Addition of Five Novel Fungal Flora to the Xylariomycetidae (Sordariomycetes, Ascomycota) in Northern Thailand
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection, Isolation, and Morphological Studies
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Phylogenetic Analyses
| Taxa | Original Code | GenBank Accession Numbers | References | |||
|---|---|---|---|---|---|---|
| ITS | LSU | RPB2 | TUB2 | |||
| Achaetomium macrosporum | CBS 532.94 | KX976574 | KX976699 | KX976797 | KX976915 | [26] |
| Alloanthostomella rubicola | MFLUCC 16-0479 T | KX533455 | KX533456 | N/A | N/A | [27] |
| Amphirosellinia nigrospora | HAS T 91092308 T | GU322457 | N/A | GQ848340 | GQ495951 | [28] |
| Anthostomella formosa | MFLUCC 14-0170 | MW240652 | MW240582 | N/A | MW820917 | [8] |
| A. helicofissa | MFLUCC 14-0173 T | MW240653 | MW240583 | KP340534 | KP406617 | [8,29] |
| A. lamiacearum | MFLU 18-0101 T | MW240669 | MW240599 | MW658648 | N/A | [8] |
| A. obesa | MFLUCC 14-0171 T | KP297405 | KP340546 | KP340533 | N/A | [29] |
| Anthostomelloides krabiensis | MFLUCC 15-0678 T | KX305927 | KX305928 | KX305929 | N/A | [30] |
| Astrocystis sublimbata | HAS T 89032207 | GU322447 | N/A | GQ844834 | GQ495940 | [28] |
| Barrmaelia macrospora | CBS 142768 T | KC774566 | KC774566 | MF488995 | MF489014 | [31,32] |
| B. moravica | CBS 142769 T | MF488987 | MF488987 | MF488996 | MF489015 | [32] |
| B. rappazii | CBS 142771 T | MF488989 | MF488989 | MF488998 | MF489017 | [32] |
| Biscogniauxia nummularia | MUCL 51395 T | KY610382 | KY610427 | KY624236 | KX271241 | [33] |
| Brunneiperidium involucratum | MFLUCC 14-0009 T | KP297399 | KP340541 | KP340527 | KP406610 | [29] |
| Camillea obularia | A TCC 28093 | KY610384 | KY610429 | KY624238 | KX271243 | [33] |
| Chaetomium elatum | CBS 374.66 | KC109758 | KC109758 | KF001820 | KC109776 | [34] |
