Animal Histoplasmosis in Europe: Review of the Literature and Molecular Typing of the Etiological Agents
Abstract
:1. Introduction
2. Materials and Methods
2.1. Literature Review
2.2. Tissue Specimens from Animal Histoplasmosis
2.3. Histoplasma Capsulatum Reference Isolates
2.4. DNA Extraction
2.5. Molecular Tests
2.6. Arf, H-anti, tub, PRP8, CYP51pA, CYP51pB in H. capulatum Whole Genome Sequences
2.7. Phylogenetic Analyses
2.7.1. Arf-H-Anti-Tub
2.7.2. Arf-H-Anti-Tub-PRP8-CYP51pA-Cyp51pB
3. Results
3.1. Literature Review
3.2. Tissue Specimens from Veterinary Histoplasmosis Cases
3.3. Results of the Multilocus Sequence Typing
3.4. Phylogenetic Analyses
3.4.1. Arf-H-Anti-Tub
3.4.2. Arf-H-Anti-Tub-PRP8-CYP51pA-Cyp51pB
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bahr, N.C.; Antinori, S.; Wheat, L.J.; Sarosi, G.A. Histoplasmosis infections worldwide: Thinking outside of the Ohio River valley. Curr. Trop. Med. Rep. 2015, 2, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Oladele, R.O.; Ayanlowo, O.O.; Richardson, M.D.; Denning, D.W. Histoplasmosis in Africa: An emerging or a neglected disease? PLoS Negl. Trop. Dis. 2018, 12, e0006046. [Google Scholar] [CrossRef] [PubMed]
- Alados, J.C.; Miranda, C.; Ortiz, F.; Cano, R. Disseminated histoplasmosis in an AIDS patient in Spain. Eur J. Clin. Microbiol. Infect. Dis. 1993, 12, 793–794. [Google Scholar] [CrossRef] [PubMed]
- Antinori, S.; Galimberti, L.; Bonaccorso, C.; Vago, L.; Nebuloni, M.; Esposito, R. A case of fatal disseminated histoplasmosis of autochthonous origin in an Italian AIDS patient. Eur. J. Clin. Microbiol. Infect. Dis. 1997, 16, 545–546. [Google Scholar] [CrossRef]
- Antinori, S.; Giacomelli, A.; Corbellino, M.; Torre, A.; Schiuma, M.; Casalini, G.; Parravicini, C.; Milazzo, L.; Gervasoni, C.; Ridolfo, A.L. Histoplasmosis Diagnosed in Europe and Israel: A Case Report and Systematic Review of the Literature from 2005 to 2020. J. Fungi 2021, 7, 481. [Google Scholar] [CrossRef]
- Ashbee, H.R.; Evans, E.G.; Viviani, M.A.; Dupont, B.; Chryssanthou, E.; Surmont, I.; Tomsikova, A.; Vachkov, P.; Enero, B.; Zala, J.; et al. Histoplasmosis in Europe: Report on an epidemiological survey from the European Confederation of Medical Mycology Working Group. Med. Mycol. 2008, 46, 57–65. [Google Scholar] [CrossRef]
- Confalonieri, M.; Aiolfi, S.; Gandola, L.; Scartabellati, A.; Colavecchio, A.; Cannatelli, G.; Mazzoni, A. [Disseminated histoplasmosis and idiopathic CD4+ T-lymphocytopenia. An autochthonous Italian case]. Presse Med. 1995, 24, 459. [Google Scholar]
- Corbelli, G.; Mazzoni, A.; Allegri, L. Two additional cases of histoplasmosis as observed in Bologna Medical Clinic. Minerva Med. 1957, 48, 3823–3836. [Google Scholar]
