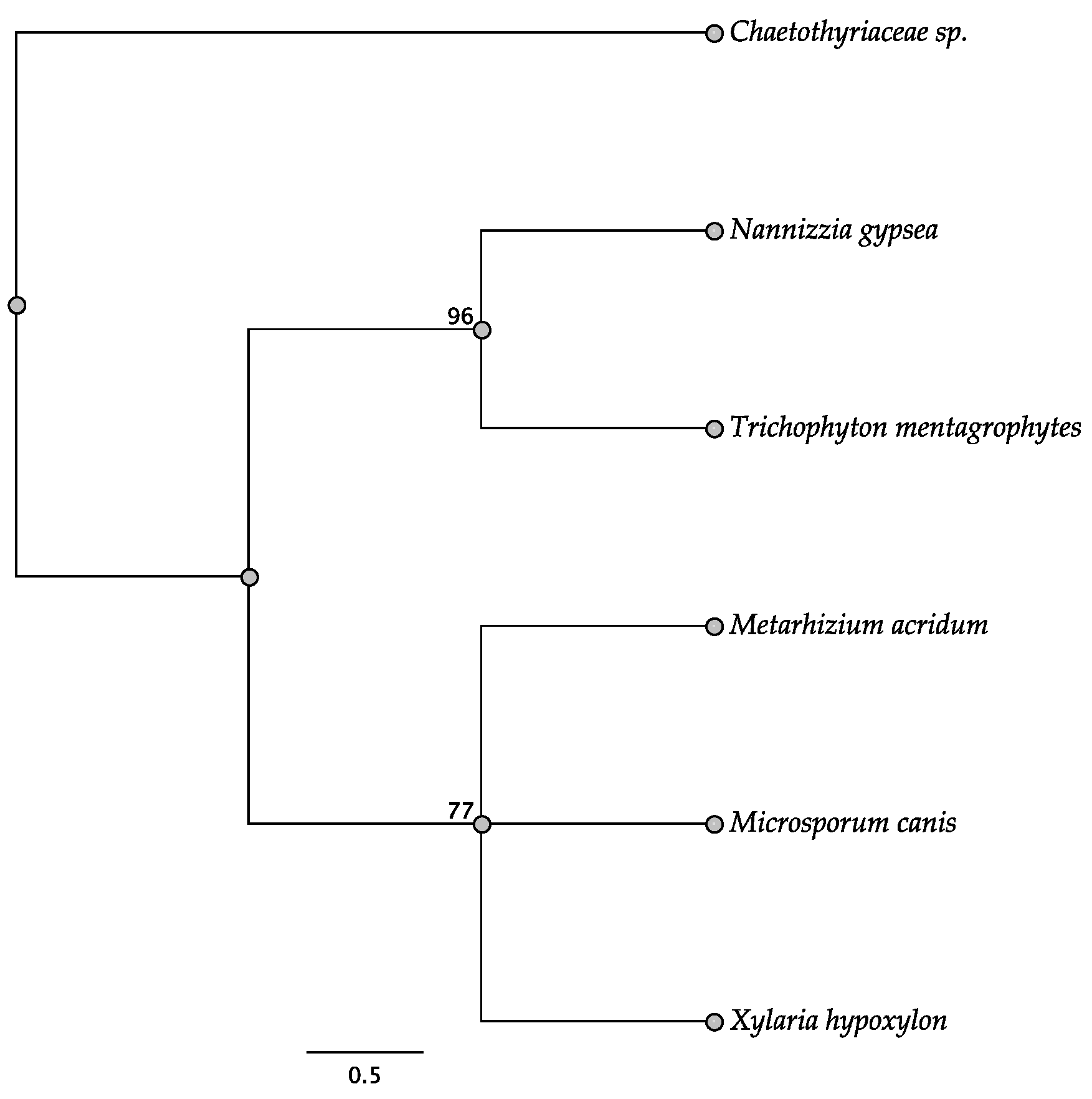
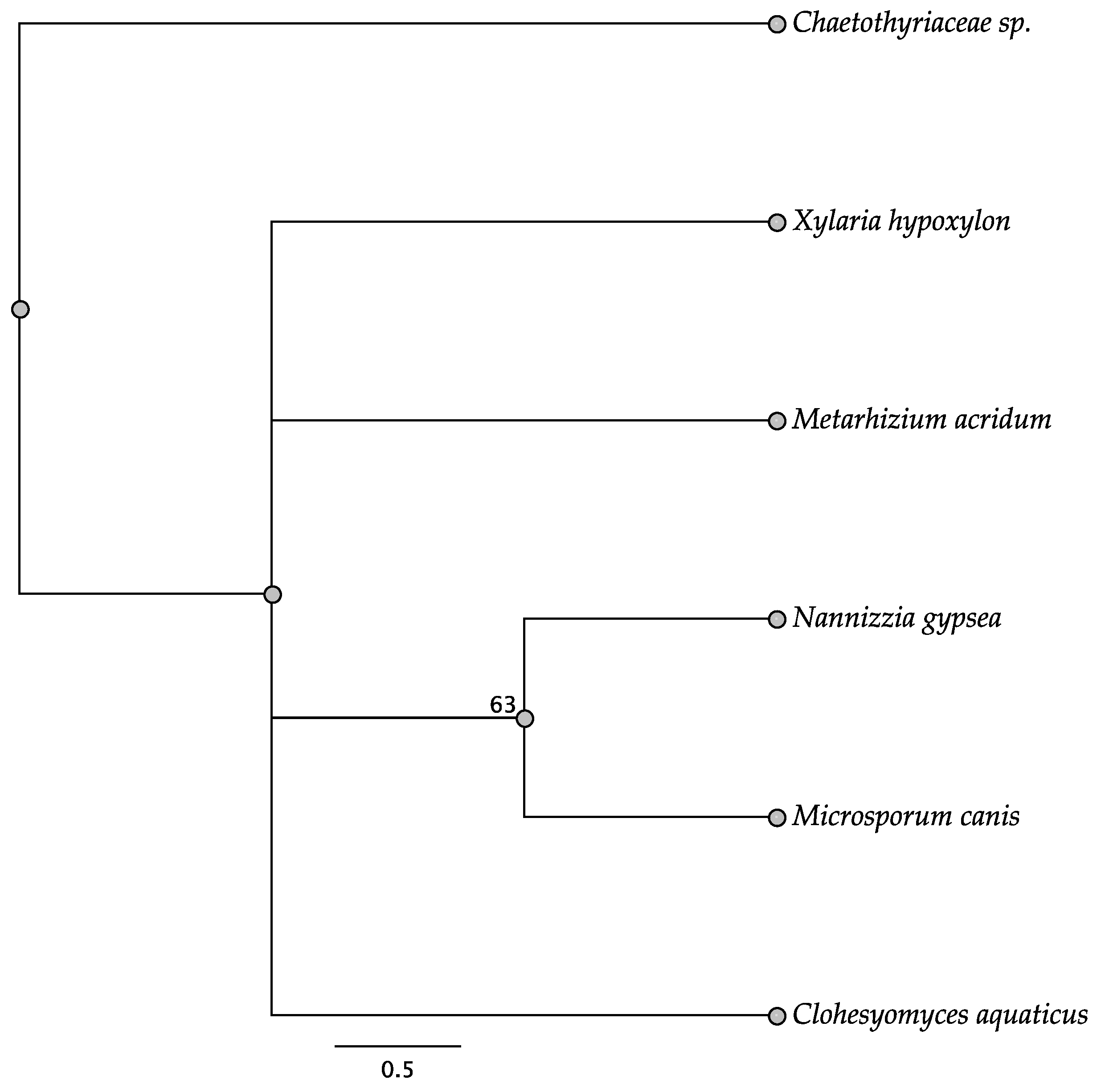
3.2. Intergenic Regions
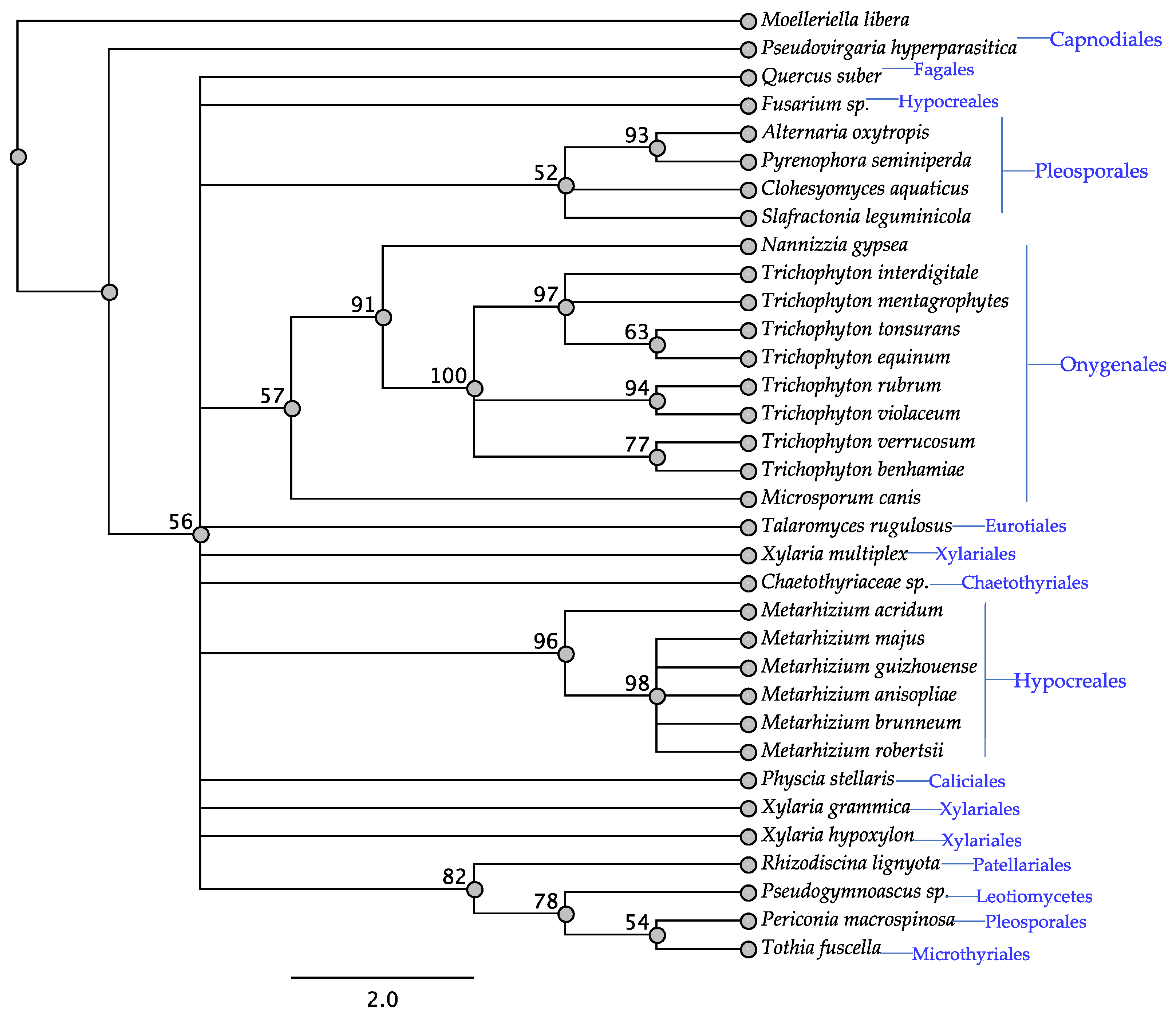
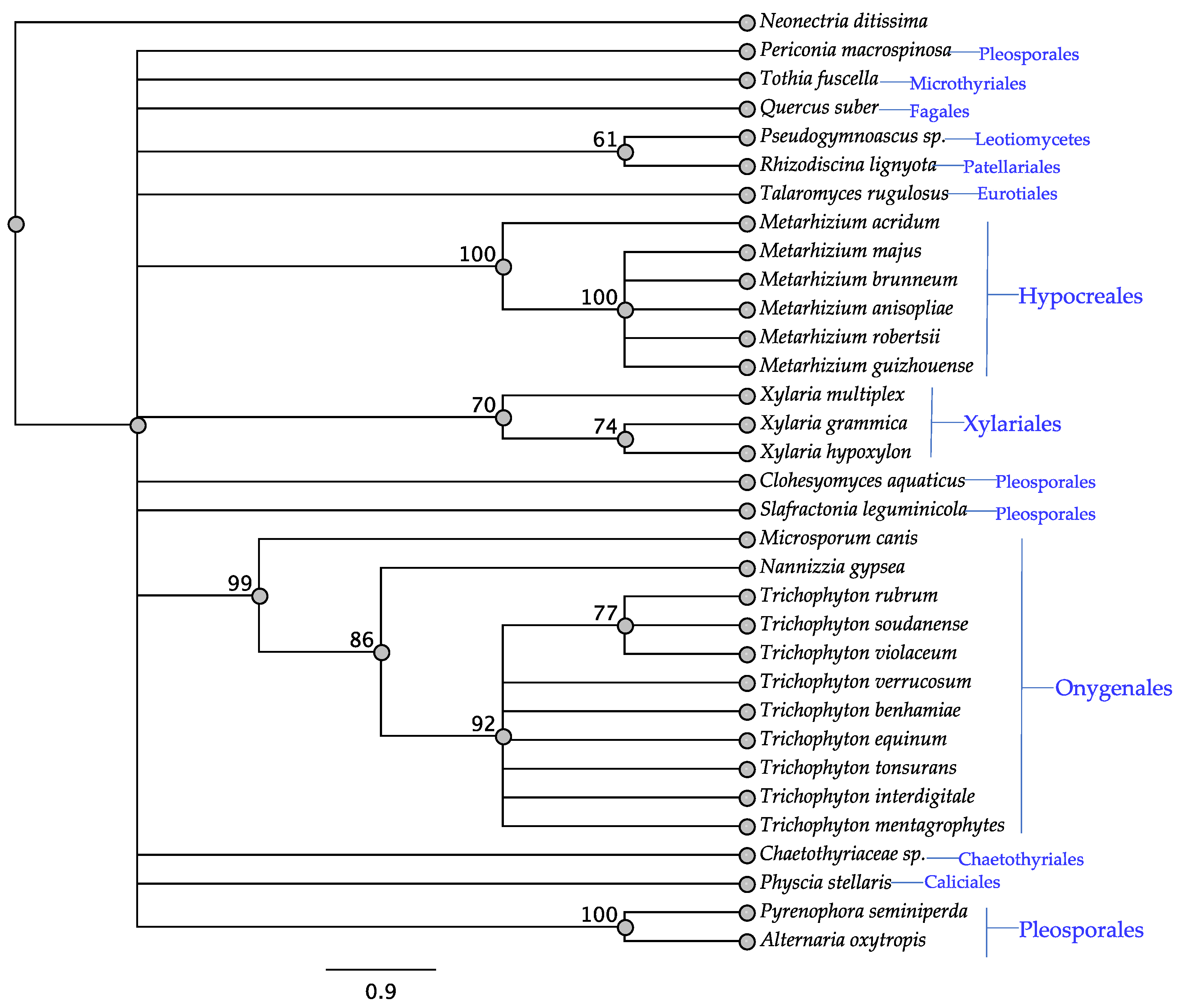
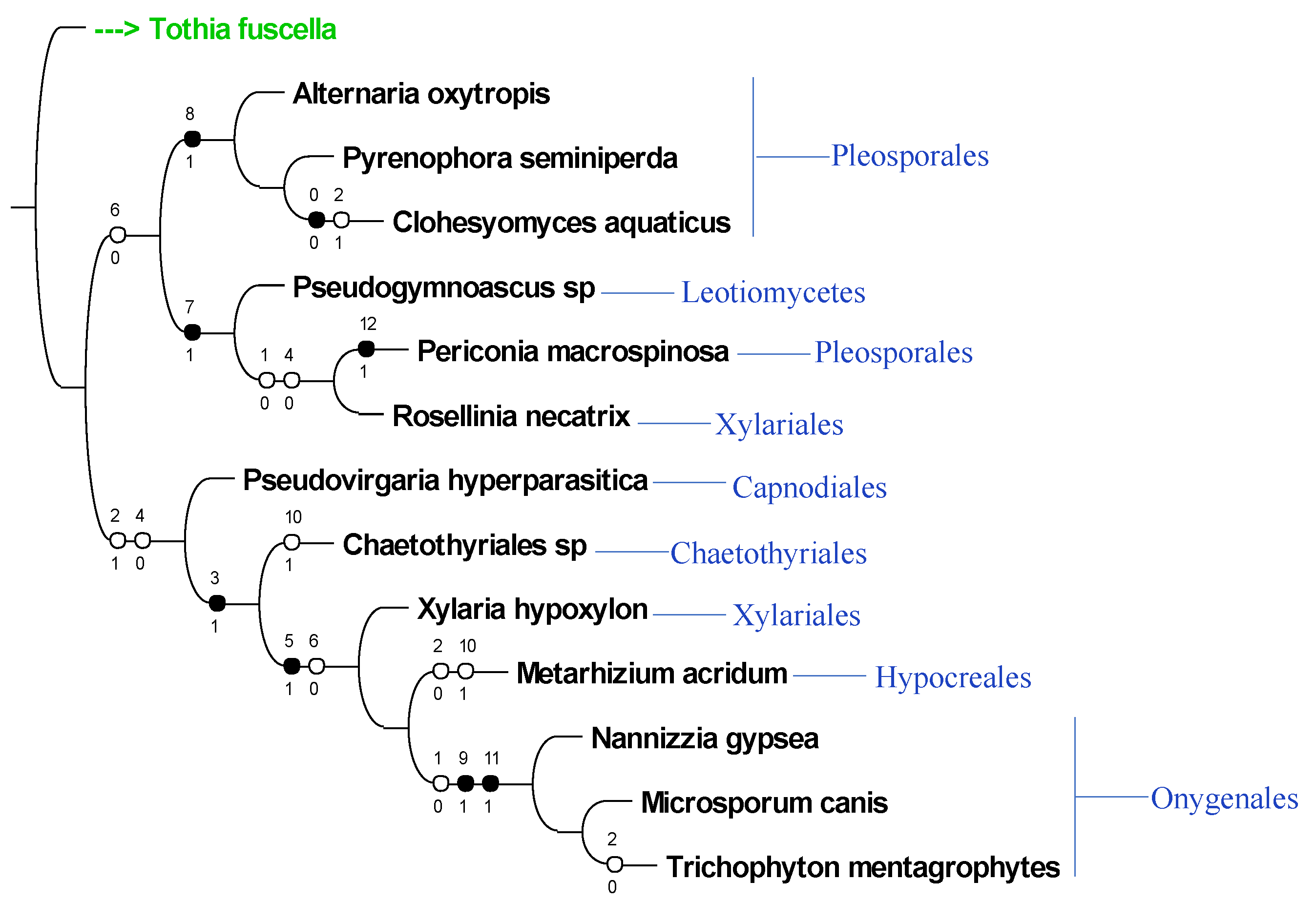
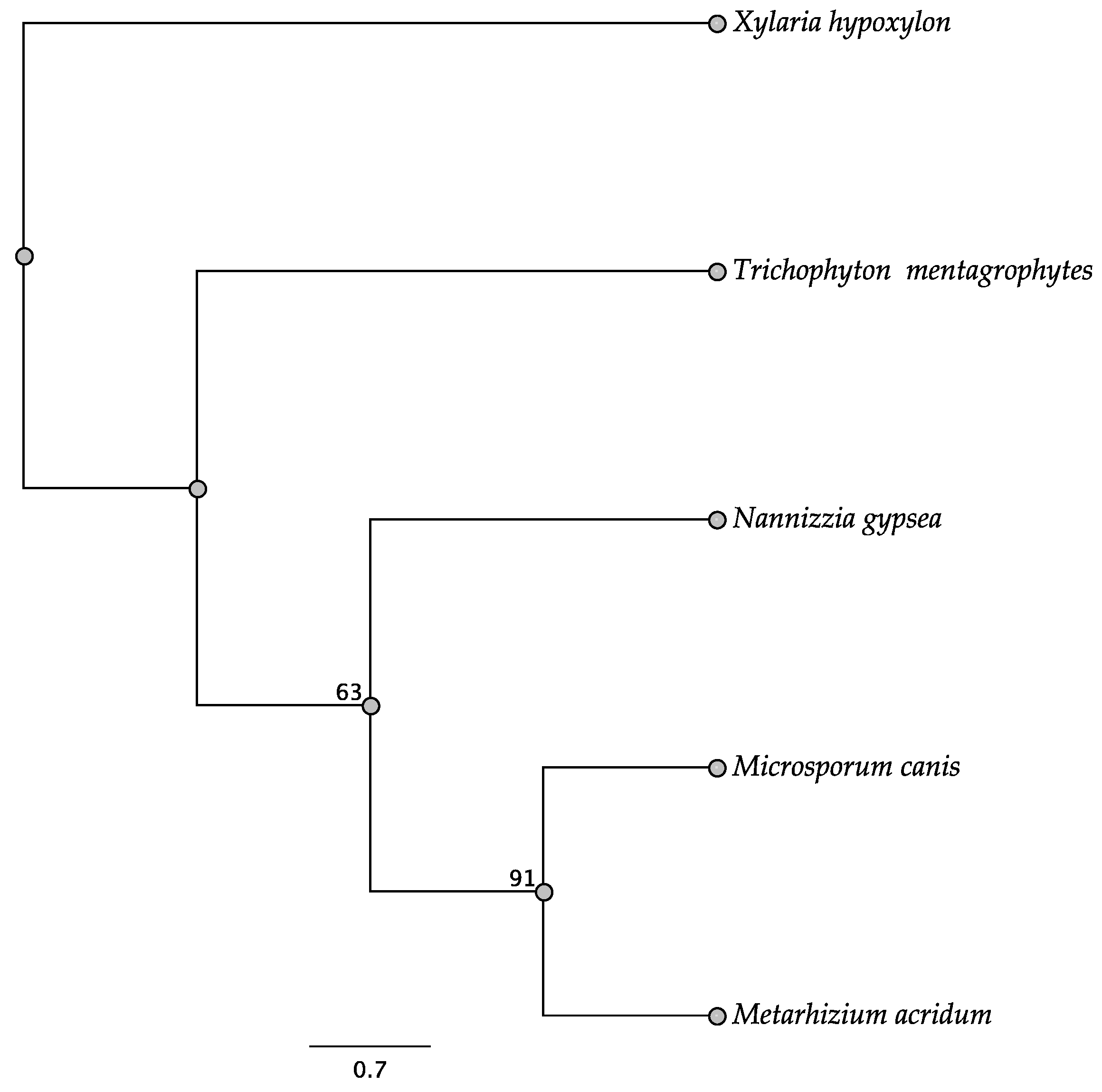
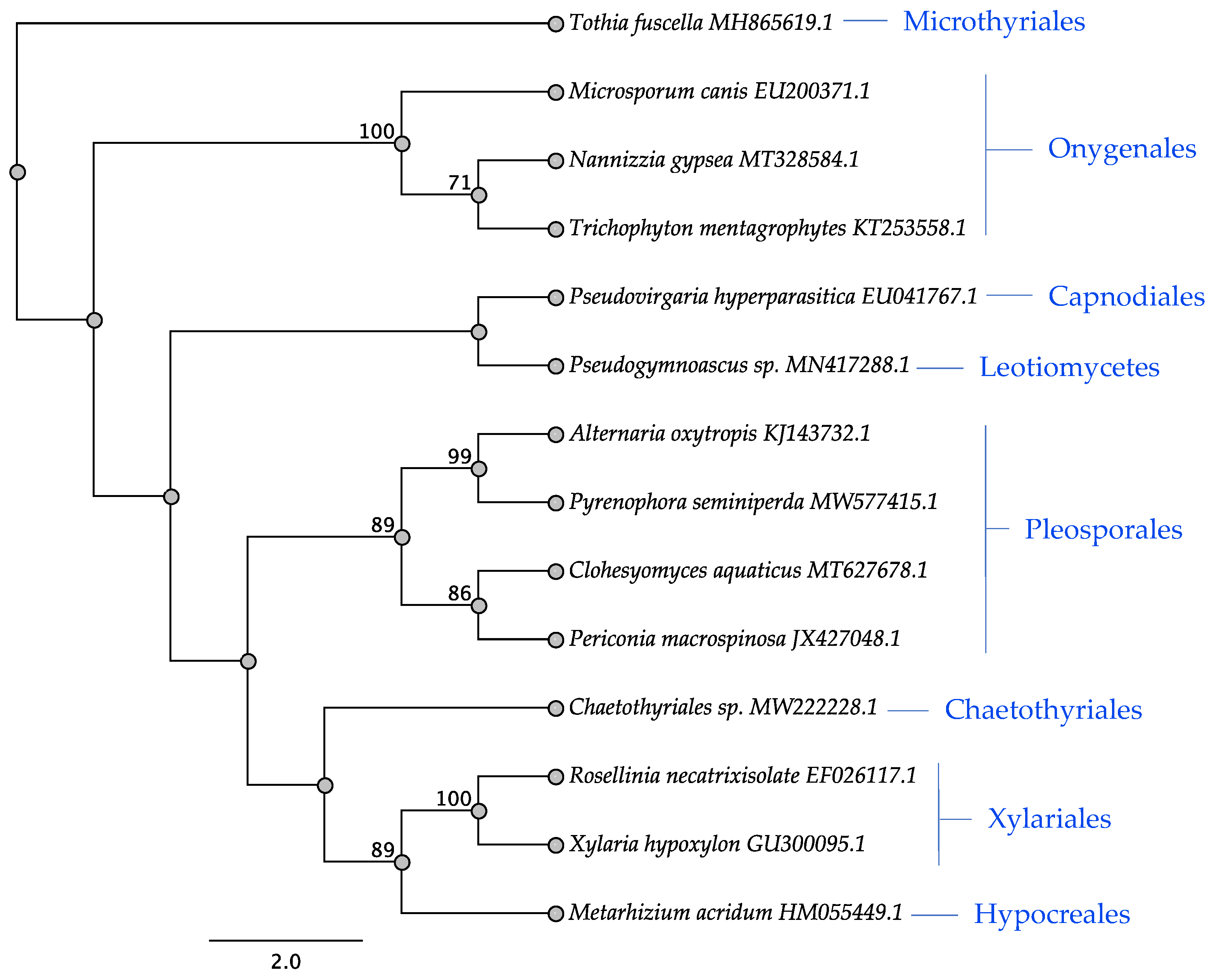
3.3. ITS Phylogeny
3.4. Open Reading Frames within the Intergenic Regions
3.5. Swainsonine
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dorling, P.R.; Huxtable, C.R.; Colegate, S.M. Inhibition of lysosomal α-mannosidase by swainsonine, an indolizidine alkaloid isolated from Swainsona canescens. Biochem. J. 1980, 191, 649–651. [Google Scholar] [CrossRef] [PubMed]
- Tulsiani, D.R.; Harris, T.M.; Touster, O. Swainsonine inhibits the biosynthesis of complex glycoproteins by inhibition of Golgi mannosidase II. J. Biol. Chem. 1982, 257, 7936–7939. [Google Scholar] [CrossRef]
- Winchester, B.; al Daher, S.; Carpenter, N.C.; Cenci di Bello, I.; Choi, S.S.; Fairbanks, A.J.; Fleet, G.W.J. The structural basis of the inhibition of human α-mannosidases by azafuranose analogues of mannose. Biochem. J. 1993, 290, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Colegate, S.M.; Dorling, P.R.; Huxtable, C.R. A spectroscopic investigation of swainsonine: An α-mannosidase inhibitor isolated from Swainsona canescens. Aust. J. Chem. 1979, 32, 2257–2264. [Google Scholar] [CrossRef]
- Cook, D.; Gardner, D.R.; Pfister, J.A. Swainsonine-containing plants and their relationship to endophytic fungi. J. Agric. Food Chem. 2014, 62, 7326–7334. [Google Scholar] [CrossRef] [PubMed]
- Grum, D.S.; Cook, D.; Baucom, D.; Mott, I.W.; Gardner, D.R.; Creamer, R.; Allen, J.G. Production of the alkaloid swainsonine by a fungal endophyte in the host Swainsona canescens. J. Nat. Prod. 2013, 76, 1984–1988. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.; Beaulieu, W.T.; Mott, I.W.; Riet-Correa, F.; Gardner, D.R.; Grum, D.; Pfister, J.; Clay., K.; Marcolongo-Pereira, C. Production of the alkaloid swainsonine by a fungal endosymbiont of the Ascomycete order Chaetothyriales in the host Ipomoea carnea. J. Agric. Food Chem. 2013, 61, 3797–3803. [Google Scholar] [CrossRef]
