Functional Characterization of the M36 Metalloprotease FgFly1 in Fusarium graminearum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Conditions for Plants and Fungi
2.2. Construction of Gene-Deletion and Complementary Mutants
2.3. Construction of Green Fluorescent Protein (GFP) Vector
2.4. Sexual Reproduction and Vegetative Growth Assays
2.5. Pathogenicity and DON Production Assays
2.6. qRT—PCR Assays
2.7. Yeast Two-Hybrid Assays
2.8. BiFC
2.9. LUC
2.10. Statistical Analyses
3. Results
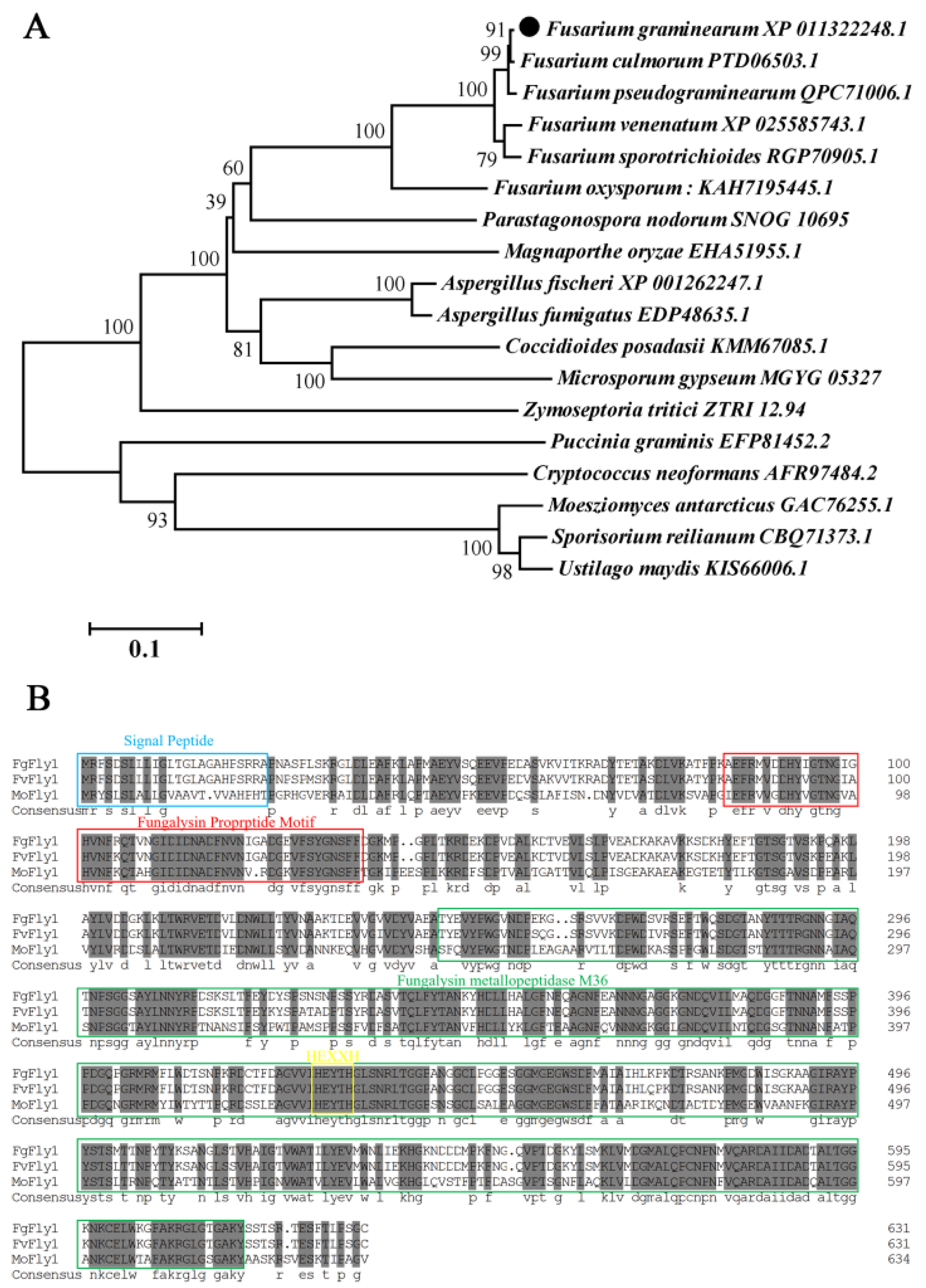
3.1. Phylogenetic Analysis and Sequence Alignment of Different Fungal Fly1 Proteins and Adcquisition of FgFly1 Deletion Mutants
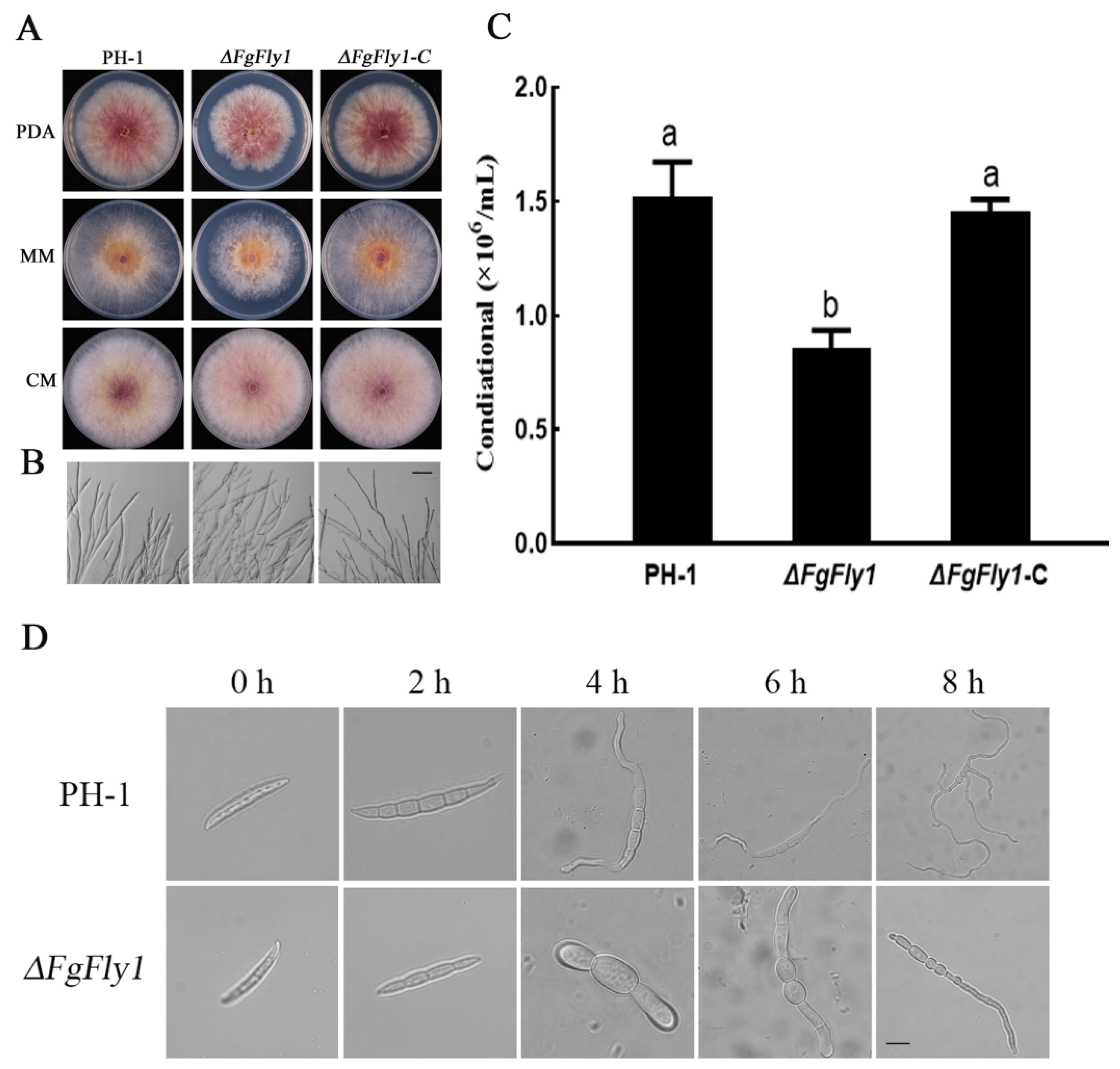
3.2. Deletion of FgFly1 Impairs Asexual Reproduction and Spore Germination of F. graminearum
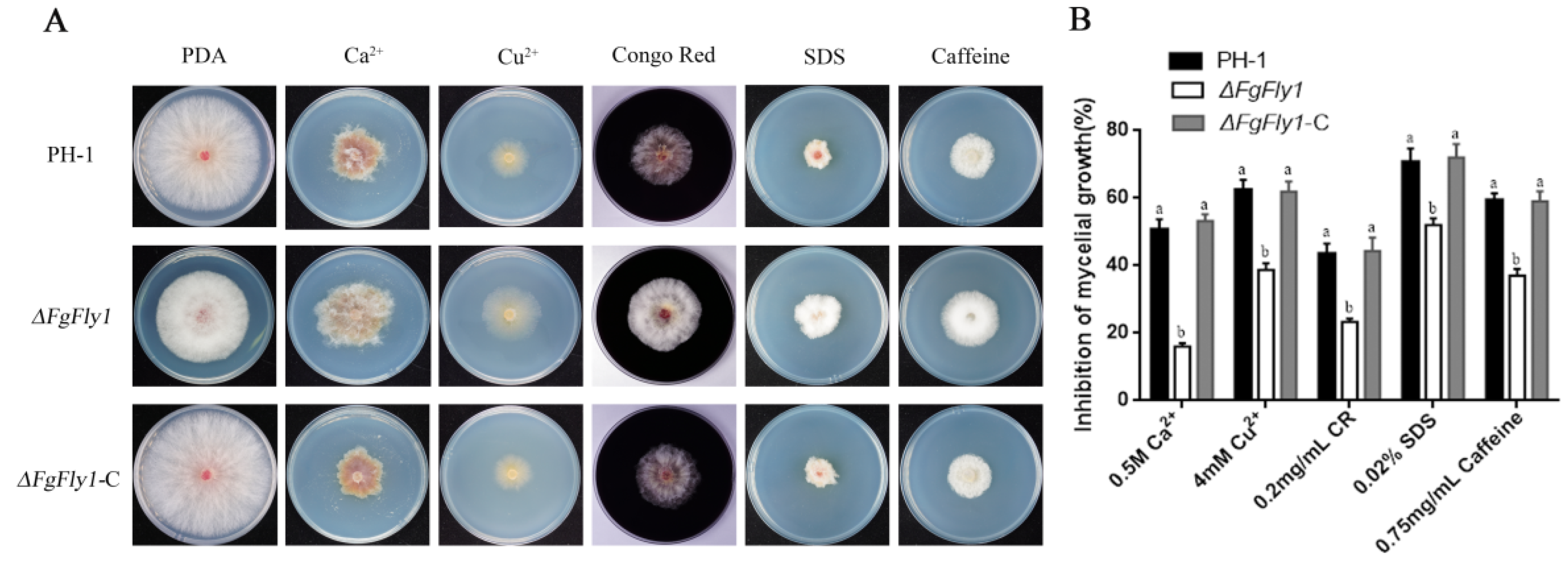
3.3. ∆FgFly1 Mutants Have Enhanced Tolerance to Metal Cation Stress and Cell-Wall-Related Stress
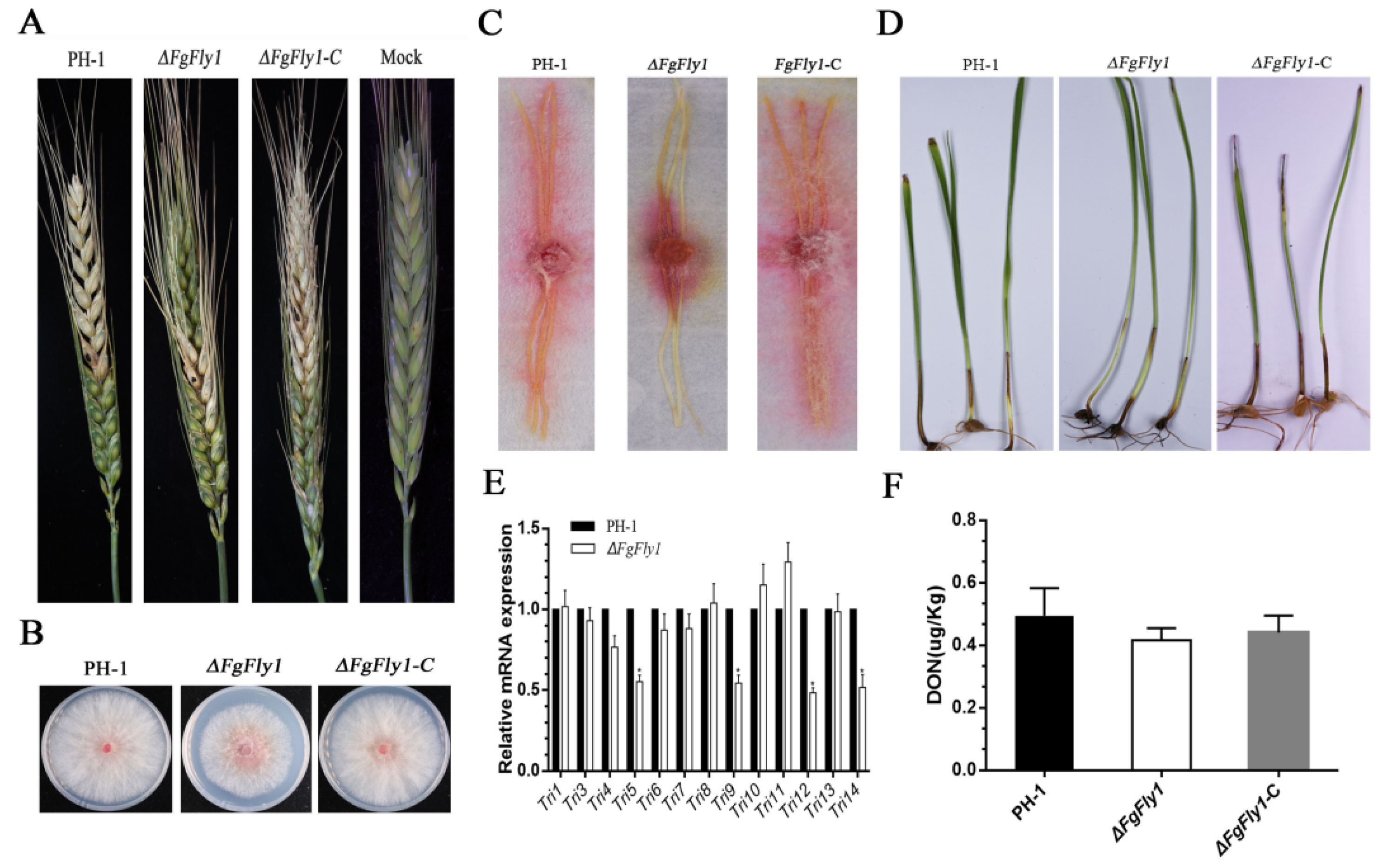
3.4. Deletion of FgFly1 Leads to Reduced Pathogenicity of F. graminearum
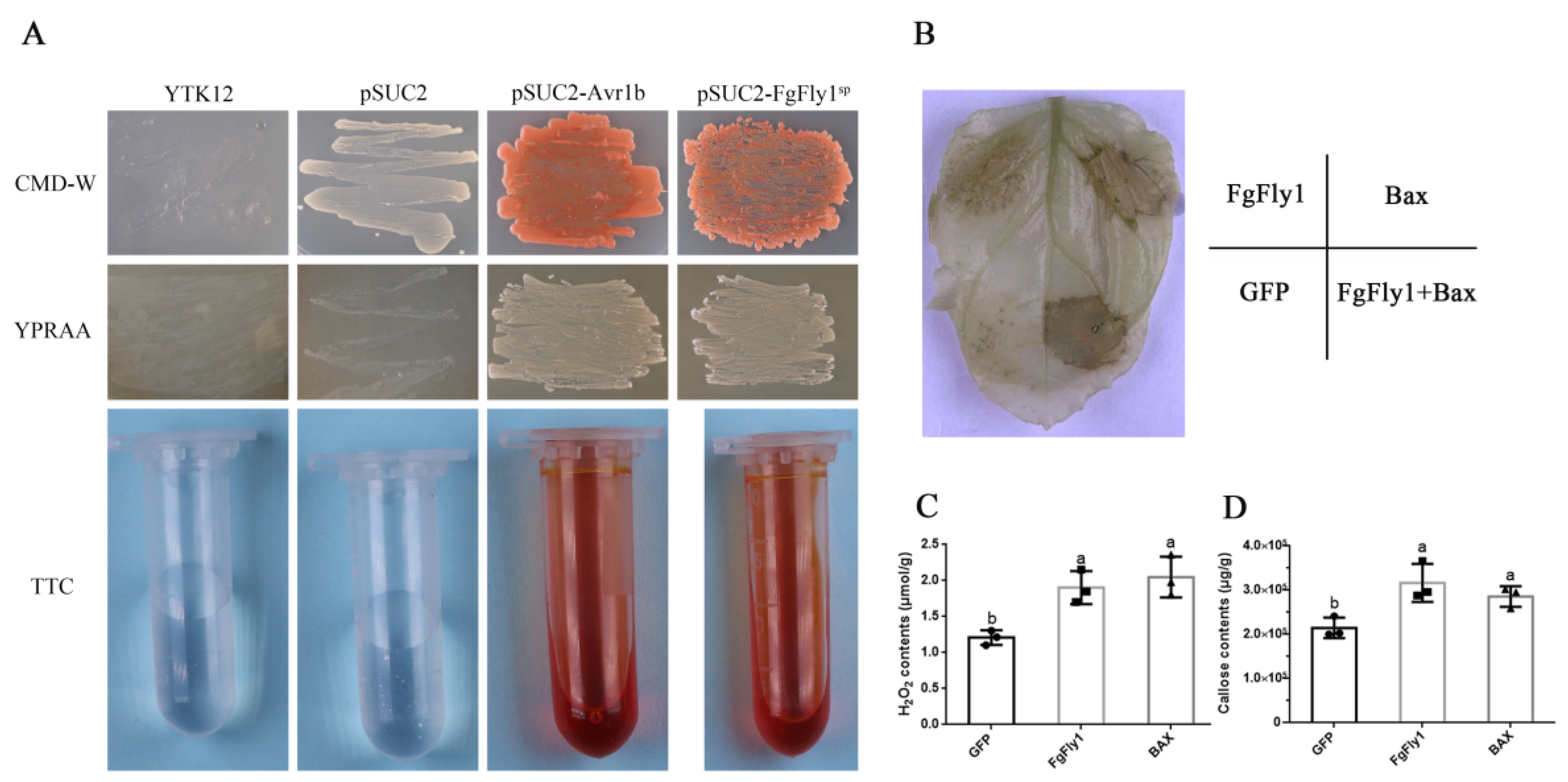
3.5. The Signal Peptide of FgFly1 Has a Secretory Function, and Overexpression Causes Allergic Necrosis in N. benthamiana
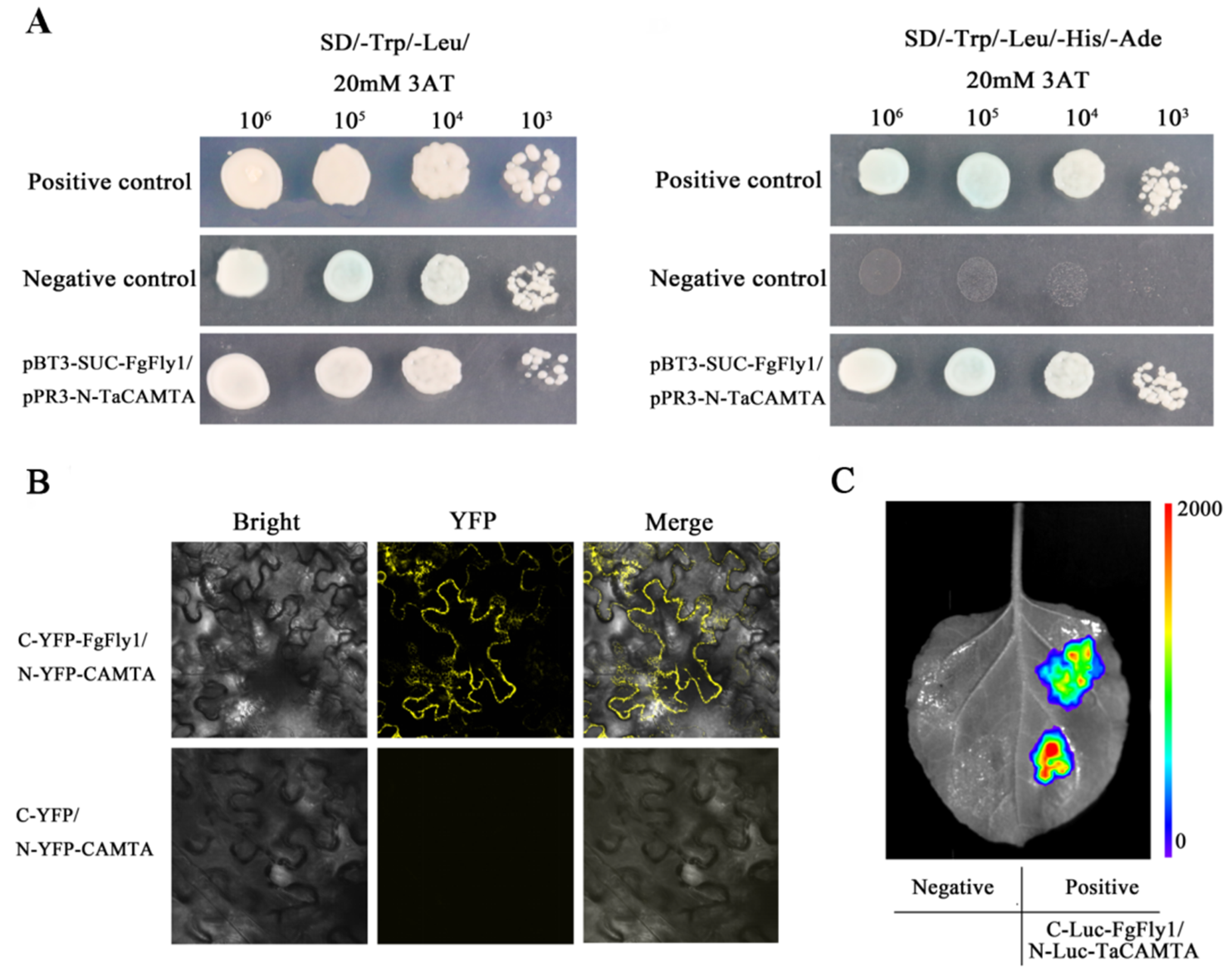
3.6. Interaction between FgFly1 and TaCAMTA
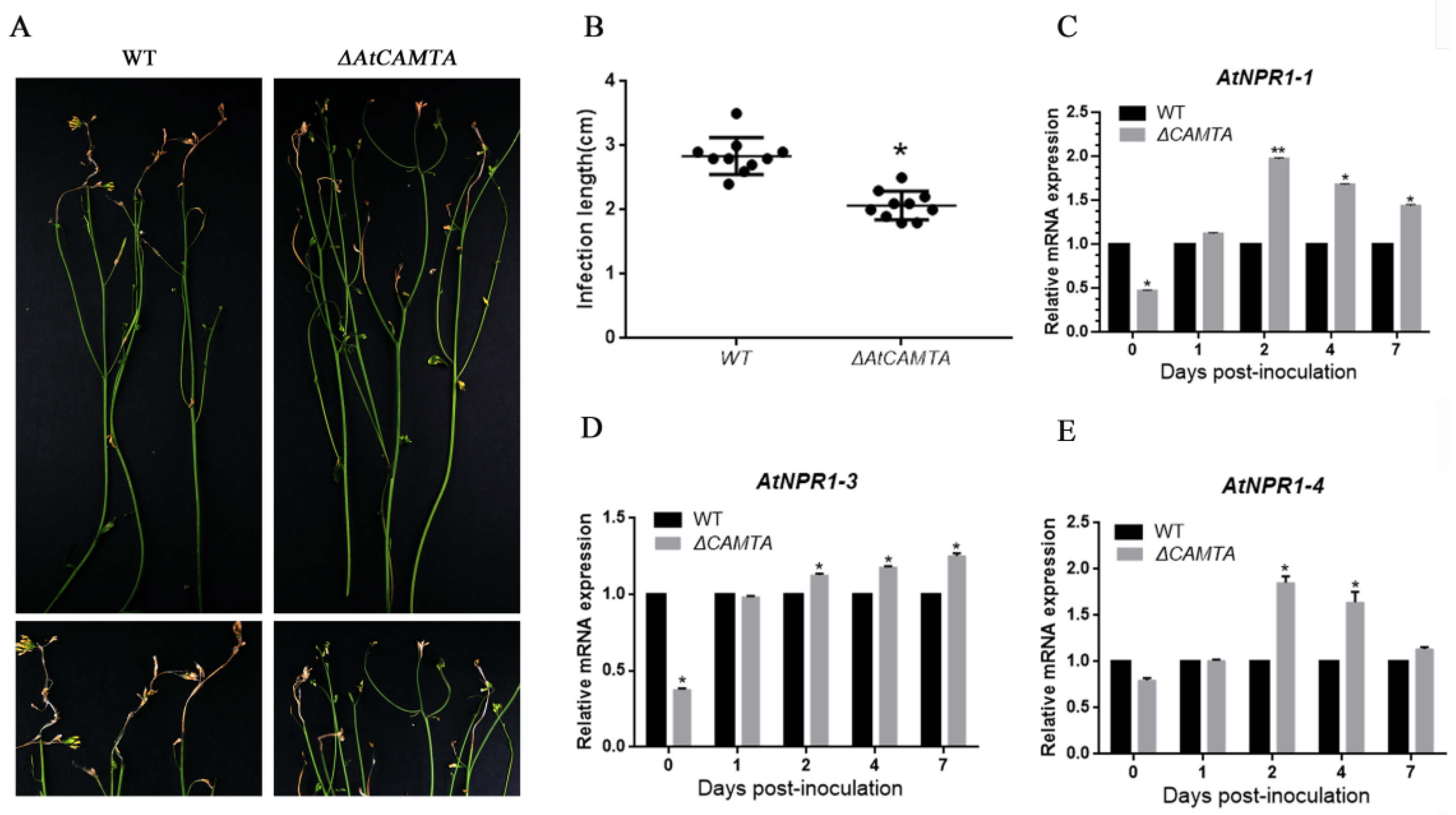
3.7. CAMTA Functional Verification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Starkey, D.E.; Ward, T.J.; Aoki, T.; Gale, L.R.; Kistler, H.; Geiser, D.M.; Suga, H.; Tóth, B.; Varga, J.; O’Donnell, K. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol. 2007, 44, 1191–1204. [Google Scholar] [CrossRef] [PubMed]
- Goswami, R.S.; Kistler, H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Shaner, G. Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 2004, 42, 135–161. [Google Scholar] [CrossRef] [PubMed]
