Genomic Characteristics and Comparative Genomics Analysis of the Endophytic Fungus Paraphoma chrysanthemicola DS-84 Isolated from Codonopsis pilosula Root
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Source, Strain Culture and Maintain
2.2. Observation of Strain Morphology by Scanning Electron Microscopy (SEM)
2.3. Fourier Transform Infrared (FTIR)
2.4. Mycelial Fatty Acid Fraction
2.5. Molecular Identification of Fungi
2.6. Effects of Carbon and Nitrogen Sources on Fungi Growth
2.7. Characterisation of the Enzyme Produced by the Fungi
2.8. Fungal Genomics Analysis
3. Result
3.1. Strain Identification
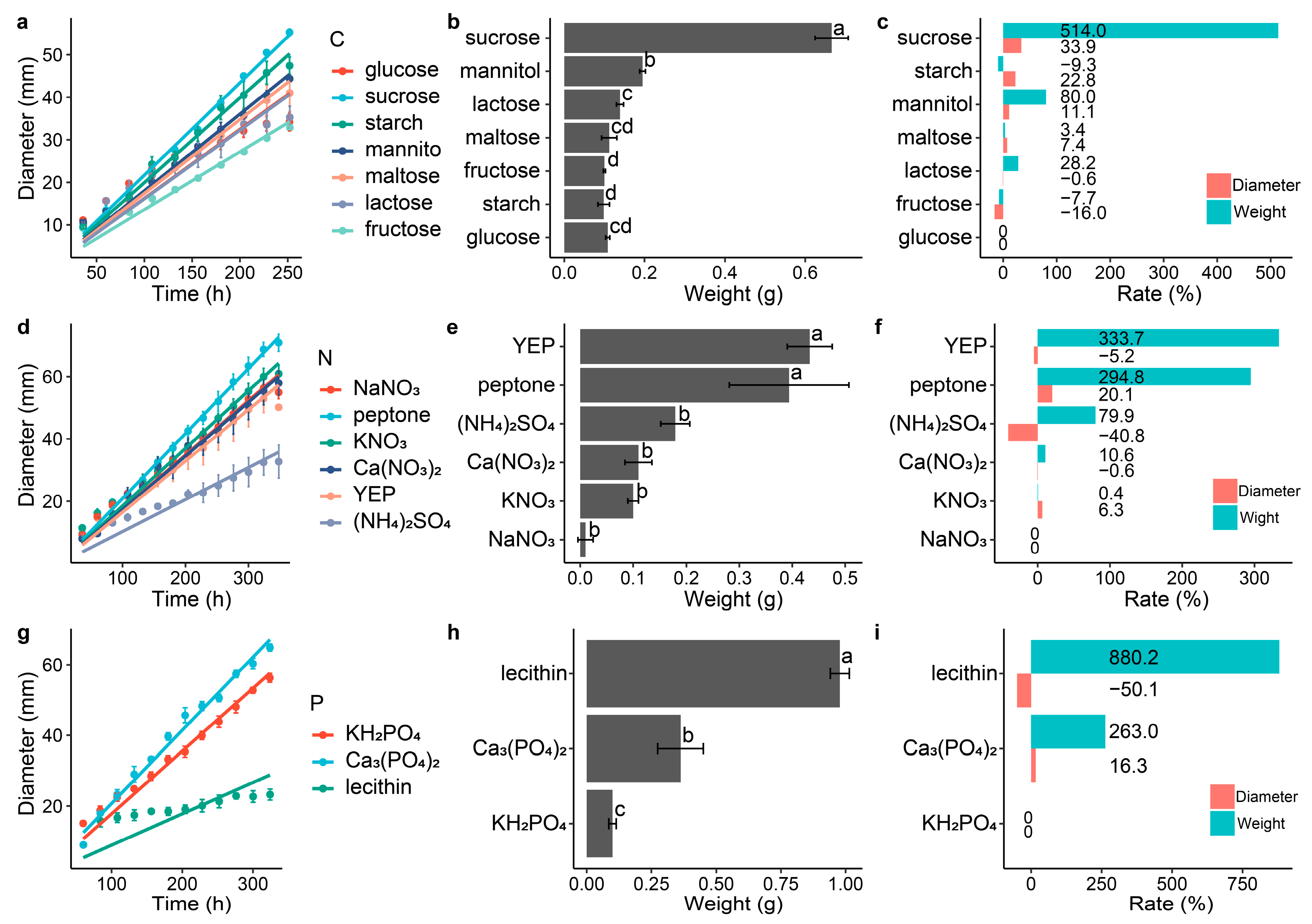
3.2. Effect of Different Carbon, Nitrogen and Phosphorus Sources on the Morphology of DS-84
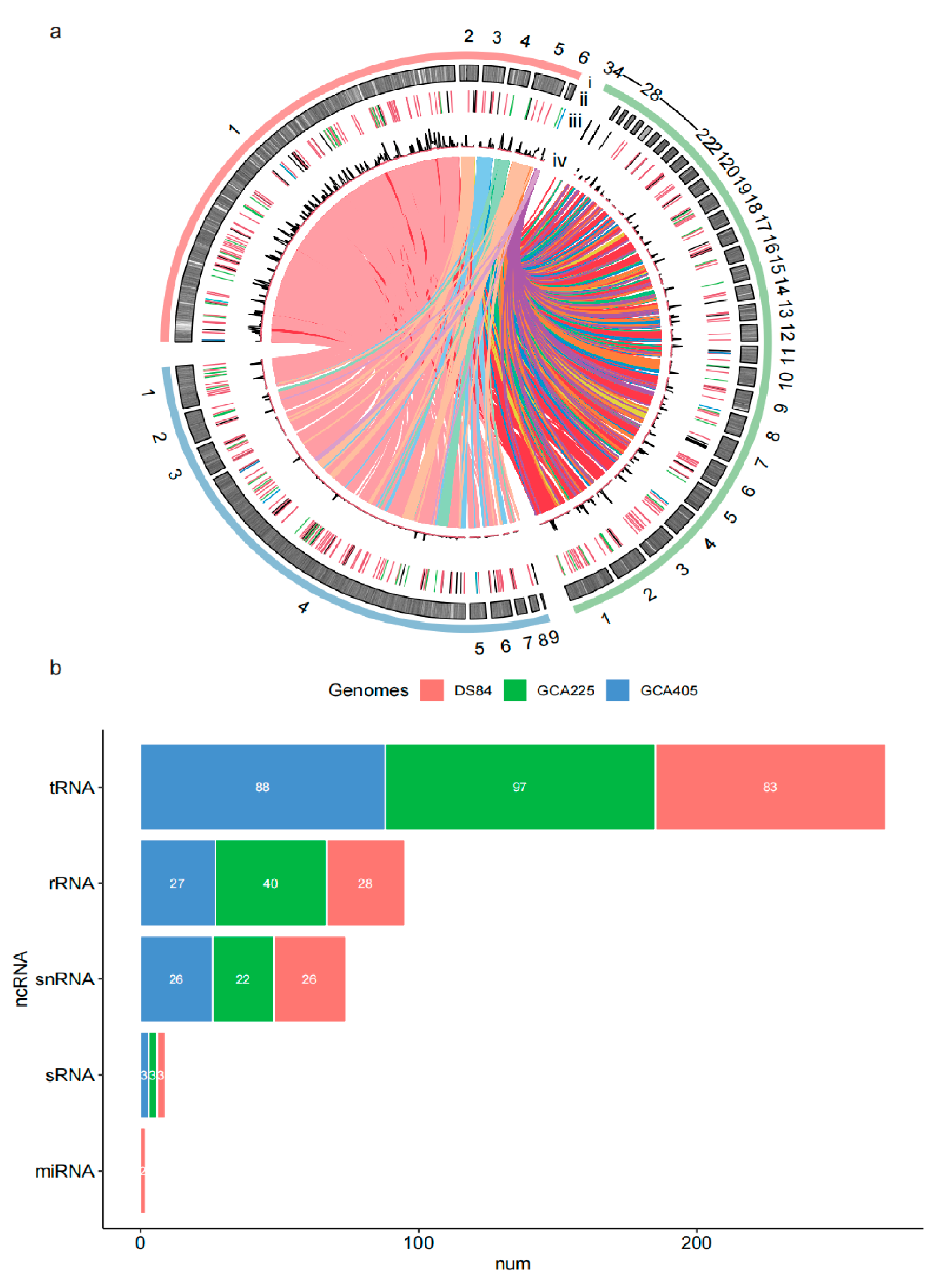
3.3. Overall Analysis of the Genome of DS-84
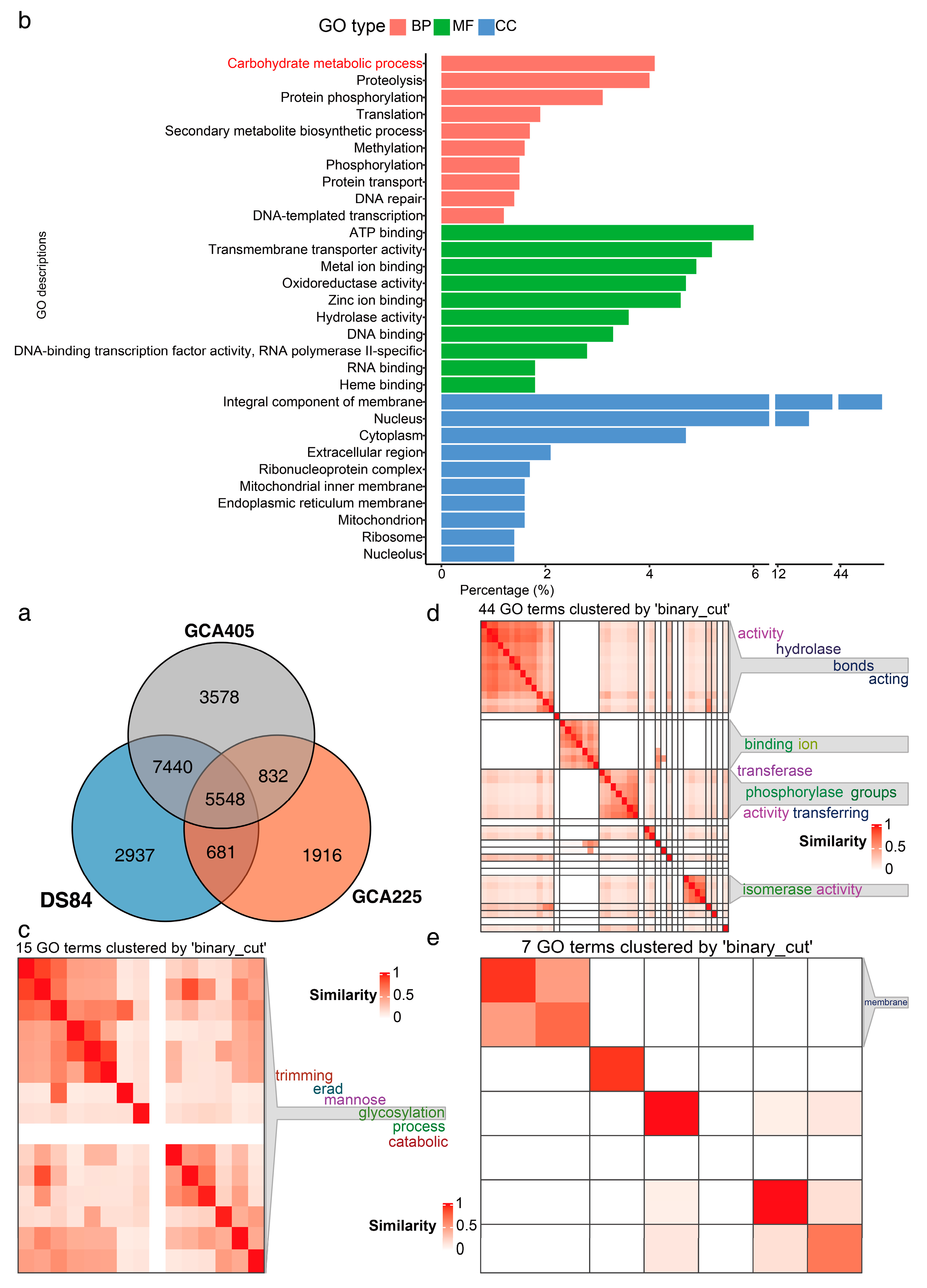
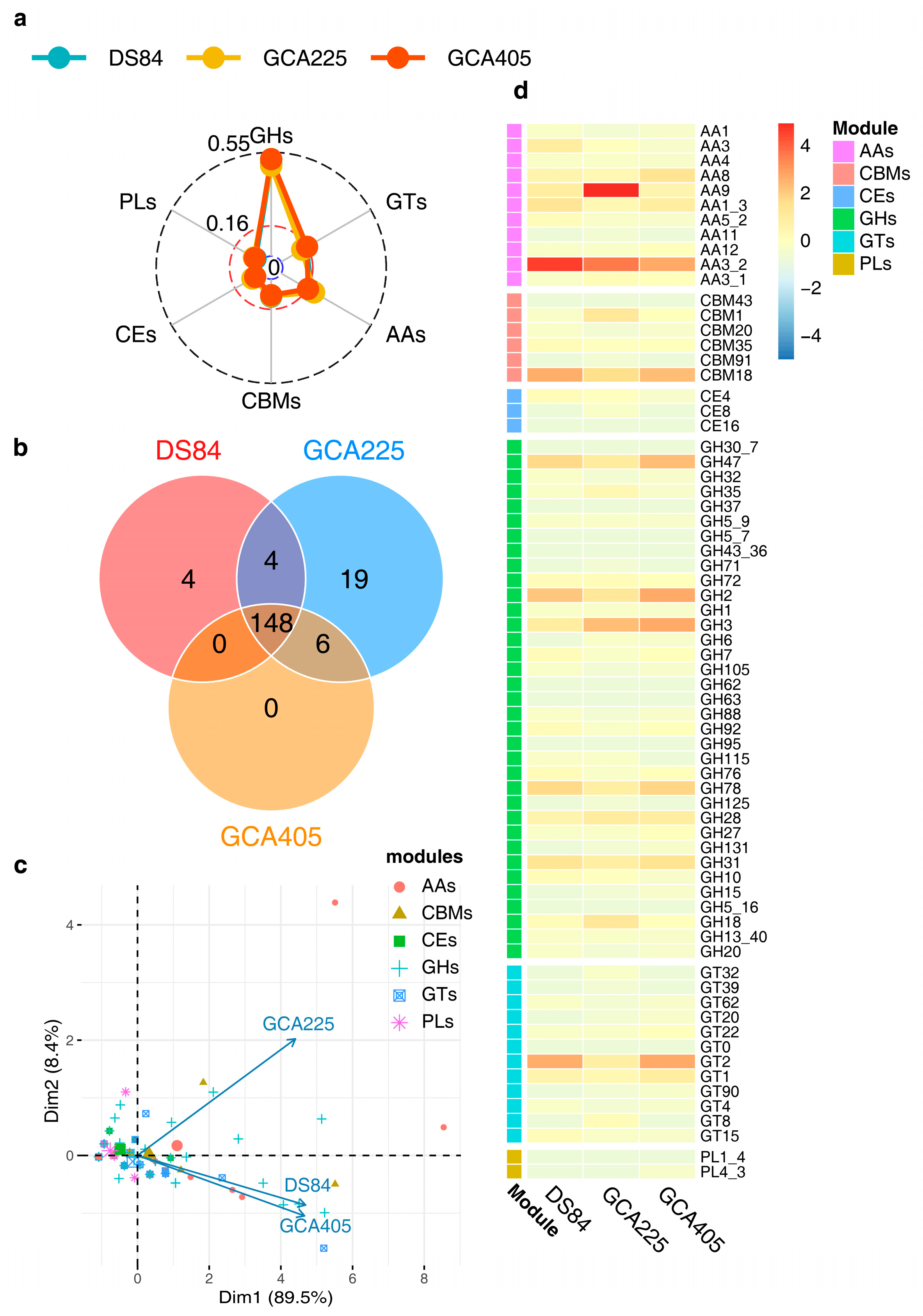
3.4. Functional Annotation of Genes in Three Genomes
3.5. Protein Advanced Annotation
4. Discussion
4.1. The Results Proved That DS-84 Is P. chrysanthemicola
4.2. Monosaccharide Preference of P. chrysanthemicola
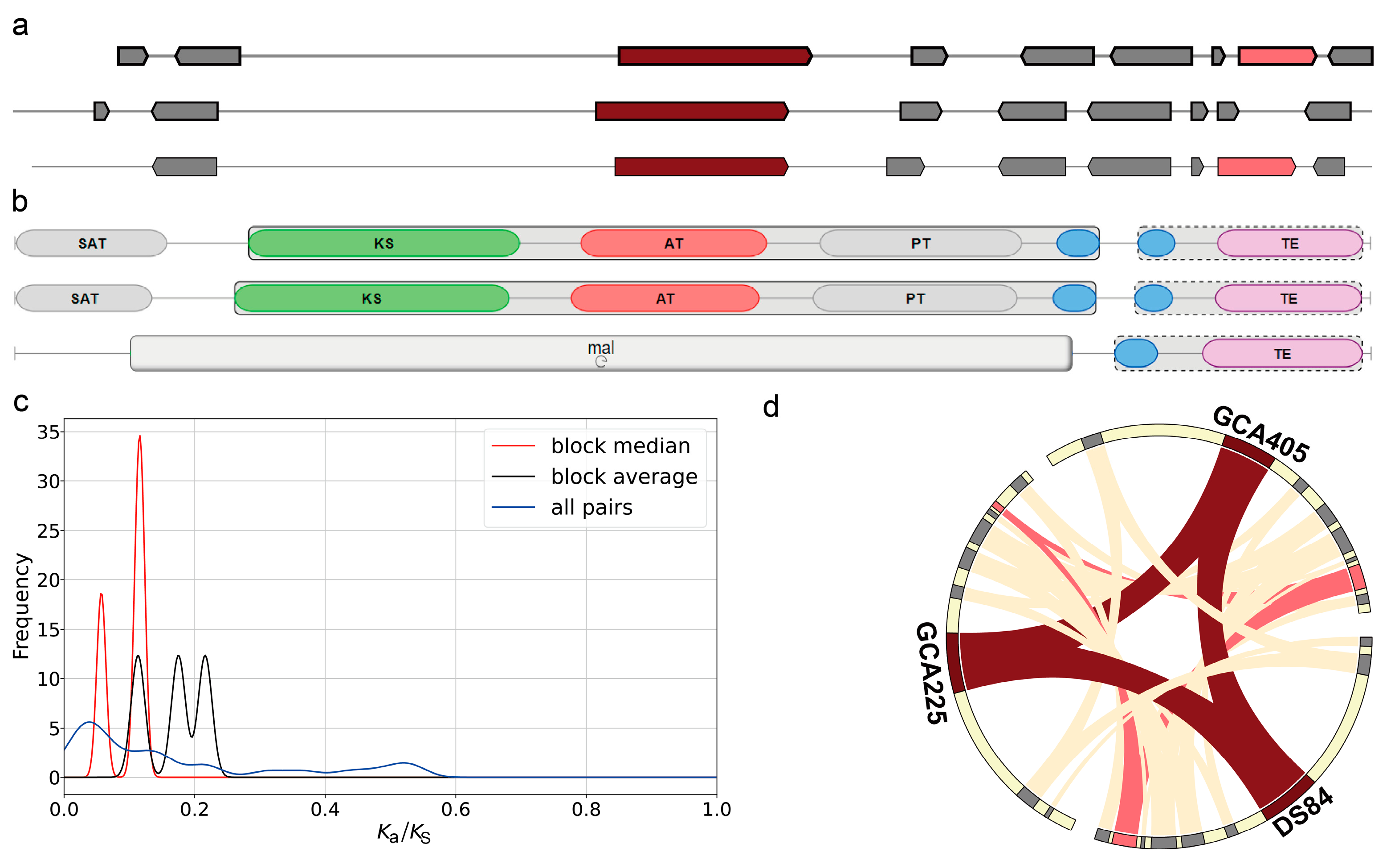
4.3. The Important Secondary Metabolite Gene Cluster of P. chrysanthemicola
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arnold, A.E.; Lutzoni, F. Diversity and host range of foliar fungal endophytes: Are tropical leaves biodiversity hotspots? Ecology 2007, 88, 541–549. [Google Scholar] [CrossRef]
- Stone, J. Endophytic fungi in grasses and woody plants. Systematics, ecology and evolution. For. Sci. 1997, 43, 458–459. [Google Scholar] [CrossRef]
- Zheng, R.; Jiang, H. Rhizomucor endophyticus sp. nov., an endophytic zygomycetes from higher plants. Mycotaxon 1995, 56, 455–466. [Google Scholar]
- Clay, K. Fungal endophytes of grasses: A defensive mutualism between plants and fungi. Ecology 1988, 69, 10–16. [Google Scholar] [CrossRef]
- Omomowo, O.I.; Babalola, O.O. Bacterial andfungal endophytes: Tiny giants with immense beneficial potential for plant growth and sustainable agricultural productivity. Microorganisms 2019, 7, 481. [Google Scholar] [CrossRef] [PubMed]
- Adeleke, B.S.; Babalola, O.O. The endosphere microbial communities, a great promise in agriculture. Int. Microbiol. 2021, 24, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Omoarelojie, L.O.; Van Staden, J. Plant-endophytic fungi interactions: A strigolactone perspective. S. Afr. J. Bot. 2020, 134, 280–284. [Google Scholar] [CrossRef]