| Circinotrichum circinatum | CBS 148326 | ON400743 | ON400796 | ON399328 | N/A | [35] |
| C. maculiforme | CBS 140016 T | KR611874 | KR611895 | ON399338 | N/A | [35] |
| Clypeosphaeria mamillana | CBS 140735 T | KT949897 | KT949897 | MF489001 | MH704637 | [32,36,37] |
| Collodiscula leigongshanensis | GZUH 0107 T | KP054281 | KP054282 | KR002588 | KR002587 | [38] |
| Coniocessia maxima | CBS 593.74 T | GU553332 | MH878275 | N/A | N/A | [39,40] |
| C. nodulisporioides | CBS 281.77 T | MH861061 | MH872831 | N/A | N/A | [40] |
| Creosphaeria sassafras | S TMA 14087 | KY610411 | KY610468 | KY624265 | KX271258 | [33] |
| Digitodochium amoenum | CBS 147285 T | ON869303 | ON869303 | ON808481 | ON808525 | [41] |
| Emarcea castanopsidicola | CBS 117105 T | AY603496 | MK762717 | MK791285 | MK776962 | [12,42] |
| E. eucalyptigena | CBS 139908 T | KR476733 | MK762718 | MK791286 | MK776963 | [12,43] |
| Entalbostroma erumpens | ICMP 21152 T | KX258206 | N/A | KX258204 | KX258205 | [44] |
| Entoleuca mammata | J.D.R. 100 | GU300072 | N/A | GQ844782 | GQ470230 | [28] |
| Entosordaria perfidiosa | CBS 142773 T | MF488993 | MF488993 | MF489003 | MF489021 | [32] |
| E. quercina | CBS 142774 T | MF488994 | MF488994 | MF489004 | MF489022 | [32] |
| Graphostroma platystomum | CBS 270.87 T | JX658535 | DQ836906 | KY624296 | HG934108 | [33,45,46,47] |
| Gyrothrix encephalarti | CBS 146684 T | MT373376 | MT373358 | ON399342 | N/A | [35,48] |
| G. eucalypti | CBS 146023 T | MN562109 | MN567617 | ON399346 | N/A | [35,49] |
| G. podosperma | MFLUCC 16-0243 T | KX505957 | KX505958 | KX789496 | KX789495 | [27] |
| G. podosperma | CBS 148804 | ON400756 | ON400810 | ON399343 | N/A | [35] |
| G. verticillata | CBS 148806 | ON400759 | ON400813 | ON399318 | N/A | [35] |