- Pestka, J.J.; Smolinski, A.T. Deoxynivalenol: Toxicology and Potential Effects on Humans. J. Toxicol. Environ. Health Part B 2005, 8, 39–69. [Google Scholar] [CrossRef]
- McMullen, M.; Jones, R.; Gallenberg, D. Scab of Wheat and Barley: Are Emerging Disease. Plant Dis. 1997, 82, 1340–1348. [Google Scholar] [CrossRef] [Green Version]
- Sobrova, P.; Adam, V.; Vasatkova, A.; Beklova, M.; Zeman, L.; Kizek, R. Deoxynivalenol and its toxicity. Interdiscip. Toxicol. 2010, 3, 94–99. [Google Scholar] [CrossRef]
- McMullen, M.; Bergstrom, G.; De Wolf, E.; Dill-Macky, R.; Hershman, D.; Shaner, G.; Van Sanford, D. A Unified Effort to Fight an Enemy of Wheat and Barley: Fusarium Head Blight. Plant Dis. 2012, 96, 1712–1728. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Duplessis, S.; Cuomo, C.A.; Lin, Y.-C.; Aerts, A.; Tisserant, E.; Veneault-Fourrey, C.; Joly, D.; Hacquard, S.; Amselem, J.; Cantarel, B.L.; et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA 2011, 108, 9166–9171. [Google Scholar] [CrossRef] [Green Version]
- Saitoh, H.; Fujisawa, S.; Mitsuoka, C.; Ito, A.; Hirabuchi, A.; Ikeda, K.; Irieda, H.; Yoshino, K.; Yoshida, K.; Matsumura, H.; et al. Large-Scale Gene Disruption in Magnaporthe oryzae Identifies MC69, a Secreted Protein Required for Infection by Monocot and Dicot Fungal Pathogens. PLoS Pathog. 2012, 8, e1002711. [Google Scholar] [CrossRef] [Green Version]
- Gan, P.; Ikeda, K.; Irieda, H.; Narusaka, M.; O’Connell, R.J.; Narusaka, Y.; Takano, Y.; Kubo, Y.; Shirasu, K. Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytol. 2013, 197, 1236–1249. [Google Scholar] [CrossRef]