- Braun, K.; Romero, J.; Liddell, C.; Creamer, R. Production of swainsonine by fungal endophytes of locoweed. Mycol. Res. 2003, 107, 980–988. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhao, Q.; Wang, J.; Wang, J.; Wang, Y.; Song, Y.; Li, Q. Swainsonine-producing fungal endophytes from major locoweed species in China. Toxicon 2010, 56, 330–338. [Google Scholar] [CrossRef] [PubMed]
- . Schneider, M.J.; Ungemach, F.S.; Broquist, H.P.; Harris, T.M. (1S, 2R, 8R, 8aR)-1, 2, 8-trihydroxyoctahydroindolizine (swainsonine), an α-mannosidase inhibitor from Rhizoctonia Leguminicola. Tetrahedron 1983, 39, 29–32. [Google Scholar] [CrossRef]
- Patrick, M.; Adlard, M.W.; Keshavarz, T. Production of an indolizidine alkaloid, swainsonine by the filamentous fungus, Metarhizium anisopliae. Biotechnol. Lett. 1993, 15, 997–1000. [Google Scholar] [CrossRef]
- Cook, D.; Donzelli, B.G.; Creamer, R.; Baucom, D.L.; Gardner, D.R.; Pan, J.; Moore, N.; Krasnoff, S.; Jaromczyk, J.; Schardl, C.L. Swainsonine biosynthesis genes in diverse symbiotic and pathogenic fungi. G3 Genes Genomes Genet. 2017, 7, 1791–1797. [Google Scholar] [CrossRef] [Green Version]
- Oldrup, E.; McLain-Romero, J.; Padilla, A.; Moya, A.; Gardner, D.; Creamer, R. Localization of endophytic Undifilum fungi in locoweed seed and influence of environmental parameters on a locoweed in vitro culture system. Botany 2010, 88, 512–521. [Google Scholar] [CrossRef] [Green Version]
- Baucom, D.L.; Romero, M.; Belfon, R.; Creamer, R. Two new species of Undifilum, fungal endophytes of Astragalus (locoweeds) in the United States. Botany 2012, 90, 866–875. [Google Scholar] [CrossRef] [Green Version]