| Hansfordia pruni | CBS 194.56 T | MK442585 | MH869122 | KU684307 | N/A | [40,50,51] |
| H. pulvinata | CBS 144422 | MK442587 | MK442527 | N/A | N/A | [50] |
| Hypocreodendron sanguineum | J.D.R. 169 T | GU322433 | N/A | GQ844819 | GQ487710 | [28] |
| Induratia apiospora | A TCC 60639 T | OP862879 | OP862881 | OP879469 | OP879468 | [52] |
| Kretzschmaria deusta | CBS 163.93 | KC477237 | KY610458 | KY624227 | KX271251 | [33,53] |
| K. guyanensis | HAS T 89062903 | GU300079 | N/A | GQ844792 | GQ478214 | [28] |
| Kretzschmariella culmorum | J.D.R. 88 | KX430043 | N/A | KX430045 | KX430046 | [44] |
| Linosporopsis ischnotheca | CBS 145761 T | MN818952 | MN818952 | MN820708 | MN820715 | [54] |
| L. ochracea | CBS 145999 T | MN818958 | MN818958 | MN820714 | MN820721 | [54] |
| Lopadostoma dryophilum | CBS 133213 T | KC774570 | KC774570 | KC774526 | MF489023 | [31,32] |
| L. quercicola | CBS 133212 T | KC774610 | KC774610 | KC774558 | N/A | [31] |
| L. turgidum | CBS 133207 T | KC774618 | KC774618 | KC774563 | MF489024 | [31] |
| Magnostiolata mucida | MFLU 19-2133 T | MW240673 | MW240603 | MW658652 | MW775618 | [8] |
| Melanographium phoenicis | MFLUCC 18-1481 T | MN482677 | MN482678 | N/A | N/A | [55] |
| M. smilacis | MFLU 21-0075 T | MZ538514 | MZ538548 | N/A | N/A | [56] |
| Microdochium lycopodinum | CBS 125585 T | JF440979 | JF440979 | KP859125 | KP859080 | [57,58] |
| M. phragmitis | CBS 285.71 T | KP859013 | KP858949 | KP859122 | KP859077 | [58] |
| Muscodor albus | 9_6 | HM034857 | HM034865 | N/A | HM034844 | [59] |
| M. albus | MON T 620 T | AF324336 | N/A | N/A | N/A | [60] |
| M. brasiliensis | LGMF1255 | KY924493 | N/A | N/A | N/A | [61] |
| M. brasiliensis | LGMF1256 T | KY924494 | N/A | N/A | N/A | [61] |
| M. brunneascosporus | CMUB 40020 T | OR507145 | OR507158 | OR504420 | OR519978 | This study |
