Putative Core Transcription Factors Affecting Virulence in Aspergillus flavus during Infection of Maize
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Growth Conditions
2.2. Kernel Infection Assays
2.3. RNA Extraction and Library Preparation for RNA Sequencing
2.4. RNA Sequencing, Alignment and Data Analysis
2.5. Whole Genome Network Co-Expression Analysis (WGNCA) and Gene Selection
2.6. Construct Assembly and Fungal Transformation
2.7. Growth Rate and Spore Production by Transcription Factor Knockout Strains
2.8. Aflatoxin and Cyclopiazonic Acid Analysis
2.9. Menadione-Induced Oxidative Stress Response
3. Results
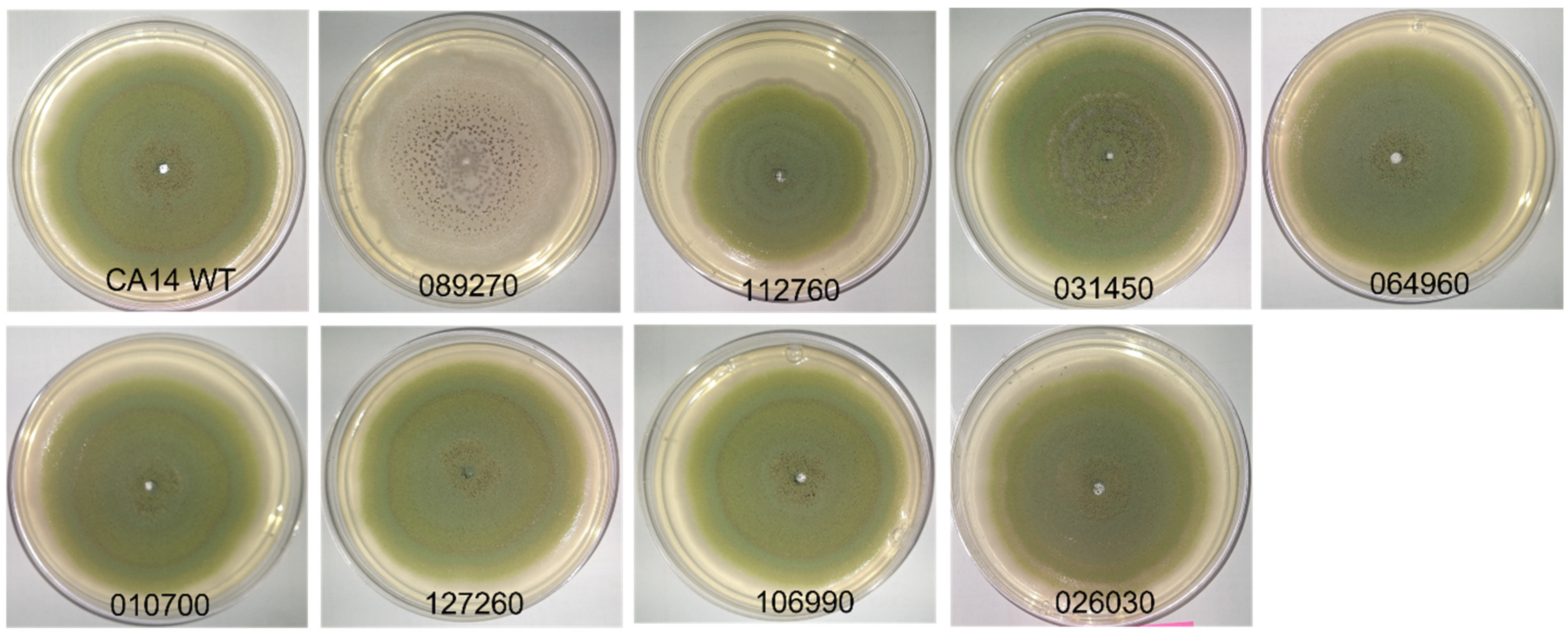
3.1. Morphology of Transcription Factor Knockout Strains
3.1.1. General Morphology
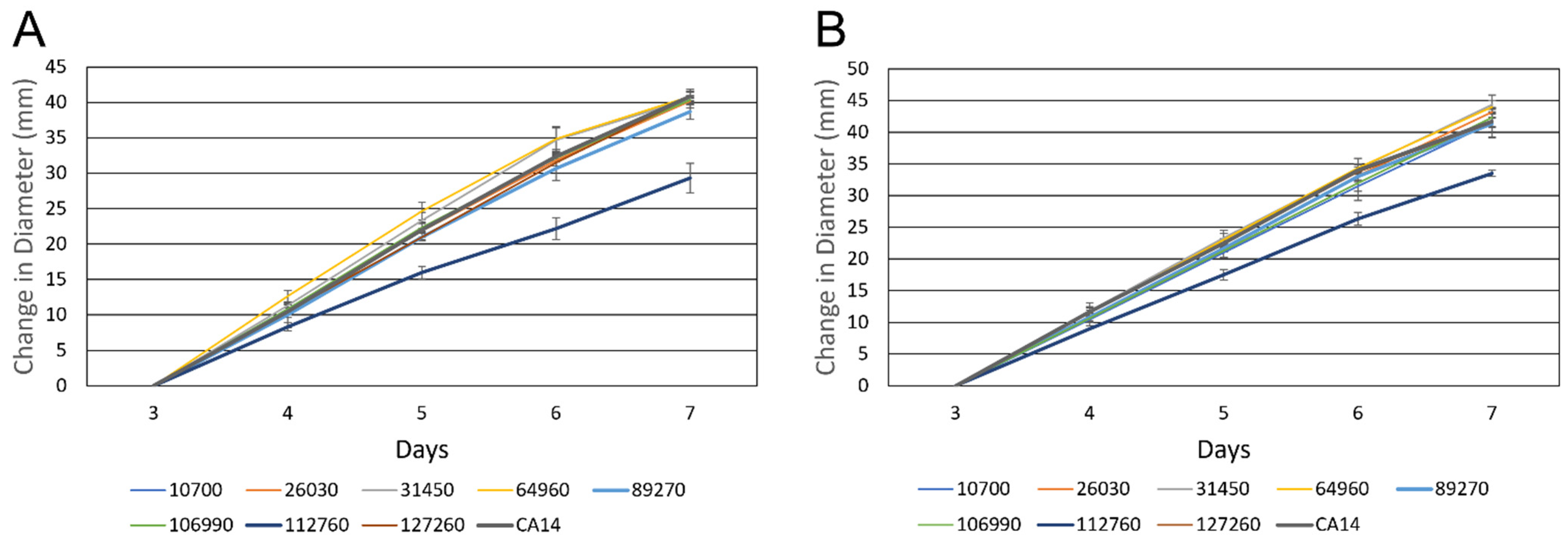
3.1.2. Growth Rate of Transcription Factor Knockout Strains
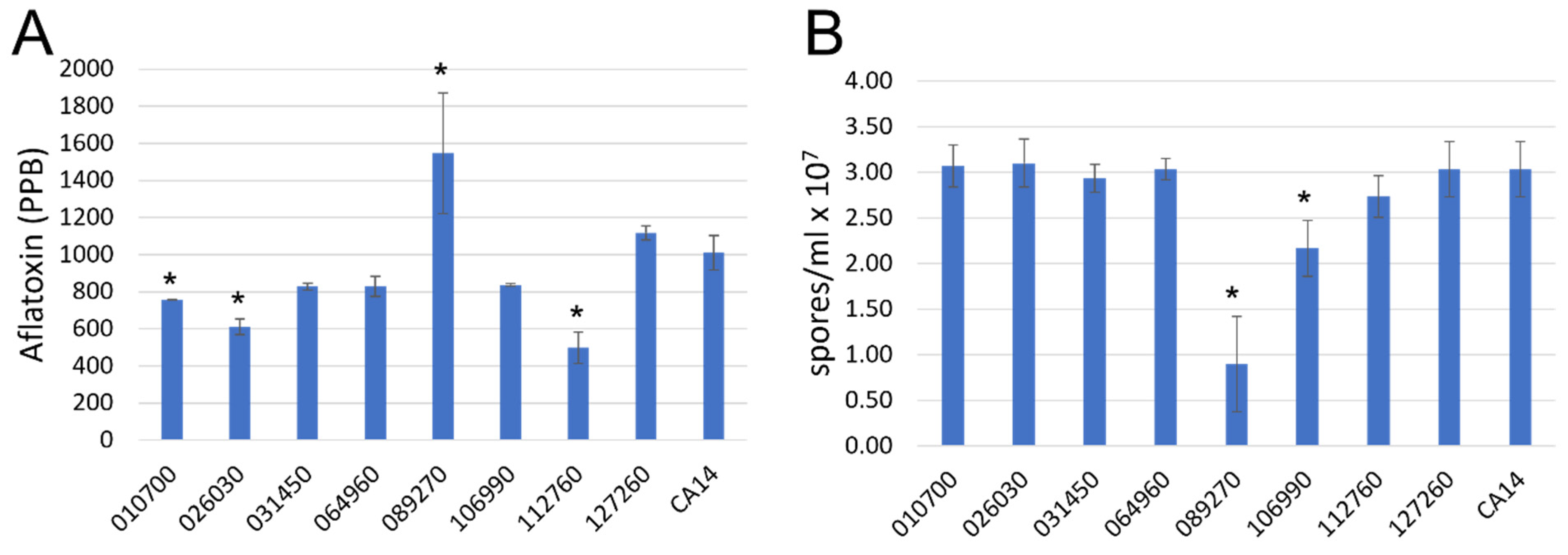
3.1.3. Aflatoxin, Spore Production on Agar Medium
3.2. Ultra-High Pressure Liquid Chromatography (UPLC) Analysis of Aflatoxin and Cyclopiazonic Acid Levels
3.3. Menadione Oxidative Stress Assays
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Klich, M.A. Aspergillus flavus: The Major Producer of Aflatoxin. Mol. Plant Pathol. 2007, 8, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Medina, A.; Gilbert, M.K.; Mack, B.M.; Obrian, G.R.; Rodríguez, A.; Bhatnagar, D.; Payne, G.; Magan, N. Interactions Between Water Activity and Temperature on the Aspergillus flavus Transcriptome and Aflatoxin B1 Production. Int. J. Food Microbiol. 2017, 256, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, N.J.; Bowers, E.; Hurburgh, C.; Wu, F. Potential Economic Losses to the US Corn Industry from Aflatoxin Contamination. Food Addit. Contam. Part A 2016, 33, 540–550. [Google Scholar] [CrossRef] [Green Version]