- Giraldo, M.; Valent, B. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 2013, 11, 800–814. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Van der Hoorn, R.A.L.; Terauchi, R.; Kamoun, S. Emerging Concepts in Effector Biology of Plant-Associated Organisms. Mol. Plant-Microbe Interact. 2009, 22, 115–122. [Google Scholar] [CrossRef] [Green Version]
- Kolattukudy, P.E. Biopolyester Membranes of Plants: Cutin and Suberin. Science 1980, 208, 990–1000. [Google Scholar] [CrossRef]
- Kolattukudy, P.E. Enzymatic penetration of the plant cuticle by fungal pathogens. Ann. Rev. Phytopathol. 1985, 23, 223–250. [Google Scholar] [CrossRef]
- Kolattukudy, P.E.; Lee, J.D.; Rogers, L.M.; Zimmerman, P.; Ceselski, S.; Fox, B.; Stein, B.; Copelan, E.A. Evidence for possible involvement of an elastolytic serine protease in aspergillosis. Infect. Immun. 1993, 61, 2357–2368. [Google Scholar] [CrossRef] [Green Version]
- Markaryan, A.; Morozova, I.; Yu, H.; Kolattukudy, P.E. Purification and characterization of an elastinolytic metalloproteas from Aspergillus fumigatus and immunoelectron microscopic evidence of secretion of this enzyme by the fungus invading the murine lung. Infect. Immun. 1994, 62, 2149–2157. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Feng, J.; Kong, J.; Ma, G.R. Research Progress on Metalloproteinases from Microorganisms. Prog. Biotechnol. 2002, 22, 61–63. [Google Scholar]
- Kothary, M.H.; Chase, T.; Macmillan, J.D. Correlation of elastase production by some strains of Aspergillus fumigatus with ability to cause pulmonary invasive aspergillosis in mice. Infect. Immun. 1984, 43, 320–325. [Google Scholar] [CrossRef] [Green Version]
- Monod, M.; Paris, S.; Sanglard, D.; Jaton-Ogay, K.; Bille, J.; Latgé, J.P. Isolation and characterization of a secreted metalloprotease of Aspergillus fumigatus. Infect. Immun. 1993, 61, 4099–4104. [Google Scholar] [CrossRef] [Green Version]