- Aly, A.H.; Debbab, A.; Proksch, P. Fungal endophytes: Unique plant inhabitants with great promises. Appl. Microbiol. Biotechnol. 2011, 90, 1829–1845. [Google Scholar] [CrossRef]
- Guo, C.J.; Wang, C.C. Recent advances in genome mining of secondary metabolites in Aspergillus terreus. Front. Microbiol. 2014, 5, 717. [Google Scholar] [CrossRef]
- Chu, L.; Huang, J.; Muhammad, M.; Deng, Z.; Gao, J. Genome mining as a biotechnological tool for the discovery of novel marine natural products. Crit. Rev. Biotechnol. 2020, 40, 571–589. [Google Scholar] [CrossRef]
- Lee, N.; Hwang, S.; Kim, J.; Cho, S.; Palsson, B.; Cho, B.K. Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput. Struct. Biotechnol. J. 2020, 18, 1548–1556. [Google Scholar] [CrossRef]
- Sagita, R.; Quax, W.J.; Haslinger, K. Current State and Future Directions of Genetics and Genomics of Endophytic Fungi for Bioprospecting Efforts. Front. Bioeng. Biotechnol. 2021, 9, 649906. [Google Scholar] [CrossRef]
- Ampt, E.A.; van Ruijven, J.; Raaijmakers, J.M.; Termorshuizen, A.J.; Mommer, L. Linking ecology and plant pathology to unravel the importance of soil-borne fungal pathogens in species-rich grasslands. Eur. J. Plant Pathol. 2018, 154, 141–156. [Google Scholar] [CrossRef]
- Cao, S.; Liang, Q.W.; Nzabanita, C.; Li, Y.Z. Paraphoma root rot of alfalfa (Medicago sativa) in Inner Mongolia, China. Plant Pathol. 2019, 69, 231–239. [Google Scholar] [CrossRef]
- Ge, X.; Zhou, R.; Yuan, Y.; Xu, H.; Fu, J.; Li, H. Identification and characterization of Paraphoma chrysanthemicola causing leaf spot disease on Atractylodes japonica in China. J. Phytopathol. 2016, 164, 372–377. [Google Scholar] [CrossRef]
- Ampt, E.A.; Francioli, D.; van Ruijven, J.; Gomes, S.I.F.; Maciá-Vicente, J.G.; Termorshuizen, A.J.; Bakker, L.M.; Mommer, L. Deciphering the interactions between plant species and their main fungal root pathogens in mixed grassland communities. J. Ecol. 2022, 110, 3039–3052. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Xu, M.; Ye, Q.; Gao, H.; He, X. Improved tolerance of Artemisia ordosica to drought stress via dark septate endophyte (DSE) symbiosis. J. Fungi 2022, 8, 730. [Google Scholar] [CrossRef]
- Chu, H.; Wang, H.; Zhang, Y.; Li, Z.; Wang, C.; Dai, D.; Tang, M. Inoculation with ectomycorrhizal fungi and dark septate endophytes contributes to the resistance of Pinus spp. to pine wilt disease. Front. Microbiol. 2021, 12, 687304. [Google Scholar] [CrossRef]