- Brown, R.L.; Menkir, A.; Chen, Z.-Y.; Bhatnagar, D.; Yu, J.; Yao, H.; Cleveland, T.E. Breeding Aflatoxin-Resistant Maize Lines Using Recent Advances in Technologies—A Review. Food Addit. Contam. Part A 2013, 30, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.K.; Accinelli, C.; Shier, W.T. Biological Control of Aflatoxin Contamination in U.S. Crops and the Use of Bioplastic Formulations of Aspergillus flavus Biocontrol Strains to Optimize Application Strategies. J. Agric. Food Chem. 2017, 65, 7081–7087. [Google Scholar] [CrossRef]
- Udomkun, P.; Wiredu, A.N.; Nagle, M.; Müller, J.; Vanlauwe, B.; Bandyopadhyay, R. Innovative Technologies to Manage Aflatoxins in Foods and Feeds and the Profitability of Application—A Review. Food Control 2017, 76, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, R.; Rajasekaran, K.; Cary, J.W. RNA Interference (RNAi) as a Potential Tool for Control of Mycotoxin Contamination in Crop Plants: Concepts and Considerations. Front. Plant Sci. 2017, 8, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, M.K.; Majumdar, R.; Rajasekaran, K.; Chen, Z.-Y.; Wei, Q.; Sickler, C.M.; Lebar, M.D.; Cary, J.W.; Frame, B.R.; Wang, K. RNA Interference-Based Silencing of the Alpha-Amylase (amy1) Gene in Aspergillus flavus Decreases Fungal Growth and Aflatoxin Production in Maize Kernels. Planta 2018, 247, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Thonberg, H.; Wang, J.; Wahlestedt, C.; Liang, Z. A Systematic Analysis of the Silencing Effects of an Active siRNA at all Single-Nucleotide Mismatched Target Sites. Nucleic Acids Res. 2005, 33, 1671–1677. [Google Scholar] [CrossRef] [Green Version]
- Dahlgren, C.; Zhang, H.-Y.; Du, Q.; Grahn, M.; Norstedt, G.; Wahlestedt, C.; Liang, Z. Analysis of siRNA Specificity on Targets with Double-Nucleotide Mismatches. Nucleic Acids Res. 2008, 36, e53. [Google Scholar] [CrossRef]
- Castano-Duque, L.; Gilbert, M.K.; Mack, B.M.; Lebar, M.D.; Carter-Wientjes, C.H.; Sickler, C.M.; Cary, J.W.; Rajasekaran, K. Flavonoids Modulate the Accumulation of Toxins from Aspergillus flavus in Maize Kernels. Front. Plant Sci. 2021, 12, 761446. [Google Scholar] [CrossRef]