- Monod, M.; Togni, G.; Rahalison, L.; Frenk, E. Isoation and characterisation of an extracellular alkaline protease of Aspergillus fumigatus. J. Med. Microbiol. 1991, 35, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, M.V.; Sirakova, T.; Kolattukudy, P.E. Isolation, characterization, and cloning of cDNA and the gene for an elastinolytic serine proteinase from Aspergillus flavus. Infect. Immun. 1994, 62, 79–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirakova, T.D.; Markaryan, A.; Kolattukudy, P.E. Molecular cloning and sequencing of the cDNA and gene for a novel elastinolytic metalloproteinase from Aspergillus fumigatus and its expression in Escherichia coli. Infect. Immun. 1994, 62, 4208–4218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Bond, J.S. Families of metalloendopeptidases and their relationships. FEBS Lett. 1992, 312, 110–114. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.-W.; Chen, X.-L. Extracellular metalloproteases from bacteria. Appl. Microbiol. Biotechnol. 2011, 92, 253–262. [Google Scholar] [CrossRef]
- Zhang, Y.; Bak, D.D.; Heid, H.; Geider, K. Molecular characterization of a protease secreted by Erwinia amylovora. J. Mol. Biol. 1999, 289, 1239–1251. [Google Scholar] [CrossRef]
- Vargas, W.A.; Martín, J.M.S.; Rech, G.E.; Rivera, L.P.; Benito, E.P.; Díaz-Mínguez, J.M.; Thon, M.R.; Sukno, S.A. Plant Defense Mechanisms Are Activated during Biotrophic and Necrotrophic Development of Colletotricum graminicola in Maize. Plant Physiol. 2012, 158, 1342–1358. [Google Scholar] [CrossRef] [Green Version]
- Sanz-Martín, J.M.; Pacheco-Arjona, J.R.; Bello-Rico, V.; Vargas, W.A.; Monod, M.; Díaz-Mínguez, J.M.; Thon, M.R.; Sukno, S.A. A highly conserved metalloprotease effector enhances virulence in the maize anthracnose fungus Colletotrichum graminicola. Mol. Plant Pathol. 2016, 17, 1048–1062. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; McAdams, S.A.; Bryan, G.T.; Hershey, H.P.; Valent, B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000, 19, 4004–4014. [Google Scholar] [CrossRef]