- Hou, L.; Li, X.; He, X.; Zuo, Y.; Zhang, D.; Zhao, L. Effect of dark septate endophytes on plant performance of Artemisia ordosica and associated soil microbial functional group abundance under salt stress. Appl. Soil Ecol. 2021, 165, 103998. [Google Scholar] [CrossRef]
- Zhang, H.H.; Tang, M.; Chen, H.; Wang, Y.J. Effects of a dark-septate endophytic isolate LBF-2 on the medicinal plant Lycium barbarum L. J. Microbiol. 2012, 50, 91–96. [Google Scholar] [CrossRef]
- Ban, Y.; Tang, M.; Chen, H.; Xu, Z.; Zhang, H.; Yang, Y. The response of dark septate endophytes (DSE) to heavy metals in pure culture. PLoS ONE 2012, 7, e47968. [Google Scholar] [CrossRef]
- Boerema, G.H.; Gruyter, J.D.; Noordeloos, M.E.; Hamers, M.E.C. Phoma Identification Manual. Differentiation of Specific and Infra-specific Taxa in Culture; CABI: Oxfordshire, UK; Cambridge, MA, USA, 2004; pp. 1–404. [Google Scholar]
- De-Gruyter, J.; Woudenberg, J.H.; Aveskamp, M.M.; Verkley, G.J.; Groenewald, J.Z.; Crous, P.W. Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia 2010, 102, 1066–1081. [Google Scholar] [CrossRef]
- Koitabashi, M.; Noguchi, M.T.; Sameshima-Yamashita, Y.; Hiradate, S.; Suzuki, K.; Yoshida, S.; Watanabe, T.; Shinozaki, Y.; Tsushima, S.; Kitamoto, H.K. Degradation of biodegradable plastic mulch films in soil environment by phylloplane fungi isolated from gramineous plants. AMB Express 2012, 2, 40. [Google Scholar] [CrossRef]
- Woudenberg, J.H.; Groenewald, J.Z.; Binder, M.; Crous, P.W. Alternaria redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Jiang, X.; Zhou, B.; Zhao, B.; Ma, M.; Guan, D.; Li, J.; Chen, S.; Cao, F.; Shen, D.; et al. Thirty four years of nitrogen fertilization decreases fungal diversity and alters fungal community composition in black soil in northeast China. Soil Biol. Biochem. 2016, 95, 135–143. [Google Scholar] [CrossRef]
- Toghueo, R.M.K.; Zabalgogeazcoa, I.; Vázquez de Aldana, B.R.; Boyom, F.F. Enzymatic activity of endophytic fungi from the medicinal plants Terminalia catappa, Terminalia mantaly and Cananga odorata. S. Afr. J. Bot. 2017, 109, 146–153. [Google Scholar] [CrossRef]
- Chin, C.S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef]
- Myers, E.W.; Sutton, G.G.; Delcher, A.L.; Dew, I.M.; Fasulo, D.P.; Flanigan, M.J.; Kravitz, S.A.; Mobarry, C.M.; Reinert, K.H.; Remington, K.A.; et al. A whole-genome assembly of Drosophila. Science 2000, 287, 2196–2204. [Google Scholar] [CrossRef]