- Farina, C.; Gnecchi, F.; Michetti, G.; Parma, A.; Cavanna, C.; Nasta, P. Imported and autochthonous histoplasmosis in Bergamo province, Northern Italy. Scand. J. Infect. Dis 2000, 32, 271–274. [Google Scholar]
- Gandola, L.; Confalonieri, M.; Aiolfi, S.; Scartabellati, A.; Patrini, G.; Ghio, L.; Mauri, F. Histoplasmosis in an HIV-negative Italian man with mycosis fungoides. Panminerva Med. 1992, 34, 93–95. [Google Scholar]
- Mantovani, A. Histoplasmosis in Europe. Ann. Soc. Belg Med. Trop 1972, 52, 421–433. [Google Scholar] [PubMed]
- Schmiedel, Y.; Büchi, A.E.; Berezowska, S.; Pöllinger, A.; Mühlethaler, K.; Funke-Chambour, M. Autochthonous Case of Pulmonary Histoplasmosis, Switzerland. Emerg Infect. Dis 2021, 27, 966–969. [Google Scholar] [CrossRef] [PubMed]
- Symmers, W.S. Histoplasmosis contracted in Britain: A case of histoplasmic lymphadenitis following clinical recovery from sarcoidosis. Br. Med. J. 1956, 2, 786–790. [Google Scholar] [CrossRef] [PubMed]
- Farina, C.; Rizzi, M.; Ricci, L.; Gabbi, E.; Caligaris, S.; Goglio, A. Imported and autochthonous histoplasmosis in Italy: New cases and old problems. Rev. Iberoam Micol 2005, 22, 169–171. [Google Scholar] [CrossRef]
- Sotgiu, G.; Mazzoni, A.; Mantovani, A.; Ajello, L.; Palmer, J. Histoplasma capsulatum: Occurence in soil from the Emilia-Romagna region of Italy. Science 1965, 147, 624. [Google Scholar] [CrossRef]
- Alteras, I. First Romanian isolation of Histoplasma capsulatum from the soil. Dermatol. Int. 1966, 5, 69–71. [Google Scholar] [CrossRef]
- Gomez, L.F.; Torres, I.P.; Jimenez, A.M.; McEwen, J.G.; de Bedout, C.; Pelaez, C.A.; Acevedo, J.M.; Taylor, M.L.; Arango, M. Detection of Histoplasma capsulatum in Organic Fertilizers by Hc100 Nested Polymerase Chain Reaction and Its Correlation with the Physicochemical and Microbiological Characteristics of the Samples. Am. J. Trop. Med. Hyg. 2018, 98, 1303–1312. [Google Scholar] [CrossRef]
- Weeks, R.J.; Padhye, A.; Ajello, L. Histoplasma capsulatum Variety Farciminosum: A New Combination for Histoplasma farciminosum. Mycologia 1985, 77, 964–970. [Google Scholar] [CrossRef]
- Kasuga, T.; Taylor, J.W.; White, T.J. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus Histoplasma capsulatum Darling. J. Clin. Microbiol. 1999, 37, 653–663. [Google Scholar] [CrossRef]
- Kasuga, T.; White, T.J.; Koenig, G.; McEwen, J.; Restrepo, A.; Castaneda, E.; Da Silva Lacaz, C.; Heins-Vaccari, E.M.; De Freitas, R.S.; Zancope-Oliveira, R.M.; et al. Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol. Ecol. 2003, 12, 3383–3401. [Google Scholar] [CrossRef]