- Nierman, W.C.; Yu, J.; Fedorova-Abrams, N.D.; Losada, L.; Cleveland, T.E.; Bhatnagar, D.; Bennett, J.W.; Dean, R.; Payne, G.A. Genome Sequence of Aspergillus flavus NRRL 3357, a Strain that Causes Aflatoxin Contamination of Food and Feed. Genome Announc. 2015, 3, e00168-15. [Google Scholar] [CrossRef] [Green Version]
- Brown, N.A.; Goldman, G.H. The Contribution of Aspergillus fumigatus Stress Responses to Virulence and Antifungal Resistance. J. Microbiol. 2016, 54, 243–253. [Google Scholar] [CrossRef]
- Chang, P.-K.; Scharfenstein, L.L.; Ehrlich, K.C.; Wei, Q.; Bhatnagar, D.; Ingber, B.F. Effects of laeA Deletion on Aspergillus flavus Conidial Development and Hydrophobicity May Contribute to Loss of Aflatoxin Production. Fungal Biol. 2011, 116, 298–307. [Google Scholar] [CrossRef]
- Kafer, E. The Anthranilate Synthetase Enzyme Complex and the Trifunctional trpC Gene of Aspergillus. Can. J. Genet. Cytol. 1977, 19, 723–738. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shelest, E. Transcription Factors in Fungi: TFome Dynamics, Three Major Families, and Dual-Specificity TFs. Front. Genet. 2017, 8, 53. [Google Scholar] [CrossRef] [Green Version]
- Jin, F.-J.; Wang, B.-T.; Wang, Z.-D.; Jin, L.; Han, P. CRISPR/Cas9-Based Genome Editing and its Application in Aspergillus Species. J. Fungi 2022, 8, 467. [Google Scholar] [CrossRef]
- Chang, P.-K.; Scharfenstein, L.L.; Wei, Q.; Bhatnagar, D. Development and Refinement of a High-Efficiency Gene-Targeting System for Aspergillus flavus. J. Microbiol. Methods 2010, 81, 240–246. [Google Scholar] [CrossRef]
- Chang, P.K.; Scharfenstein, L.L.; Abbas, H.K.; Bellaloui, N.; Accinelli, C.; Ebelhar, M.W. Prevalence of NRRL21882-Like (Afla-Guard®) Aspergillus flavus on Sesame Seeds Grown in Research Fields in the Mississippi Delta. Biocontrol Sci. Technol. 2020, 30, 1090–1099. [Google Scholar] [CrossRef]
- Paré, A.; Kim, M.; Juarez, M.T.; Brody, S.; McGinnis, W. The Functions of Grainy Head-Like Proteins in Animals and Fungi and the Evolution of Apical Extracellular Barriers. PLoS ONE 2012, 7, e36254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulder, H.J.; Saloheimo, M.; Penttilä, M.; Madrid, S.M. The Transcription Factor HACA Mediates the Unfolded Protein Response in Aspergillus niger, and Up-Regulates its Own Transcription. Mol. Genet. Genom. 