- Koren, S.; Schatz, M.C.; Walenz, B.P.; Martin, J.; Howard, J.T.; Ganapathy, G.; Wang, Z.; Rasko, D.A.; McCombie, W.R.; Jarvis, E.D.; et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol. 2012, 30, 693–700. [Google Scholar] [CrossRef]
- Chen, K.T.; Shen, H.T.; Lu, C.L. Multi-CSAR: A multiple reference-based contig scaffolder using algebraic rearrangements. BMC Syst. Biol. 2018, 12, 139. [Google Scholar] [CrossRef]
- Lomsadze, A.; Ter-Hovhannisyan, V.; Chernoff, Y.O.; Borodovsky, M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005, 33, 6494–6506. [Google Scholar] [CrossRef] [PubMed]
- Ter-Hovhannisyan, V.; Lomsadze, A.; Chernoff, Y.O.; Borodovsky, M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008, 18, 1979–1990. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, D. RepeatMasker. Biotech Softw. Internet Rep. 2000, 1, 36–39. [Google Scholar] [CrossRef]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 2020, 117, 9451–9457. [Google Scholar] [CrossRef] [PubMed]
- Gary, B. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rodland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef]
- Li, W.; Jaroszewski, L.; Godzik, A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 2002, 18, 77–82. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Hernandez-Plaza, A.; Szklarczyk, D.; Botas, J.; Cantalapiedra, C.P.; Giner-Lamia, J.; Mende, D.R.; Kirsch, R.; Rattei, T.; Letunic, I.; Jensen, L.J.; et al. EggNOG 6.0: Enabling comparative genomics across 12 535 organisms. Nucleic Acids Res. 2023, 51, D389–D394. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Sun, P.; Jiao, B.; Yang, Y.; Shan, L.; Li, T.; Li, X.; Xi, Z.; Wang, X.; Liu, J. WGDI: A user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. Mol. Plant 2022, 15, 1841–1851. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, R.A.M.; Chen, Z.J. Ggplot2: Elegant graphics for data analysis (2nd ed.). Meas.-Interdiscip. Res. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Hubschmann, D. Simplify Enrichment: A Bioconductor Package for Clustering and Visualizing Functional Enrichment Results. Genom. Proteom. Bioinform. 2023, 21, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for Interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- De Gruyter, J.; Aveskamp, M.M.; Woudenberg, J.H.; Verkley, G.J.; Groenewald, J.Z.; Crous, P.W. Molecular phylogeny of Phoma and allied anamorph genera: Towards a reclassification of the Phoma complex. Mycol. Res. 2009, 113, 508–519. [Google Scholar] [CrossRef]