| M. brunneascosporus | MFLU 23-0406 | OR507146 | OR507159 | N/A | N/A | This study |
| M. camphorae | NFCCI 3236 T | KC481681 | N/A | N/A | N/A | [62] |
| M. cinnanomi | BCC 38842 T | GQ848369 | N/A | N/A | N/A | [63] |
| M. coffeanum | COAD 1842 T | KM514680 | N/A | KP862881 | N/A | [64] |
| M. coffeanum | MFLUCC 13-0159 | MK634693 | MK634694 | MK644942 | MK644943 | [65] |
| M. coffeanum | COAD 1900 | KP862879 | N/A | KP862880 | N/A | [64] |
| M. coffeanum | CMUB 40022 | OR507147 | OR507160 | N/A | N/A | This study |
| M. crispans | MON T 2347 T | EU195297 | N/A | N/A | N/A | [66] |
| M. equiseti | JCM 18233 T | JX089322 | N/A | N/A | N/A | [67] |
| M. fengyangensis | CGMCC 2863 | HM034855 | HM034861 | HM034851 | HM034842 | [59] |
| M. fengyangensis | CGMCC 2862 T | HM034856 | HM034859 | HM034849 | HM034843 | [59] |
| M. ghoomensis | NFCCI 3234 T | KF537625 | N/A | N/A | N/A | [68] |
| M. kashayum | NFCCI 2947 T | KC481680 | N/A | N/A | N/A | [69] |
| M. lamphunensis | CMUB 40021 T | OR507148 | OR507161 | OR504421 | OR519979 | This study |
| M. lamphunensis | MFLU 23-0408 | OR507149 | OR507162 | N/A | N/A | This study |
| M. musae | JCM 18230 T | JX089323 | N/A | N/A | N/A | [67] |
| M. oryzae | JCM 18231 T | JX089321 | N/A | N/A | N/A | [67] |
| M. roseus | MON T 2098 T | AH010859 | N/A | N/A | N/A | [70] |
| Muscodor sp. | SMH 1255 | MN250031 | AY780069 | N/A | AY780119 | [12,71] |
| M. strobelii | NFCCI 2907 T | JQ409999 | N/A | N/A | N/A | [72] |
| M. suthepensis | JCM 18232 T | JN558830 | N/A | N/A | N/A | [67] |
| M. sutura | MSUB 2380 T | JF938595 | N/A | N/A | N/A | [73] |
| M. thailandica | HKAS 102323 | MK762708 | MK762715 | MK791284 | MK776961 | [12] |
| M. thailandica | MFLUCC 17-2669 T | MK762707 | MK762714 | MK791283 | MK776960 | [12] |