- Teixeira Mde, M.; Patane, J.S.; Taylor, M.L.; Gomez, B.L.; Theodoro, R.C.; de Hoog, S.; Engelthaler, D.M.; Zancope-Oliveira, R.M.; Felipe, M.S.; Barker, B.M. Worldwide Phylogenetic Distributions and Population Dynamics of the Genus Histoplasma. PLoS Negl. Trop. Dis. 2016, 10, e0004732. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, V.E.; Marquez, R.; Turissini, D.A.; Goldman, W.E.; Matute, D.R. Genome Sequences Reveal Cryptic Speciation in the Human Pathogen Histoplasma capsulatum. MBio 2017, 8. [Google Scholar] [CrossRef]
- Arunmozhi Balajee, S.; Hurst, S.F.; Chang, L.S.; Miles, M.; Beeler, E.; Hale, C.; Kasuga, T.; Benedict, K.; Chiller, T.; Lindsley, M.D. Multilocus sequence typing of Histoplasma capsulatum in formalin-fixed paraffin-embedded tissues from cats living in non-endemic regions reveals a new phylogenetic clade. Med. Mycol. 2013, 51, 345–351. [Google Scholar] [CrossRef]
- Eisenberg, T.; Seeger, H.; Kasuga, T.; Eskens, U.; Sauerwald, C.; Kaim, U. Detection and characterization of Histoplasma capsulatum in a German badger (Meles meles) by ITS sequencing and multilocus sequencing analysis. Med. Mycol. 2013, 51, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, A.; von Bomhard, W.; Antweiler, E.; Tintelnot, K. Molecular identification of fungal pathogens in nodular skin lesions of cats. Med. Mycol. 2015, 53, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Murata, Y.; Sano, A.; Ueda, Y.; Inomata, T.; Takayama, A.; Poonwan, N.; Nanthawan, M.; Mikami, Y.; Miyaji, M.; Nishimura, K.; et al. Molecular epidemiology of canine histoplasmosis in Japan. Med. Mycol. 2007, 45, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Theodoro, R.C.; Scheel, C.M.; Brandt, M.E.; Kasuga, T.; Bagagli, E. PRP8 intein in cryptic species of Histoplasma capsulatum: Evolution and phylogeny. Infect. Genet. Evol. 2013, 18, 174–182. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. J. Clin. Epidemiol. 2009, 62, 1006–1012. [Google Scholar] [CrossRef]
- Donnelly, J.P.; Chen, S.C.; Kauffman, C.A.; Steinbach, W.J.; Baddley, J.W.; Verweij, P.E.; Clancy, C.J.; Wingard, J.R.; Lockhart, S.R.; Groll, A.H.; et al. Revision and Update of the Consensus Definitions of Invasive Fungal Disease From the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin. Infect. Dis. 2019. [Google Scholar] [CrossRef]
- Wilmes, D.; McCormick-Smith, I.; Lempp, C.; Mayer, U.; Schulze, A.B.; Theegarten, D.; Hartmann, S.; Rickerts, V. Detection of Histoplasma DNA from Tissue Blocks by a Specific and a Broad-Range Real-Time PCR: Tools to Elucidate the Epidemiology of Histoplasmosis. J. Fungi 2020, 6, 319. [Google Scholar] [CrossRef]