- Yang, T.; Poovaiah, B.W. An Early Ethylene Up-regulated Gene Encoding a Calmodulin-binding Protein Involved in Plant Senescence and Death. J. Biol. Chem. 2000, 275, 38467–38473. [Google Scholar] [CrossRef] [Green Version]
- Bouché, N.; Scharlat, A.; Snedden, W.; Bouchez, D.; Fromm, H. A Novel Family of Calmodulin-binding Transcription Activators in Multicellular Organisms. J. Biol. Chem. 2002, 277, 21851–21861. [Google Scholar] [CrossRef] [Green Version]
- Choi, M.S.; Kim, M.C.; Yoo, J.H.; Moon, B.C.; Koo, S.C.; Park, B.O.; Lee, J.H.; Koo, Y.D.; Han, H.J.; Lee, S.Y.; et al. Isolation of a Calmodulin-binding Transcription Factor from Rice (Oryza sativa L.). J. Biol. Chem. 2005, 280, 40820–40831. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Peng, H.; Whitaker, B.D.; Conway, W.S. Characterization of a calcium/calmodulin-regulated SR/CAMTA gene family during tomato fruit development and ripening. BMC Plant Biol. 2012, 12, 19. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Poovaiah, B. A Calmodulin-binding/CGCG Box DNA-binding Protein Family Involved in Multiple Signaling Pathways in Plants. J. Biol. Chem. 2002, 277, 45049–45058. [Google Scholar] [CrossRef] [Green Version]
- Galon, Y.; Nave, R.; Boyce, J.M.; Nachmias, D.; Knight, M.R.; Fromm, H. Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 2008, 582, 943–948. [Google Scholar] [CrossRef] [Green Version]
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.S.N.; Poovaiah, B.W. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154–1158. [Google Scholar] [CrossRef]
- Harris, S.D.; Du, M.; Schardl, C.L.; Nuckles, E.M.; Vaillancourt, L.J. Morphogenesis in germinating Fusarium graminearum macroconidia. Mycologia 2005, 97, 880–887. [Google Scholar] [CrossRef]