| M. tigerensis | NFCCI 3172 T | JQ409998 | N/A | N/A | N/A | [74] |
| M. vitigenus | MON T P-15 T | AY100022 | N/A | N/A | N/A | [75] |
| M. vitigenus | CE-QCA-O1100 | KC771512 | N/A | N/A | N/A | Unpublished |
| M. yucatanensis | MEXU 25511 T | FJ917287 | N/A | N/A | N/A | [76] |
| M. yucatanensis | CDA744 | KU094056 | N/A | N/A | N/A | Unpublished |
| M. yunnanensis | CGMCC 3.18908 T | MG866046 | MG866038 | MG866059 | MG866066 | [77] |
| M. ziziphi | MFLUCC 17-2662 T | MK762705 | MK762712 | MK791281 | MK776958 | [12] |
| M. ziziphi | HKAS 102300 | MK762706 | MK762713 | MK791282 | MK776959 | [12] |
| Nemania abortiva | BISH 467 T | GU292816 | N/A | GQ844768 | GQ470219 | [28] |
| N. macrocarpa | WSP 265 T | GU292823 | MH874423 | GQ844776 | GQ470226 | [28,40] |
| N. primolutea | HAS T 91102001 T | EF026121 | N/A | GQ844767 | EF025607 | [28] |
| Neoanthostomella bambusicola | MFLU 18-0796 T | MW240657 | MW240587 | MW658641 | MW775610 | [8] |
| N. fici | MFLU 19-2765 T | MW114390 | MW114445 | MW177711 | N/A | [78] |
| N. pseudostromatica | MFLU 15-1190 T | KU940158 | KU863146 | N/A | N/A | [79] |
| Neogyrothrix oleae | CBS 146068 | MN562137 | MN567644 | N/A | N/A | [49] |
| N. oleae | CBS 146069 T | MN562136 | MN567643 | N/A | N/A | [49] |
| Nigropunctata bambusicola | MFLU 19-2134 | MW240662 | MW240592 | MW658644 | N/A | [8] |
| N. bambusicola | MFLU 19-2145 T | MW240664 | MW240594 | MW658646 | N/A | [8] |
| N. hydei | CMUB 40018 T | OR507150 | OR507163 | OR504422 | N/A | This study |
| N. hydei | MFLU 23-0410 | OR507151 | OR507164 | N/A | N/A | This study |
| N. nigrocircularis | MFLU 19-2130 T | MW240661 | MW240591 | N/A | MW775612 | [8] |
| N. saccata | MFLU 19-2144 T | MW240663 | MW240593 | MW658645 | MW775613 | This study |
| N. saccata | MFLU 18-0804 | MW240658 | MW240588 | MW658642 | MW775611 | This study |