- Ralphs, M.H.; Mickelsen, L.V.; Turner, D.L. Cattle grazing white locoweed: Diet selection patterns of native and introduced cattle. Rangel. Ecol. Manag. J. Range Manag. Arch. 1987, 40, 333–335. [Google Scholar] [CrossRef]
- Cook, D.; Ralphs, M.; Welch, K.; Stegelmeier, B. Locoweed poisoning in livestock. Rangelands 2009, 31, 16–21. [Google Scholar] [CrossRef] [Green Version]
- Alhawatema, M.S.; Gebril, S.; Cook, D.; Creamer, R. RNAi-mediated down-regulation of a melanin polyketide synthase (pks1) gene in the fungus Slafractonia leguminicola. World J. Microbiol. Biotechnol. 2017, 33, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Leger, R.J.S.; Wang, C. Genetic engineering of fungal biocontrol agents to achieve greater efficacy against insect pests. Appl. Microbiol. Biotechnol. 2010, 85, 901–907. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Hong, S.; Chen, B.; Yin, Y.; Tang, G.; Hu, F.; Zhang, H.; Wang, C. Unveiling of swainsonine biosynthesis via a multibranched pathway in fungi. ACS Chem. Biol. 2020, 15, 2476–2484. [Google Scholar] [CrossRef] [PubMed]
- Noor, A.I.; Neyaz, M.; Cook, D.; Creamer, R. Molecular characterization of a fungal ketide synthase gene among swainsonine-producing Alternaria species in the USA. Curr. Microbiol. 2020, 77, 2554–2563. [Google Scholar] [CrossRef] [PubMed]
- Creamer, R.; Hille, D.B.; Neyaz, M.; Nusayr, T.; Schardl, C.L.; Cook, D. Genetic relationships in the toxin-producing fungal endophyte, Alternaria oxytropis using polyketide synthase and non-ribosomal peptide synthase genes. J. Fungi 2021, 7, 538. [Google Scholar] [CrossRef] [PubMed]
- Chooi, Y.H.; Tang, Y. Navigating the fungal polyketide chemical space: From genes to molecules. J. Org. Chem. 2012, 77, 9933–9953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boettger, D.; Hertweck, C. Molecular diversity sculpted by fungal PKS–NRPS hybrids. ChemBioChem 2013, 14, 28–42. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.D.; Johnson, L.; Itoh, Y.; Kodama, M.; Otani, H.; Kohmoto, K. Cloning and characterization of a cyclic peptide synthetase gene from Alternaria alternata apple pathotype whose product is involved in AM-toxin synthesis and pathogenicity. Mol. Plant-Microbe Interact. 2000, 13, 742–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, G.; Schörgendorfer, K.; Schneider-Scherzer, E.; Leitner, E. The peptide synthetase catalyzing cyclosporine production in Tolypocladium niveum is encoded by a giant 45.8-kilobase open reading frame. Curr. Genet. 1994, 26, 120–125. [Google Scholar] [CrossRef]
- Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Ed. 2009, 48, 4688–4716. [Google Scholar] [CrossRef] [PubMed]
- Lind, A.L.; Wisecaver, J.H.; Lameiras, C.; Wiemann, P.; Palmer, J.M.; Keller, N.P.; Rodrigues, F.; Goldman, G.; Rokas, A. Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLoS Biol. 2017, 15, e2003583. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, D.P.; Kroken, S.; Pryor, B.M.; Arnold, A.E. Interkingdom gene transfer of a hybrid NPS/PKS from bacteria to filamentous ascomycota. PLoS ONE 2011, 6, e28231. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardner, D.R.; Molyneux, R.J.; Ralphs, M.H. Analysis of swainsonine: Extraction methods, detection, and measurement in populations of locoweeds (Oxytropis spp.). J. Agric. Food Chem. 2001, 49, 4573–4580. [Google Scholar] [CrossRef] [PubMed]
- Gardner, D.R.; Cook, D. A comparison of alternative sample preparation procedures for the analysis of swainsonine using LC-MS/MS. Phytochem. Anal. 2011, 22, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Neyaz, M.; Gardner, D.R.; Creamer, R.; Cook, D. Localization of the Swainsonine-Producing Chaetothyriales Symbiont in the Seed and Shoot Apical Meristem in Its Host Ipomoea carnea. Microorganisms 2022, 10, 545. [Google Scholar] [CrossRef] [PubMed]