2004, 271, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Mulder, H.J.; Nikolaev, I. HacA-Dependent Transcriptional Switch Releases hacA mRNA From a Translational Block Upon Endoplasmic Reticulum Stress. Eukaryot. Cell 2009, 8, 665–675. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.; Krishnan, K.; Richie, D.L.; Aimanianda, V.; Hartl, L.; Grahl, N.; Powers-Fletcher, M.V.; Zhang, M.; Fuller, K.K.; Nierman, W.C.; et al. HacA-Independent functions of the ER Stress Sensor IreA Synergize With the Canonical UPR to Influence Virulence Traits in Aspergillus fumigatus. PLoS Pathog. 2011, 7, e1002330. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Xie, J.; Liu, X.; Wang, B.; Pan, L. Functional and Transcriptomic Analysis of the Key Unfolded Protein Response Transcription Factor HacA in Aspergillus oryzae. Gene 2016, 593, 143–153. [Google Scholar] [CrossRef]
- Bitencourt, T.A.; Lang, E.A.S.; Sanches, P.R.; Peres, N.T.A.; Oliveira, V.M.; Fachin, A.L.; Rossi, A.; Martinez-Rossi, N.M. HacA Governs Virulence Traits and Adaptive Stress Responses in Trichophyton rubrum. Front. Microbiol. 2020, 11, 193. [Google Scholar] [CrossRef] [Green Version]
- Kimata, Y.; Ishiwata-Kimata, Y.; Yamada, S.; Kohno, K. Yeast Unfolded Protein Response Pathway Regulates Expression of Genes for Anti-Oxidative Stress and for Cell Surface Proteins. Genes Cells 2005, 11, 59–69. [Google Scholar] [CrossRef]
- Zhao, Q.; Pei, H.; Zhou, X.; Zhao, K.; Yu, M.; Han, G.; Fan, J.; Tao, F. Systematic Characterization of bZIP Transcription Factors Required for Development and Aflatoxin Generation by High-Throughput Gene Knockout in Aspergillus flavus. J. Fungi 2022, 8, 356. [Google Scholar] [CrossRef]
- Torres, M.A.; Jones, J.D.; Dangl, J.L. Reactive Oxygen Species Signaling in Response to Pathogens. Plant. Physiol. 2009, 141, 373–378. [Google Scholar] [CrossRef]