- Simon, S.; Veron, V.; Boukhari, R.; Blanchet, D.; Aznar, C. Detection of Histoplasma capsulatum DNA in human samples by real-time polymerase chain reaction. Diagn. Microbiol Infect. Dis. 2010, 66, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Rickerts, V.; Khot, P.D.; Myerson, D.; Ko, D.L.; Lambrecht, E.; Fredricks, D.N. Comparison of quantitative real time PCR with Sequencing and ribosomal RNA-FISH for the identification of fungi in formalin fixed, paraffin-embedded tissue specimens. BMC Infect. Dis. 2011, 11, 202. [Google Scholar] [CrossRef] [PubMed]
- Revankar, S.G.; Fu, J.; Rinaldi, M.G.; Kelly, S.L.; Kelly, D.E.; Lamb, D.C.; Keller, S.M.; Wickes, B.L. Cloning and characterization of the lanosterol 14alpha-demethylase (ERG11) gene in Cryptococcus neoformans. Biochem. Biophys. Res. Commun. 2004, 324, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Wheat, L.J.; Connolly, P.; Smedema, M.; Durkin, M.; Brizendine, E.; Mann, P.; Patel, R.; McNicholas, P.M.; Goldman, M. Activity of newer triazoles against Histoplasma capsulatum from patients with AIDS who failed fluconazole. J. Antimicrob. Chemother. 2006, 57, 1235–1239. [Google Scholar] [CrossRef] [PubMed]
- Amos, B.; Aurrecoechea, C.; Barba, M.; Barreto, A.; Basenko, E.Y.; Bazant, W.; Belnap, R.; Blevins, A.S.; Bohme, U.; Brestelli, J.; et al. VEuPathDB: The eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 2022, 50, D898–D911. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Rapp, J.; Löfqvist, A.; Breuer, E.; Rang, H. Sproßpilze als Ursache von Hautgranulomen beim Dachs in Süddeutschland. Tierärztl. Umschau 1992, 47, 451–454. [Google Scholar]
- Wohlsein, P.; Bauder, B.; Kuttin, E.S.; Kaufman, L.; Seeliger, F.; von Keyserlingk, M. [Histoplasmosis in two badgers (Meles meles) in northern Germany]. Dtsch. Tierarztl. Wochenschr. 2001, 108, 273–276. [Google Scholar]
- Jacobsen, B.; Baumgartner, W.; Bialek, R. Disseminated histoplasmosis in a European hedgehog (Erinaceus europaeus) in Northern Germany. Mycoses 2011, 54, 538–541. [Google Scholar] [CrossRef]
- Richter, M.; Hauser, B.; Kaps, S.; Spiess, B.M. Keratitis due to Histoplasma spp. in a horse. Vet. Ophthalmol 2003, 6, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Burgisser, H.; Fankhauser, R.; Kaplan, W.; Klingler, K.; Scholer, H.J. Mycoses in a badger in Switzerland: Histologically histoplasmosis. Pathol. Microbiol. 1961, 24, 794–802. [Google Scholar] [PubMed]
- Akdesir, E.; Origgi, F.C.; Wimmershoff, J.; Frey, J.; Frey, C.F.; Ryser-Degiorgis, M.P. Causes of mortality and morbidity in free-ranging mustelids in Switzerland: Necropsy data from over 50 years of general health surveillance. BMC Vet. Res. 2018, 14, 195. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N.M.; Favrot, C.; Monod, M.; Grest, P.; Rech, K.; Wilhelm, S. A case in Europe of feline histoplasmosis apparently limited to the skin. Vet. Dermatol. 2013, 24, 635–638. [Google Scholar] [CrossRef]