- Rokas, A.; Mead, M.E.; Steenwyk, J.L.; Raja, H.A.; Oberlies, N.H. Biosynthetic gene clusters and the evolution of fungal chemodiversity. Nat. Prod. Rep. 2020, 37, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Manning, V.A.; Pandelova, I.; Dhillon, B.; Wilhelm, L.J.; Goodwin, S.B.; Berlin, A.M.; Figueroa, M.; Ciuffetti, L.M. Comparative genomics of a plant-pathogenic fungus, Pyrenophora tritici-repentis, reveals transduplication and the impact of repeat elements on pathogenicity and population divergence. G3 Genes Genomes Genet. 2013, 3, 41–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Classification | Genebank Accessions | |||||||
|---|---|---|---|---|---|---|---|---|
| Genus and Species | Order | swnK | swnH2 | swnN | swnH1 | swnR | swnA | swnT |
| Pyrenophora seminiperda * | Pleosporales | RMZ73569 | RMZ73568 | RMZ73566 | RMZ73567 | RMZ73565 | - | - |
| Periconia macrospinosa | Pleosporales | PVI03268 | PVI03269 | PVI03272 | PVI03273 | PVI03271 | - | - |
| Slafractonia leguminicola * | Pleosporales | AQV04236 | AQV04235 | AQV04233 | AQV04234 | AQV04232 | - | AQV04231 |
| Alternaria oxytropis * | Pleosporales | AQV04230 | AQV04229 | AQV04227 | AQV04228 | AQV04226 | - | - |
| Clohesyomyces aquaticus | Pleosporales | ORY11783 | ORY11779 | ORY11781 | ORY11780 | ORY11782 | - | - |
| Microsporum canis | Onygenales | XP_002850891 | XP_002850892 | XP_002850894 | XP_002850896 | XP_002850890 | XP_002850893 | XP_002850895 |
| Nannizzia gypsea * | Onygenales | XP_003176907 | XP_003176906 | XP_003176904 | XP_003176902 | XP_003176908 | XP_003176905 | XP_003176903 |
| Trichophyton benhamiae * | Onygenales | XP_003014124 | XP_003014123 | DAA76506 | XP_003014119 | XP_003016302 | XP_003014122 | XP_003014120 |
| Trichophyton verrucosum | Onygenales | XP_003020763 | XP_003020762 | XP_003020760 | XP_003020758 | XP_003022179 | XP_003020761 | XP_003020759 |
| Trichophyton violaceum | Onygenales | OAL75151 | OAL75150 | OAL75148 | OAL75146 | OAL69278 | OAL75149 | OAL75147 |