- Yu, J.-H.; Hamari, Z.; Han, K.-H.; Seo, J.-A.; Reyes-Domínguez, Y.; Scazzocchio, C. Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004, 41, 973–981. [Google Scholar] [CrossRef]
- Proctor, R.H.; Hohn, T.M.; Mccormick, S.P. Reduced virulence of Gibberella zeae caused by disruption of a trichthecine toxin biosynthetic gene. Mol. Plant Microbe Interact. 1994, 8, 593–601. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Ren, X.; Wang, X.; Wan, Q.; Ding, K.; Chen, L. FgRad50 Regulates Fungal Development, Pathogenicity, Cell Wall Integrity and the DNA Damage Response in Fusarium graminearum. Front. Microbiol. 2020, 10, 2970. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Liu, X.; Yin, Y.; Ma, Z. Involvement of a Velvet Protein FgVeA in the Regulation of Asexual Development, Lipid and Secondary Metabolisms and Virulence in Fusarium graminearum. PLoS ONE 2011, 6, e28291. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Gu, Q.; Yun, Y.; Yin, Y.; Xu, J.; Shim, W.; Ma, Z. The TOR signaling pathway regulates vegetative development and virulence in Fusarium graminearum. New Phytol. 2014, 203, 219–232. [Google Scholar] [CrossRef]
- Seong, K.; Hou, Z.; Tracy, M.; Kistler, H.C.; Xu, J.-R. Random Insertional Mutagenesis Identifies Genes Associated with Virulence in the Wheat Scab Fungus Fusarium graminearum. Phytopathology 2005, 95, 744–750. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.-D.; Kerppola, T.K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 2003, 21, 539–545. [Google Scholar] [CrossRef]
- Zhou, Z.; Bi, G.; Zhou, J.-M. Luciferase Complementation Assay for Protein-Protein Interactions in Plants. Curr. Protoc. Plant Biol. 2018, 3, 42–50. [Google Scholar] [CrossRef]