- De Gruyter, J.; Woudenberg, J.H.; Aveskamp, M.M.; Verkley, G.J.; Groenewald, J.Z.; Crous, P.W. Redisposition of phoma-like anamorphs in Pleosporales. Stud. Mycol. 2013, 75, 1–36. [Google Scholar] [CrossRef]
- Moslemi, A.; Ades, P.K.; Crous, P.W.; Groom, T.; Scott, J.B.; Nicolas, M.E.; Taylor, P.W.J. Paraphoma chlamydocopiosa sp. nov. and Paraphoma pye sp. nov., two new species associated with leaf and crown infection of pyrethrum. Plant Pathol. 2017, 67, 124–135. [Google Scholar] [CrossRef]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. Bioessays 2006, 28, 799–808. [Google Scholar] [CrossRef]
- Salman, A.; Tsror, L.; Pomerantz, A.; Moreh, R.; Mordechai, S.; Huleihel, M. FTIR spectroscopy for detection and identification of fungal phytopathogenes. Spectroscopy 2010, 24, 261–267. [Google Scholar] [CrossRef]
- Salman, A.; Lapidot, I.; Pomerantz, A.; Tsror, L.; Shufan, E.; Moreh, R.; Mordechai, S.; Huleihel, M. Identification of fungal phytopathogens using Fourier transform infrared-attenuated total reflection spectroscopy and advanced statistical methods. J. Biomed. Opt. 2012, 17, 017002. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B.; List, F.B.C.A.; Bolchacova, E.; et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Tripathi, A.; Rai, A.; Dubey, S.C.; Akhtar, J.; Kumar, P. DNA barcode, multiplex PCR and qPCR assay for diagnosis of pathogens infecting pulse crops to facilitate safe exchange and healthy conservation of germplasm. Arch. Microbiol. 2021, 203, 2575–2589. [Google Scholar] [CrossRef]
- Aveskamp, M.M.; Verkley, G.J.; de Gruyter, J.; Murace, M.A.; Perello, A.; Woudenberg, J.H.; Groenewald, J.Z.; Crous, P.W. DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia 2009, 101, 363–382. [Google Scholar] [CrossRef]
- Zeyen, R.J.; Kruger, W.M.; Lyngkjær, M.F.; Carver, T.L.W. Differential effects of D-mannose and 2-deoxy-D-glucose on attempted powdery mildew fungal infection of inappropriate and appropriate G ramineae. Physiol. Mol. Plant Pathol. 2002, 61, 315–323. [Google Scholar] [CrossRef]
- Fleet, C.; Breuil, C.; Uzunovic, A. Nutrient consumption and pigmentation of deep and surface colonizing sapstaining fungi in Pinus contorta. Holzforschung 2001, 55, 340–346. [Google Scholar] [CrossRef]