| Nigropunctata sp. | HKAS 122747 | OQ158966 | OQ170888 | N/A | N/A | Unpublished |
| N. thailandica | MFLU 19-2118 T | MW240659 | MW240589 | MW658643 | N/A | [8] |
| N. thailandica | HKAS 106975 | MW240660 | MW240590 | N/A | N/A | [8] |
| Occultitheca rosae | HKAS 102393 T | MW240672 | MW240602 | MW658651 | MW775617 | [8] |
| Paraxylaria rosacearum | TASM 6132 T | MG828941 | MG829050 | N/A | N/A | [80] |
| Peglionia verticiclada | CBS 127654 T | ON400763 | ON400815 | ON399352 | N/A | [35] |
| Pirozynkiomyces brasiliensis | CBS 112314 T | ON400767 | ON400819 | ON399341 | N/A | [35] |
| Podosordaria mexicana | WSP 176 | GU324762 | N/A | GQ853039 | GQ844840 | [28] |
| Poronia punctata | CBS 656.78 T | KT281904 | KY610496 | KY624278 | KX271281 | [33,81] |
| Pseudoanthostomella delitescens | MFLUCC 16-0477 | KX533451 | KX533452 | KX789491 | KX789490 | [27] |
| P. pini-nigrae | MFLUCC 16-0478 T | KX533453 | KX533454 | KX789492 | N/A | [27] |
| Pseudoceratocladium polysetosum | FMR 10750 T | KY853430 | KY853490 | ON399348 | N/A | [35,82] |
| P. polysetosum | CBS 126092 | MH864077 | MH875534 | ON399347 | N/A | [35,40] |
| Pseudocircinotrichum papakurae | CBS 101373 | KR611876 | KR611897 | N/A | N/A | [35] |
| P. papakurae | CBS 140221 | ON400768 | ON400820 | ON399349 | N/A | [35] |
| Rosellinia buxi | J.D.R. 99 | GU300070 | N/A | GQ844780 | GQ470228 | [28] |
| R. necatrix | HAS T 89062904 | EF026117 | KF719204 | GQ844779 | EF025603 | [28] |
| Sarcoxylon compunctum | CBS 359.61 | KT281903 | KT281898 | KY624230 | KX271255 | [33,81] |
| Selenodriella brasiliana | CBS 140227 T | ON400769 | ON400821 | ON399356 | N/A | [35] |
| S. cubensis | CBS 683.96 T | KP859053 | KP858990 | N/A | N/A | [58] |
| Spiririma gaudefroyi | CBS 147284 T | ON869320 | ON869320 | ON808497 | ON808541 | [41] |