- Seong, K.Y.; Pasquali, M.; Zhou, X.; Song, J.; Hilburn, K.; McCormick, S.; Dong, Y.; Xu, J.R.; Kistler, H.C. Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol. Microbiol. 2009, 72, 354–367. [Google Scholar] [CrossRef]
- Huang, Z.; Li, H.; Zhou, Y.; Bao, Y.; Duan, Z.; Wang, C.; Powell, C.A.; Chen, B.; Zhang, M.; Yao, W. Predication of the Effector Proteins Secreted by Fusarium sacchari Using Genomic Analysis and Heterogenous Expression. J. Fungi 2022, 8, 59. [Google Scholar] [CrossRef]
- Kim, Y.; Gilmour, S.J.; Chao, L.; Park, S.; Thomashow, M.F. Arabidopsis CAMTA Transcription Factors Regulate Pipecolic Acid Biosynthesis and Priming of Immunity Genes. Mol. Plant 2020, 13, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Tanaka, K.; Poovaiah, B.W. Calmodulin-binding transcription activator AtSR1/CAMTA3 fine-tunes plant immune response by transcriptional regulation of the salicylate receptor NPR1. Plant Cell Environ. 2021, 44, 3140–3154. [Google Scholar] [CrossRef] [PubMed]
- Gehrig, H.; Schüßler, A.; Kluge, M. Geosiphon pyriforme, a fungus forming endocytobiosis with Nostoc (Cyanobacteria), is an ancestral member of the glomales: Evidence by SSU rRNA Analysis. J. Mol. Evol. 1996, 43, 71–81. [Google Scholar] [CrossRef]
- Randall, T.A.; Dwyer, R.A.; Huitema, E.; Beyer, K.; Cvitanich, C.; Kelkar, H.; Fong, A.M.V.A.; Gates, K.; Roberts, S.; Yatzkan, E.; et al. Large-Scale Gene Discovery in the Oomycete Phytophthora infestans Reveals Likely Components of Phytopathogenicity Shared with True Fungi. Mol. Plant-Microbe Interact. 2005, 18, 229–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talbot, N.J. On the Trail of a Cereal Killer: Exploring the Biology of Magnaporthe grisea. Annu. Rev. Microbiol. 2003, 57, 177–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dow, M.; Newman, M.-A.; Von Roepenack, E. The Induction and Modulation of Plant Defense Responses by Bacterial Lipopolysaccharides. Annu. Rev. Phytopathol. 2000, 38, 241–261. [Google Scholar] [CrossRef] [PubMed]