| Trichophyton soudanense | Onygenales | EZF69148 | EZF69147 | EZF69143 | EZF69142 | EZF69504 | EZF69146 | EZF69144 |
| Trichophyton rubrum * 118892 | Onygenales | XP_003238870 | XP_003238869 | XP_003238867 | XP_003238865 | XP_003238559 | XP_003238868 | XP_003238866 |
| Trichophyton tonsurans | Onygenales | EGD97139 | EGD97140 | EGD97142 | EGD97144 | EGD97826 | EGD97141 | EGD97143 |
| Trichophyton equinum * | Onygenales | EGE01982 | EGE01983 | EGE01985 | EGE01987 | EGE02341 | EGE01984 | EGE01986 |
| Trichophyton interdigitale * H6 | Onygenales | EZF30502 | EZF30503 | EZF30348 | EZF30351 | EZF32734 | KDB22224 | EZF30349 |
| Trichophyton mentagrophytes | Onygenales | GBF63673 | GBF63672 | GBF63670 | GBF63668 | - | GBF63671 | GBF63669 |
| Metarhizium acridum CQM | Hypocreales | XP_007815889 | XP_007815888 | XP_007815891 | XP_007815893 | XP_007815890 | XP_007815894 | XP_007815892 |
| Metarhizium anisopliae 53293 | Hypocreales | KJK74452 | KJK74453 | KJK74450 | KJK74448 | KJK74451 | KJK74447 | KJK74449 |
| Metarhizium brunneum * 3297 | Hypocreales | XP_014543166 | XP_014543167 | XP_014543164 | XP_014543162 | XP_014543165 | XP_014543161 | XP_014543163 |
| Metarhizium guizhouense * 977 | Hypocreales | KID83603 | KID83604 | KID83601 | KID83599 | KID83602 | KID83598 | KID83600 |
| Metarhizium majus * 297 | Hypocreales | KID96318 | KID96317 | KID96320 | KID96322 | KID96319 | KID96323 | KID96321 |
| Metarhizium robertsii * 23 | Hypocreales | XP_007824811 | XP_007824812 | XP_007824809 | XP_007824807 | XP_007824810 | XP_007824806 | XP_007824808 |
| Fusarium sp. NRRL 66182 | Hypocreales | KAF5022960 | KAF5022961 | KAF5022958 | KAF5022962 | KAF5022959 | - | - |
| Chaetothyriaceae sp. * | Chaetothyriales | AQV04224 | AQV04220 | AQV04221 | AQV04219 | AQV04222 | AQV04223 | - |
| Xylaria hypoxylon | Xylariales | TGJ83472 | TGJ83466 | TGJ83449 | TGJ83451 | TGJ83448 | - | TGJ83450 |