- Asada, K. The Water-Water Cycle in Chloroplasts: Scavenging of Active Oxygens and Dissipation of Excess Photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef] [PubMed]
- Campos, E.G.; Jesuino, R.S.A.; Dantas, A.D.S.; Brigido, M.; Felipe, M.S.S. Oxidative Stress Response in Paracoccidioides brasiliensis. Genet. Mol. Res. 2005, 4, 409–429. [Google Scholar] [PubMed]
- Jayashree, T.; Subramanyam, C. Oxidative Stress as a Prerequisite for Aflatoxin Production by Aspergillus parasiticus. Free Radic. Biol. Med. 2000, 29, 981–985. [Google Scholar] [CrossRef] [PubMed]
- Fountain, J.C.; Bajaj, P.; Pandey, M.; Nayak, S.N.; Yang, L.; Kumar, V.; Jayale, A.S.; Chitikineni, A.; Zhuang, W.; Scully, B.T.; et al. Oxidative Stress and Carbon Metabolism Influence Aspergillus flavus Transcriptome Composition and Secondary Metabolite Production. Sci. Rep. 2016, 6, 38747. [Google Scholar] [CrossRef] [Green Version]
- Chalivendra, S.C.; DeRobertis, C.; Chang, P.-K.; Damann, K.E. Cyclopiazonic Acid Is a Pathogenicity Factor for Aspergillus flavus and a Promising Target for Screening Germplasm for Ear Rot Resistance. Mol. Plant-Microbe Interact. 2017, 30, 361–373. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gilbert, M.K.; Mack, B.M.; Lebar, M.D.; Chang, P.-K.; Gross, S.R.; Sweany, R.R.; Cary, J.W.; Rajasekaran, K. Putative Core Transcription Factors Affecting Virulence in Aspergillus flavus during Infection of Maize. J. Fungi 2023, 9, 118. https://doi.org/10.3390/jof9010118
Gilbert MK, Mack BM, Lebar MD, Chang P-K, Gross SR, Sweany RR, Cary JW, Rajasekaran K. Putative Core Transcription Factors Affecting Virulence in Aspergillus flavus during Infection of Maize. Journal of Fungi. 2023; 9(1):118. https://doi.org/10.3390/jof9010118
Chicago/Turabian StyleGilbert, Matthew K., Brian M. Mack, Matthew D. Lebar, Perng-Kuang Chang, Stephanie R. Gross, Rebecca R. Sweany, Jeffrey W. Cary, and Kanniah Rajasekaran. 2023. "Putative Core Transcription Factors Affecting Virulence in Aspergillus flavus during Infection of Maize" Journal of Fungi 9, no. 1: 118. https://doi.org/10.3390/jof9010118