- Mantovani, A.; Mazzoni, A.; Ajello, L. Histoplasmosis in Italy. I. Isolation of Histoplasma capsulatum from dogs in the province of Bologna. Sabouraudia 1968, 6, 163–164. [Google Scholar] [CrossRef]
- Reginato, A.; Giannuzzi, P.; Ricciardi, M.; De Simone, A.; Sanguinetti, M.; Porcellato, I.; Mandara, M.T. Extradural spinal cord lesion in a dog: First case study of canine neurological histoplasmosis in Italy. Vet. Microbiol 2014, 170, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Mavropoulou, A.; Grandi, G.; Calvi, L.; Passeri, B.; Volta, A.; Kramer, L.H.; Quintavalla, C. Disseminated histoplasmosis in a cat in Europe. J. Small Anim. Pract. 2010, 51, 176–180. [Google Scholar] [CrossRef]
- Sangiorgi, C. Blastomicosi spontanea nei muridi. Pathologica 1922, 14, 493–495. [Google Scholar]
- Sinkovics, J. [Histoplasmosis in laboratory mouse]. Orv. Hetil. 1956, 97, 968–969. [Google Scholar]
- Jensen, H.E.; Bloch, B.; Henriksen, P.; Dietz, H.H.; Schonheyder, H.; Kaufman, L. Disseminated histoplasmosis in a badger (Meles meles) in Denmark. Apmis 1992, 100, 586–592. [Google Scholar] [CrossRef]
- Bauder, B.; Kubber-Heiss, A.; Steineck, T.; Kuttin, E.S.; Kaufman, L. Granulomatous skin lesions due to histoplasmosis in a badger (Meles meles) in Austria. Med. Mycol. 2000, 38, 249–253. [Google Scholar] [CrossRef]
- Farinas, F.; Flores, L.; Rodriguez, P.; Sabalete, T.; Quevedo, M.A. [Disseminated histoplasmosis in a dorcas gazelle (Gazella dorcas neglecta) kept in captivity conditions in Spain]. Rev. Iberoam Micol. 2009, 26, 152–154. [Google Scholar] [CrossRef] [PubMed]
- Akun, R.S. Histoplasmosis in a Cat. J. Am. Vet. Med. Assoc. 1950, 116, 880. [Google Scholar]
- Salfelder, K. Zur Frage des Vorkommens von Histoplasmose in Europa. Dtsch Med. Wochenschr 1965, 90, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Grosse, G.; Staib, F.; Rapp, J.; Rang, H.; Heise, W.; Kaufman, L. Pathological and epidemiological aspects of skin lesions in histoplasmosis: Observations in an AIDS patient and badgers outside endemic areas of histoplasmosis. Zentralbl. Bakteriol. 1997, 285, 531–539. [Google Scholar] [CrossRef]
- Vite-Garín, T.; Estrada-Bárcenas, D.A.; Gernandt, D.S.; Reyes-Montes, M.D.R.; Sahaza, J.H.; Canteros, C.E.; Ramírez, J.A.; Rodríguez-Arellanes, G.; Serra-Damasceno, L.; Zancopé-Oliveira, R.M.; et al. Histoplasma capsulatum Isolated from Tadarida brasiliensis Bats Captured in Mexico Form a Sister Group to North American Class 2 Clade. J. Fungi 2021, 7, 529. [Google Scholar] [CrossRef]