| Xylaria multiplex | Xylariales | KAF2965339 | KAF2965340 | KAF2965337 | KAF2965359 | KAF2965338 | - | KAF2965358 |
| Xylaria grammica | Xylariales | RWA14357 | RWA14350 | RWA14377 | RWA14352 | RWA14351 | - | - |
| Xylaria polymorpha | Xylariales | KAH8158131 | KAH8158129 | - | - | KAH8158130 | - | - |
| Rosellinia necatrix | Xylariales | GAP93000 | GAP93001 | - | - | GAP93002 | - | - |
| Pseudovirgaria hyperparasitica | Capnodiales | XP_033604919 | XP_033604918 | XP_033604921 | - | XP_033604920 | XP_033604917 | XP_033604916 |
| Tothia fuscella | Microthyriales | KAF2430781 | KAF2430780 | KAF2430777 | KAF2430776 | KAF2430778 | - | - |
| Pseudogymnoascus sp. | Leotiomycetes | KFY51099 | KFY51100 | KFY51103 | KFY51104 | KFY51102 | - | - |
| Quercus suber | Fagales | XP_023929147 | XP_023929150 | XP_023929148 | XP_023929149 | - | - | - |
| Physcia stellaris | Caliciales | KAG6993713 | KAG6993712 | KAG6993710 | KAG6993709 | KAG6993711 | KAG6993708 | - |
| Rhizodiscinia lignyota | Patellariales | KAF2103738 | KAF2103739 | KAF2103742 | KAF2103743 | KAF2103741 | - | - |
| Talaromyces rugulosus | Eurotiales | XP_035344740 | XP_035344739 | XP_035344742 | XP_035344744 | XP_035344741 | XP_035344738 | XP_035344743 |
| Character | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organism | swnH2-swnK | swnR-swnN | swnK-swnR | swnH1-swnT | swnN-swnH1 | swnT-swnN | swnT-swnH2 | swnH2-swnR | swnH1-swnH2 | swnA-swnH2 | swnA-swnH1 | swnN-swnA | swnR-swnH1 |
| Pyrenophora seminiperda A | 1162 3O/FA | 40 ----- | - | - | 606 2O/F | - | - | - | 3 ----- | - | - | - | - |
| Periconia macrospinosa A/B | 913 * 4O/FA | - | - | - | - | - | - | 1808 1O/- | - | - | - | - | 1993 6O/F |
| Alternaria oxytropis A | 1094 2O/FB | 275 1O/A | - | - | 2007 1O/F | - | - | - | 47 ----- | - | - | - | - |
| Clohesyomyces aquaticus B | - | 285 2O/FB | 651 3O/F | - | 511 1O/F | - | - | - | 45 ----- | - | - | - | - |
| Microsporum canis B | 1453 1O/F | - | 348 1O/- | 1043 ----- | - | 212 N/A | - | - | - | 1267 1O/FA | - | 674 1O/F | - |