- Kulkarni, R.K.; Nielsen, B.D. Nutritional requirements for growth of a fungus endophyte of tall fescue grass. Mycologia 2018, 78, 781–786. [Google Scholar] [CrossRef]
- Sharma, V.; Ichikawa, M.; Freeze, H.H. Mannose metabolism: More than meets the eye. Biochem. Biophys. Res. Commun. 2014, 453, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; Latge, J.P.; Munro, C.A. The fungal cell Wall: Structure, biosynthesis, and function. Microbiol. Spectr. 2017, 5, 28513415. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, N.; Wilkins, M.R.; Packer, N.; Nevalainen, H. Protein glycosylation pathways in filamentous fungi. Glycobiology 2008, 18, 626–637. [Google Scholar] [CrossRef]
- Valentine, B.; Bainbridge, B. The relevance of a study of a temperature-sensitive ballooning mutant of Aspergillus nidulans defective in mannose metabolism to our understanding of mannose as a wall component and carbon/energy source. Microbiology 1978, 109, 155–168. [Google Scholar] [CrossRef]
- Li, T.; Gong, L.; Jiang, G.; Wang, Y.; Gupta, V.K.; Qu, H.; Duan, X.; Wang, J.; Jiang, Y. Carbon Sources Influence Fumonisin Production in Fusarium proliferatum. Proteomics 2017, 17, 1700070. [Google Scholar] [CrossRef]
- Sorensen, J.L.; Giese, H. Influence of carbohydrates on secondary metabolism in Fusarium avenaceum. Toxins 2013, 5, 1655–1663. [Google Scholar] [CrossRef]
- Warit, S.; Zhang, N.; Short, A.; Walmsley, R.M.; Oliver, S.G.; Stateva, L.I. Glycosylation deficiency phenotypes resulting from depletion of GDP-mannose pyrophosphorylase in two yeast species. Mol. Microbiol. 2000, 36, 1156–1166. [Google Scholar] [CrossRef]
- Cottrell, T.R.; Griffith, C.L.; Liu, H.; Nenninger, A.A.; Doering, T.L. The pathogenic fungus Cryptococcus neoformans expresses two functional GDP-mannose transporters with distinct expression patterns and roles in capsule synthesis. Eukaryot. Cell 2007, 6, 776–785. [Google Scholar] [CrossRef]
- Kruszewska, J.S.; Saloheimo, M.; Migdalski, A.; Orlean, P.; Penttilä, M.; Palamarczyk, G. Dolichol phosphate mannose synthase from the filamentous fungus Trichoderma reesei belongs to the human and Schizosaccharomyces pombe class of the enzyme. Glycobiology 2000, 10, 983–991. [Google Scholar] [CrossRef]
- Kolaczkowski, B.M.; Jorgensen, C.I.; Spodsberg, N.; Stringer, M.A.; Supekar, N.T.; Azadi, P.; Westh, P.; Krogh, K.; Jensen, K. Analysis of fungal high-mannose structures using CAZymes. Glycobiology 2022, 32, 304–313. [Google Scholar] [CrossRef]