- Taylor, J.W.; Hann-Soden, C.; Branco, S.; Sylvain, I.; Ellison, C.E. Clonal reproduction in fungi. Proc. Natl. Acad. Sci. USA 2015, 112, 8901–8908. [Google Scholar] [CrossRef]
- Bialek, R.; Ernst, F.; Dietz, K.; Najvar, L.K.; Knobloch, J.; Graybill, J.R.; Schaumburg-Lever, G. Comparison of staining methods and a nested PCR assay to detect Histoplasma capsulatum in tissue sections. Am. J. Clin. Pathol. 2002, 117, 597–603. [Google Scholar] [CrossRef]
- Bialek, R.; Feucht, A.; Aepinus, C.; Just-Nubling, G.; Robertson, V.J.; Knobloch, J.; Hohle, R. Evaluation of two nested PCR assays for detection of Histoplasma capsulatum DNA in human tissue. J. Clin. Microbiol. 2002, 40, 1644–1647. [Google Scholar] [CrossRef]
- al-Ani, F.K. Epizootic lymphangitis in horses: A review of the literature. Rev. Sci. Tech. 1999, 18, 691–699. [Google Scholar] [CrossRef]
- Jones, K. Epizootic lymphangitis: The impact on subsistence economies and animal welfare. Vet. J. 2006, 172, 402–404. [Google Scholar] [CrossRef] [PubMed]
- Biglino, A.; De Rosa, G.; Lipani, F. Upper lobe infiltrate with cough, fever, fatigue. Eur. Respir. J. 1992, 5, 1021–1022. [Google Scholar]
- Confalonieri, M.; Nanetti, A.; Gandola, L.; Colavecchio, A.; Aiolfi, S.; Cannatelli, G.; Parigi, P.; Scartabellati, A.; Della Porta, R.; Mazzoni, A. Histoplasmosis capsulati in Italy: Autochthonous or imported? Eur. J. Epidemiol. 1994, 10, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Rosam, K.; Monk, B.C.; Lackner, M. Sterol 14α-Demethylase Ligand-Binding Pocket-Mediated Acquired and Intrinsic Azole Resistance in Fungal Pathogens. J. Fungi 2020, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Canteros, C.E.; Madariaga, M.J.; Lee, W.; Rivas, M.C.; Davel, G.; Iachini, R. [Endemic fungal pathogens in a rural setting of Argentina: Seroepidemiological study in dogs]. Rev. Iberoam Micol. 2010, 27, 14–19. [Google Scholar] [CrossRef]
- Cordeiro, R.A.; Coelho, C.G.; Brilhante, R.S.; Sidrim, J.J.; Castelo-Branco, D.S.; Moura, F.B.; Rocha, F.A.; Rocha, M.F. Serological evidence of Histoplasma capsulatum infection among dogs with leishmaniasis in Brazil. Acta Trop. 2011, 119, 203–205. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Hanzlicek, A.S. Evaluation of a novel monoclonal antibody-based enzyme immunoassay for detection of Histoplasma antigen in urine of dogs. J. Vet. Intern. Med. 2021, 35, 284–293. [Google Scholar] [CrossRef]

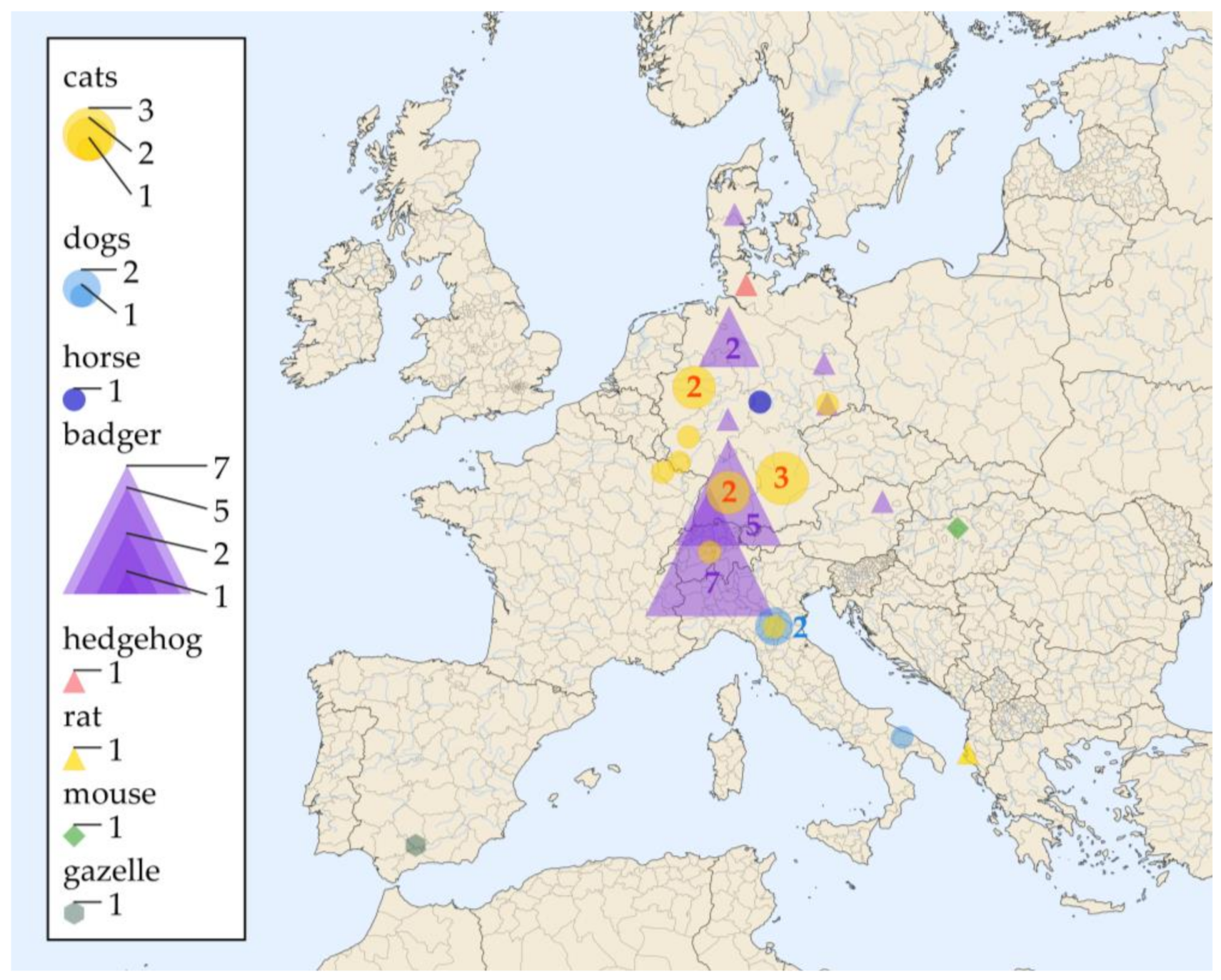


| Nr | Identificator | Living Environment: Country (State) | Animal | Positively Sampled Tissues | Diagnosis | Supplementary Diagnostic Methods | Reference |
|---|---|---|---|---|---|---|---|