- Ökmen, B.; Kemmerich, B.; Hilbig, D.; Wemhöner, R.; Aschenbroich, J.; Perrar, A.; Huesgen, P.F.; Schipper, K.; Doehlemann, G. Dual function of a secreted fungalysin metalloprotease in Ustilago maydis. New Phytol. 2018, 220, 249–261. [Google Scholar] [CrossRef] [Green Version]
- Naumann, T.A. Modification of recombinant maize ChitA chitinase by fungal chi-tinase-modifying proteins. Mol. Plant Pathol. 2011, 12, 365–372. [Google Scholar] [CrossRef]
- Yuan, P.; Du, L.; Poovaiah, B.W. Ca2+/Calmodulin-Dependent AtSR1/CAMTA3 Plays Critical Roles in Balancing Plant Growth and Immunity. Int. J. Mol. Sci. 2018, 19, 1764. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; He, M.; Liu, H.; Ding, H.; Liu, K.; Li, Y.; Cheng, P.; Li, Q.; Wang, B. Functional Characterization of the M36 Metalloprotease FgFly1 in Fusarium graminearum. J. Fungi 2022, 8, 726. https://doi.org/10.3390/jof8070726
Wang X, He M, Liu H, Ding H, Liu K, Li Y, Cheng P, Li Q, Wang B. Functional Characterization of the M36 Metalloprotease FgFly1 in Fusarium graminearum. Journal of Fungi. 2022; 8(7):726. https://doi.org/10.3390/jof8070726
Chicago/Turabian StyleWang, Xintong, Miaomiao He, Huanhuan Liu, Huiyi Ding, Kouhan Liu, Ying Li, Peng Cheng, Qiang Li, and Baotong Wang. 2022. "Functional Characterization of the M36 Metalloprotease FgFly1 in Fusarium graminearum" Journal of Fungi 8, no. 7: 726. https://doi.org/10.3390/jof8070726
APA StyleWang, X., He, M., Liu, H., Ding, H., Liu, K., Li, Y., Cheng, P., Li, Q., & Wang, B. (2022). Functional Characterization of the M36 Metalloprotease FgFly1 in Fusarium graminearum. Journal of Fungi, 8(7), 726. https://doi.org/10.3390/jof8070726