- Yoshida, T.; Kato, Y.; Asada, Y.; Nakajima, T. Filamentous fungus Aspergillus oryzae has two types of alpha-1,2-mannosidases, one of which is a microsomal enzyme that removes a single mannose residue from Man9GlcNAc2. Glycoconj. J. 2000, 17, 745–748. [Google Scholar] [CrossRef] [PubMed]
- Macheleidt, J.; Mattern, D.J.; Fischer, J.; Netzker, T.; Weber, J.; Schroeckh, V.; Valiante, V.; Brakhage, A.A. Regulation and role of fungal secondary metabolites. Annu. Rev. Genet. 2016, 50, 371–392. [Google Scholar] [CrossRef] [PubMed]
- Ancheeva, E.; Daletos, G.; Proksch, P. Bioactive secondary metabolites from endophytic fungi. Curr. Med. Chem. 2020, 27, 1836–1854. [Google Scholar] [CrossRef]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Eisenman, H.C.; Casadevall, A. Synthesis and assembly of fungal melanin. Appl. Microbiol. Biotechnol. 2012, 93, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Renshaw, J.C.; Robson, G.D.; Trinci, A.P.J.; Wiebe, M.G.; Livens, F.R.; Collison, D.; Taylor, R.J. Fungal siderophores: Structures, functions and applications. Mycol. Res. 2002, 106, 1123–1142. [Google Scholar] [CrossRef]
- Coe, S. Phyllostictine a Ring Assembly via Ring Closing Metathesis; University of Warwick: Coventry, UK, 2014. [Google Scholar]
- Li, X.; He, C.; He, X.; Su, F.; Hou, L.; Ren, Y.; Hou, Y. Dark septate endophytes improve the growth of host and non-host plants under drought stress through altered root development. Plant Soil 2019, 439, 259–272. [Google Scholar] [CrossRef]
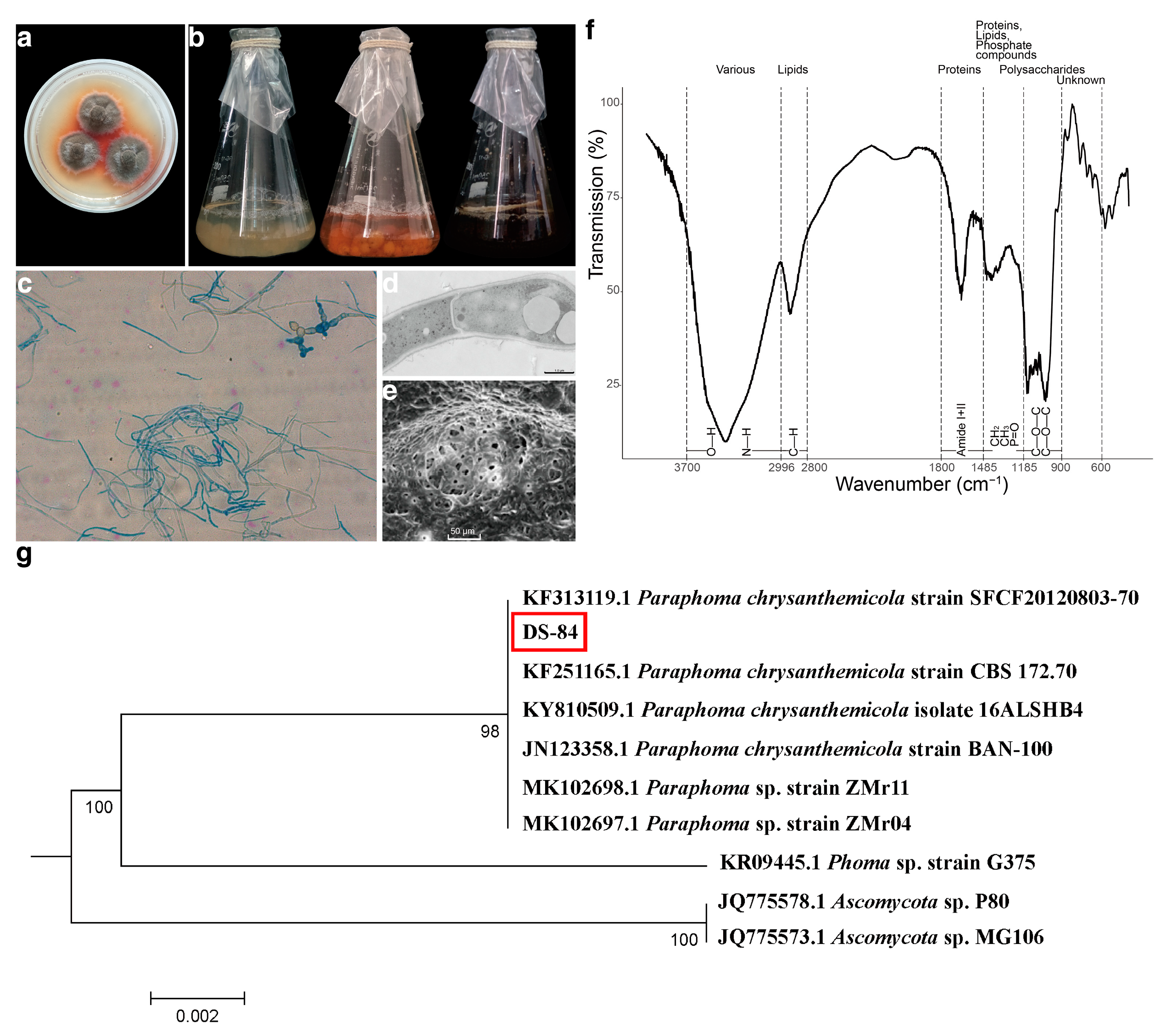
| SN | Lipid Acid | RTS a (min) | RTSA b (min) | PAS c (pA·s) | RAP d (%) | CF e | RFA f (%) | |
|---|---|---|---|---|---|---|---|---|
| 1 | Palmitic Acid | C16:0 | 34.23 | 34.27 | 704.90 | 14.93 | 0.95 | 14.16 |
| 2 | Palmitoleic Acid | C16:1 | 35.80 | 35.83 | 678.00 | 14.36 | 0.95 | 13.61 |
| 3 | Stearic Acid | C18:0 | 39.39 | 39.44 | 153.30 | 3.25 | 0.95 | 3.10 |
| 4 | Oleic Acid | C18:1n9c | 41.09 | 41.22 | 1559.30 | 33.02 | 0.95 | 31.46 |
| 5 | Linoleic Acid | C18:2ω6 | 43.96 | 44.07 | 1453.60 | 30.78 | 0.95 | 29.31 |
| 6 | Eicosanoic Acid | C20:0 | 45.87 | 45.84 | 8.70 | 0.18 | 0.96 | 0.17 |
| 7 | cis-11-Eicosenoic Acid | C20:1n9 | 47.67 | 47.69 | 32.90 | 0.7 | 0.96 | 0.67 |
| 8 | Docosanoic Acid | C22:0 | 54.31 | 54.20 | 11.50 | 0.24 | 0.96 | 0.23 |
| 9 | cis-4,7,10,13,16,19-Docosahexaenoic Acid | C22:6n3 | 76.32 | 76.26 | 7.20 | 0.15 | 0.96 | 0.14 |
| Types of Enzymes | Enzyme Production Capacity |