| 1 | 2009-I | Germany (BB) | badger | skin | histopathology | PCR (18SrDNA 2, ITS2 rDNA 3) | this study |
| 2 | 2010-I | Germany (BY) | cat | skin | histopathology | PCR (100 kDa-like protein 6, ITS2 rDNA 3), MLST (Eurasian clade) | [25] |
| 3 | 2011-I | Germany (RP) | cat | skin | histopathology | PCR (100 kDa-like protein 6, ITS2 rDNA 3), MLST (Eurasian clade) | [25] |
| 4 | 2012-I | France (Lorraine) | cat | skin | histopathology | PCR (18SrDNA 2, ITS2 rDNA 3) | this study |
| 5 | 2014-I | Germany (BW) | badger | skin, testicles | histopathology | PCR (18SrDNA 2, ITS2 rDNA 3) | this study |
| 6 | 2015-I | Germany (SN) | badger | skin | histopathology | PCR (18SrDNA 2, ITS2 rDNA 3) | this study |
| 7 | 2021-I | Germany (BW) | badger | skin, regional lymph node | histopathology | PCR (H qPCR 4) | this study |
| 8 | 2010-II | Germany (SL) | cat | skin | histopathology | PCR (18SrDNA 2, ITS2 rDNA 3) | this study |
| 9 | 2014-II | Germany (BY) | cat | skin | histopathology | PCR (28SrDNA qPCR 5) | this study |
| 10 | 2017-I | Germany (BW) | cat | skin | histopathology | PCR (H qPCR 4) | this study |
| 11 | 2018-I | Germany (BW) | cat | skin | histopathology | PCR (H qPCR 4) | this study |
| 12 | 2018-II | Germany (NW) | cat | skin | histopathology | PCR (H qPCR 4) | this study |
| 13 | 2020-I | Germany (NW) | cat | skin | histopathology | PCR (H qPCR 4) | this study |
| 14 | 2021-II | Germany (NI) | badger (A) | skin, regional lymph nodes, spleen, testicle | histopathology | immunohistology | |
| 15 | n.a. | Germany (BY) | cat | skin | histopathology | PCR (100 kDa-like protein6, ITS2 rDNA 3), MLST (Eurasian clade) | [25] |
| 16 | n.a. | Germany (BW) | badger | skin | histopathology | immunohistology | [38] |
| 17 | n.a. | Germany (BW) | badger | skin | histopathology | immunohistology | [38] |
| 18 | n.a. | Germany (BW) | badger | skin | histopathology | immunohistology | [38] |
| 19 | n.a. | Germany (NI) | badger (B) | skin, regional lymph nodes, spleen | histopathology | immunohistology | [39] |
| 20 | Meles meles | Germany (HE) | badger | skin and regional lymphnode | histopathology | PCR (ITS1-5.8S-ITS2 7), MLST (Eurasian clade) | [24] |
| 21 | n.a. | Germany (SH) | hedgehog | spleen, liver, lung, bone marrow, lymph nodes, myocardium, kidney | histopathology | PCR (100 kDa-like protein 6, 18SrDNA 2) | [40] |
| 22 | n.a. | Germany (unknown) | horse | cornea | histopathology | n.a. | [41] |
| 23 | n.a. | Switzerland (Bern) | badger | submandibular lymph node | histopathology | immunohistology | [42] |
| 24 | n.a. | Switzerland | badger | skin | histopathology | n.a. | [43] |
| 25 | n.a. | Switzerland | badger | skin | histopathology | n.a. | [43] |