| Nannizzia gypsea A | 1774 1O/- | - | 621 2O/FA | 1080 3O/FB | - | 316 2O/B | - | - | - | 976 1O/F | - | 823 3O/FB | - |
| Trichophyton mentagrophytes A | 1353 1O/- | - | - | 1014 4O/F | - | 306 ----- | - | - | - | 970 3O/F | - | 878 ----- | - |
| Metarhizium acridum A/B | 1808 6O/F | 851 1O/- | 451 2O/F | 1317 * 1O/F | - | 293 * ----- | - | - | - | - | 390 * 1O/- | - | - |
| Chaetothyriaceae sp. A | 1989 3O/F | 863 3O/FA | 320 1O/- | 2139 2O/F | - | - | 163 ----- | - | - | - | 945 3O/B | - | - |
| Xylaria hypoxylon A/B | 2581 * 3O/F | 870 * ----- | 184 * 3O/F | 1576 1O/- | - | 842 1O/F | - | - | - | - | - | - | - |
| Rosellinia necatrix A/B | 1664 2O/B | - | - | - | - | - | - | 1208 * 1O/A | - | - | - | - | - |
| Pseudovirgaria hyperparasitica A | 899 1O/F | 154 ----- | −v | - | - | - | 1910 6O/F | - | - | - | - | - | - |
| Tothia fuscella A/B | 437 3O/F | −v * | - | - | 805 * 2O/F | - | 1646 3O/F | - | - | - | - | - | - |
| Pseudogymnoascus sp. A/B | 1519 * 6O/FA | 568 2O/F | - | - | 639 1O/F | - | - | 1958 3O/F | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neyaz, M.; Das, S.; Cook, D.; Creamer, R. Phylogenetic Comparison of Swainsonine Biosynthetic Gene Clusters among Fungi. J. Fungi 2022, 8, 359. https://doi.org/10.3390/jof8040359
Neyaz M, Das S, Cook D, Creamer R. Phylogenetic Comparison of Swainsonine Biosynthetic Gene Clusters among Fungi. Journal of Fungi. 2022; 8(4):359. https://doi.org/10.3390/jof8040359
Chicago/Turabian StyleNeyaz, Marwa, Sumanjari Das, Daniel Cook, and Rebecca Creamer. 2022. "Phylogenetic Comparison of Swainsonine Biosynthetic Gene Clusters among Fungi" Journal of Fungi 8, no. 4: 359. https://doi.org/10.3390/jof8040359
APA StyleNeyaz, M., Das, S., Cook, D., & Creamer, R. (2022). Phylogenetic Comparison of Swainsonine Biosynthetic Gene Clusters among Fungi. Journal of Fungi, 8(4), 359. https://doi.org/10.3390/jof8040359