|---|---|
| Amylase | + |
| Cellulase | + |
| Lipase | − |
| Alkaline protease | + |
| Fibrinolytic enzymes | − |
| Catalase | − |
| Chitosanase | + |
| Items | DS84 | GCA225 | GCA405 |
|---|---|---|---|
| Total Length | 44,103,462 | 40,663,325 | 38,237,961 |
| Total Sequence Num | 6 | 34 | 9 |
| Total N Counts | 8200 | 0 | 47,934 |
| Total Low Case Counts | 0 | 2,311,763 | 1,754,046 |
| Total GC content | 0.5 | 0.51 | 0.52 |
| Minimum Length | 551,857 | 8623 | 134,204 |
| Maximum Length | 36,058,779 | 3,887,370 | 24,712,919 |
| Mean Length | 7,350,577 | 119,980.15 | 4,248,662.33 |
| Median Length | 2,141,636.5 | 1,259,297 | 2,559,472 |
| N50 | 36,058,779 | 1,781,809 | 24,712,919 |
| CDS | 37,077 | 15,088 | 36,812 |
| Genome accession | GCA_030544205.1 | GCA_020744225.13 | GCA_001748405.1 |
| Interspersed Repeats | DS-84 | GCA225 | GCA405 |
|---|---|---|---|
| Total interspersed repeats | 9.57% (4,221,058 bp) | 5.21% (2,116,650 bp) | 1.56% (597,717 bp) |
| DNA transposons | 2.72% (1,198,859 bp) | 1.29% (525,828 bp) | 0.26% (101,026 bp) |
| LTR elements | 2.11% (932,269 bp) | 1.77% (720,094 bp) | 0.21% (81,527 bp) |
| LINEs: | 0.23% (101,041 bp) | 0.03% (11,678 bp) | 0.01% (2178 bp) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, W.; Feng, M.; Zhu, N.; Leng, F.; Yang, M.; Wang, Y. Genomic Characteristics and Comparative Genomics Analysis of the Endophytic Fungus Paraphoma chrysanthemicola DS-84 Isolated from Codonopsis pilosula Root. J. Fungi 2023, 9, 1022. https://doi.org/10.3390/jof9101022
Sun W, Feng M, Zhu N, Leng F, Yang M, Wang Y. Genomic Characteristics and Comparative Genomics Analysis of the Endophytic Fungus Paraphoma chrysanthemicola DS-84 Isolated from Codonopsis pilosula Root. Journal of Fungi. 2023; 9(10):1022. https://doi.org/10.3390/jof9101022
Chicago/Turabian StyleSun, Wenbin, Min Feng, Ning Zhu, Feifan Leng, Mingjun Yang, and Yonggang Wang. 2023. "Genomic Characteristics and Comparative Genomics Analysis of the Endophytic Fungus Paraphoma chrysanthemicola DS-84 Isolated from Codonopsis pilosula Root" Journal of Fungi 9, no. 10: 1022. https://doi.org/10.3390/jof9101022
APA StyleSun, W., Feng, M., Zhu, N., Leng, F., Yang, M., & Wang, Y. (2023). Genomic Characteristics and Comparative Genomics Analysis of the Endophytic Fungus Paraphoma chrysanthemicola DS-84 Isolated from Codonopsis pilosula Root. Journal of Fungi, 9(10), 1022. https://doi.org/10.3390/jof9101022