| 26 | n.a. | Switzerland | badger | skin | histopathology | n.a. | [43] |
| 27 | n.a. | Switzerland | badger | skin | histopathology | n.a. | [43] |
| 28 | n.a. | Switzerland | badger | skin, subcutaneous lymph nodes, lungs | histopathology | n.a. | [43] |
| 29 | n.a. | Switzerland | badger | skin, subcutaneous lymph nodes, lungs | histopathology | n.a. | [43] |
| 30 | n.a. | Switzerland | cat | skin | histopathology | PCR (28S rDNA 8) | [44] |
| 31 | n.a. | Italy (ER) | dog | peribronchial lymph nodes | culture | n.a. | [45] |
| 32 | n.a. | Italy (ER) | dog | peribronchial lymph nodes | culture | n.a. | [45] |
| 33 | n.a. | Italy (Apulia) | dog | epidural spinal cord | histopathology | immunohistology, PCR (ITS1-5.8S-ITS2 8) | [46] |
| 34 | n.a. | Italy (ER) | cat | lung, abdominal mass | histopathology | immunohistology | [47] |
| 35 | n.a. | Albania (Vlorë) | rat | spleen, liver | histopathology | n.a. | [48] |
| 36 | n.a. | Hungary | laboratory mouse | liver, peritoneal liquid | histopathology | n.a. | [49] |
| 37 | n.a. | Denmark (NJ) | badger | skin, liver, kidney, lymph node | histopathology | immunohistology | [50] |
| 38 | n.a. | Austria (NOE) | badger | skin, regional lymph nodes | histopathology | immunohistology | [51] |
| 39 | n.a. | Spain (Andalusia) | Gazelle 1 | lung, intestines, spleen, kidneys, myocardium, liver | culture, histopathology | n.a. | [52] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilmes, D.; Mayer, U.; Wohlsein, P.; Suntz, M.; Gerkrath, J.; Schulze, C.; Holst, I.; von Bomhard, W.; Rickerts, V. Animal Histoplasmosis in Europe: Review of the Literature and Molecular Typing of the Etiological Agents. J. Fungi 2022, 8, 833. https://doi.org/10.3390/jof8080833
Wilmes D, Mayer U, Wohlsein P, Suntz M, Gerkrath J, Schulze C, Holst I, von Bomhard W, Rickerts V. Animal Histoplasmosis in Europe: Review of the Literature and Molecular Typing of the Etiological Agents. Journal of Fungi. 2022; 8(8):833. https://doi.org/10.3390/jof8080833
Chicago/Turabian StyleWilmes, Dunja, Ursula Mayer, Peter Wohlsein, Michael Suntz, Jasmin Gerkrath, Christoph Schulze, Ina Holst, Wolf von Bomhard, and Volker Rickerts. 2022. "Animal Histoplasmosis in Europe: Review of the Literature and Molecular Typing of the Etiological Agents" Journal of Fungi 8, no. 8: 833. https://doi.org/10.3390/jof8080833
APA StyleWilmes, D., Mayer, U., Wohlsein, P., Suntz, M., Gerkrath, J., Schulze, C., Holst, I., von Bomhard, W., & Rickerts, V. (2022). Animal Histoplasmosis in Europe: Review of the Literature and Molecular Typing of the Etiological Agents. Journal of Fungi, 8(8), 833. https://doi.org/10.3390/jof8080833