| Sordaria fimicola | CBS 723.96 | MH862606 | MH874231 | DQ368647 | N/A | [40,83] |
| Stilbohypoxylon elaeicola | HAS T 94082615 | GU322440 | N/A | GQ844827 | GQ495933 | [28] |
| Xenoanthostomella calami | MFLUCC 14-0617A T | ON650684 | ON650706 | N/A | ON745964 | [84] |
| X. calami | MFLUCC 14-0617B | ON650685 | ON650707 | N/A | ON745965 | [84] |
| X. chromolaenae | MFLUCC 17-1484 T | MN638863 | MN638848 | MN648729 | N/A | [55] |
| X. chromolaenae | CBS 148702 | ON400784 | ON400841 | N/A | N/A | [35] |
| X. chromolaenae | MFLU 18-0840 | MW240668 | MW240598 | N/A | N/A | [8] |
| X. cycadis | CBS 137969 T | KJ869121 | KJ869178 | ON399350 | N/A | [35] |
| X. cycadis | CPC 25749 | ON400786 | ON400843 | N/A | N/A | [35] |
| X. olivacea | CBS 101185 | ON400787 | ON400844 | ON399351 | N/A | [35] |
| X. parvispora | CMUB 40019 T | OR507143 | OR507156 | OR504419 | OR519977 | This study |
| X. parvispora | MFLU 23-0409 | OR507144 | OR507157 | N/A | N/A | This study |
| Xenoanthostomella sp. | MFLUCC 23-0098 | OR438208 | OR438209 | N/A | N/A | Unpublished |
| Xenoanthostomella sp. | MFLUCC 23-182 | OR438210 | N/A | N/A | N/A | Unpublished |
| Xylaria adscendens | J.D.R. 865 | GU322432 | N/A | GQ844818 | GQ487709 | [28] |
| X. arbuscula | CBS 126415 | KY610394 | KY610463 | KY624287 | KX271257 | [33] |
| X. bambusicola | WSP 205 T | EF026123 | N/A | GQ844802 | AY951762 | [28] |
| X. cubensis | J.D.R. 860 | GU991523 | N/A | GQ848365 | GQ502700 | [28] |
| X. discolor | HAS T 131023 T | JQ087405 | N/A | JQ087411 | JQ087414 | [28] |
| X. hypoxylon | CBS 122620 T | AM993141 | KM186301 | KM186302 | KM186300 | [29,85] |
3. Results
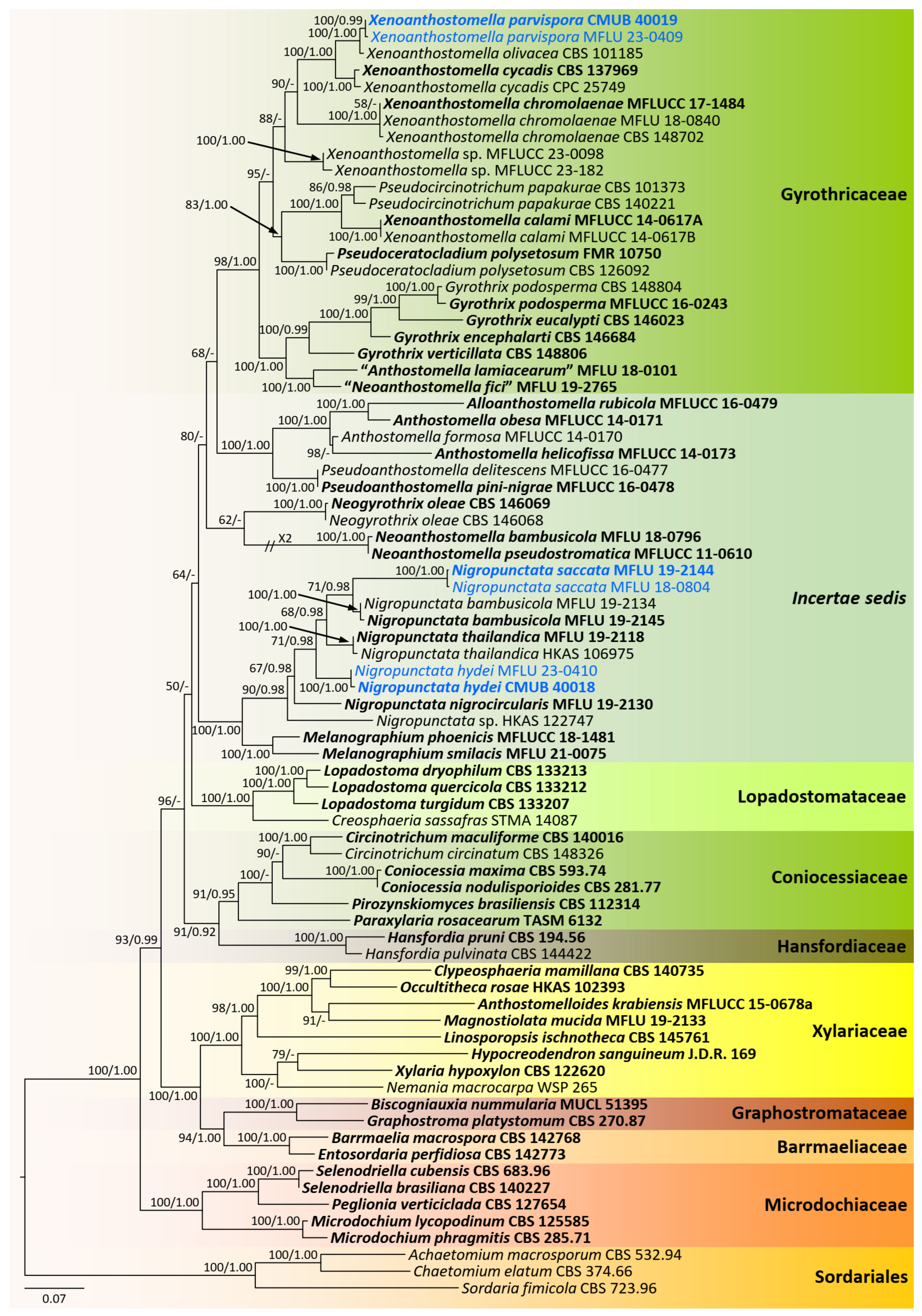
3.1. Phylogenetic Analyses
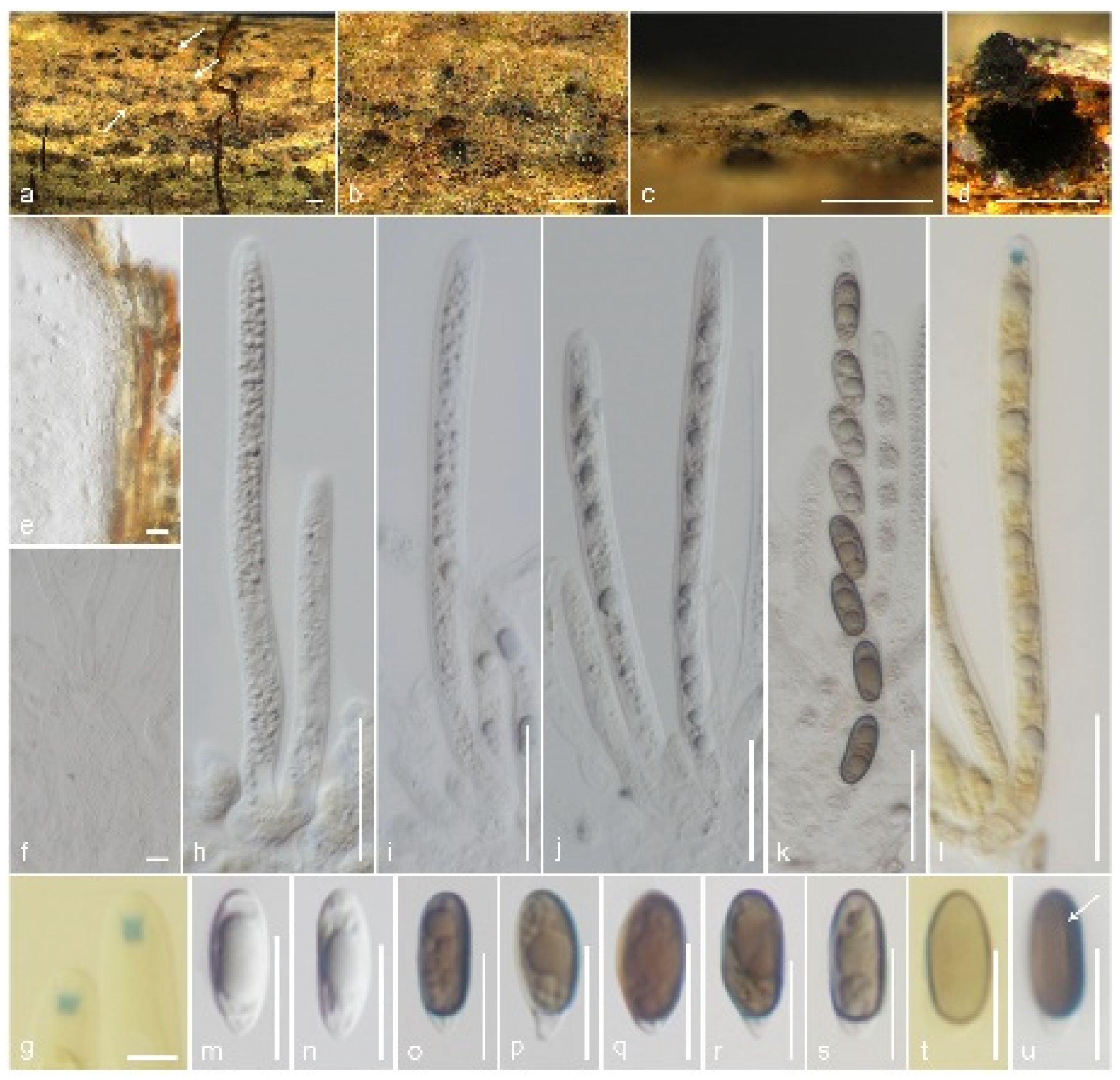
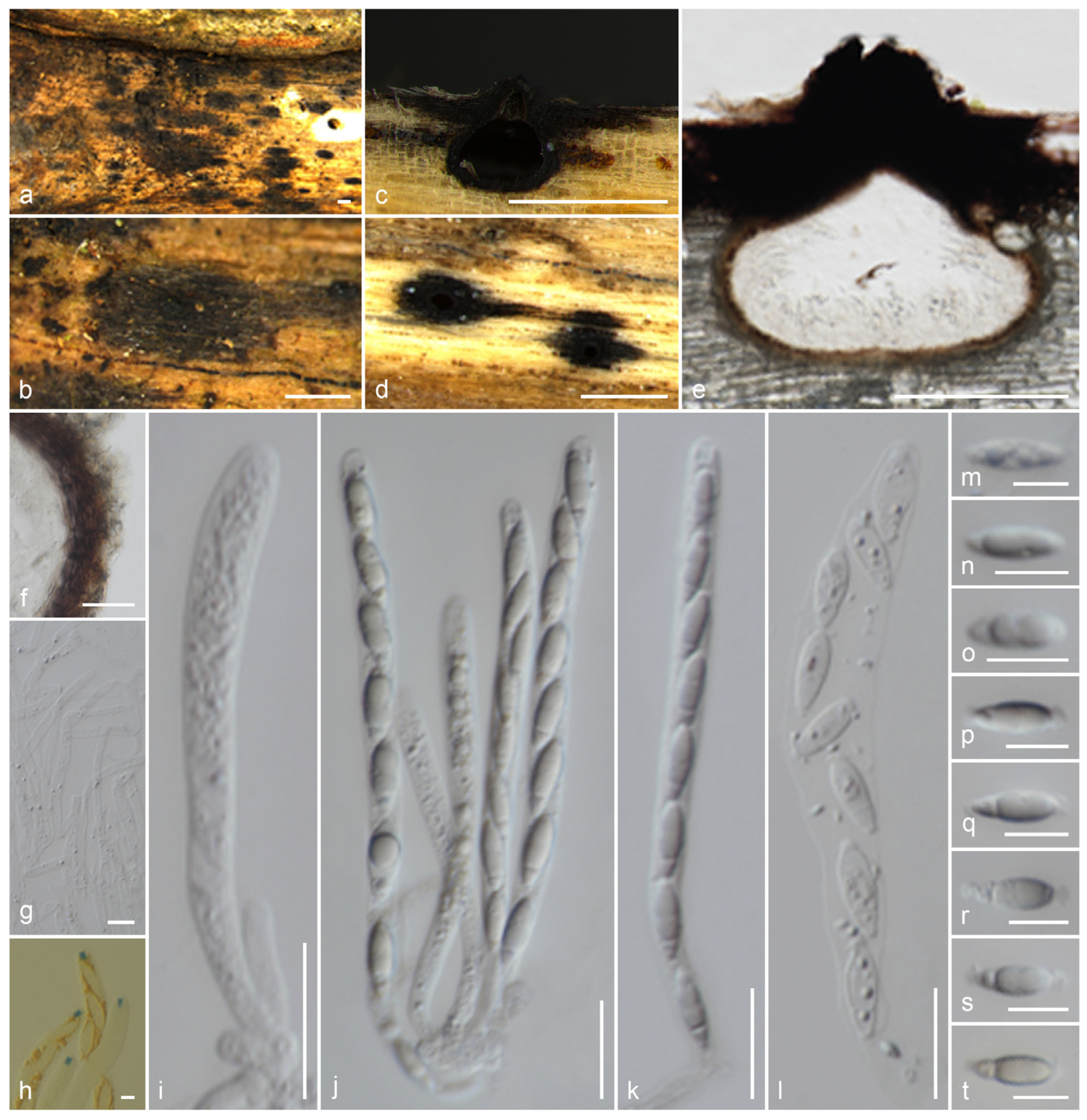
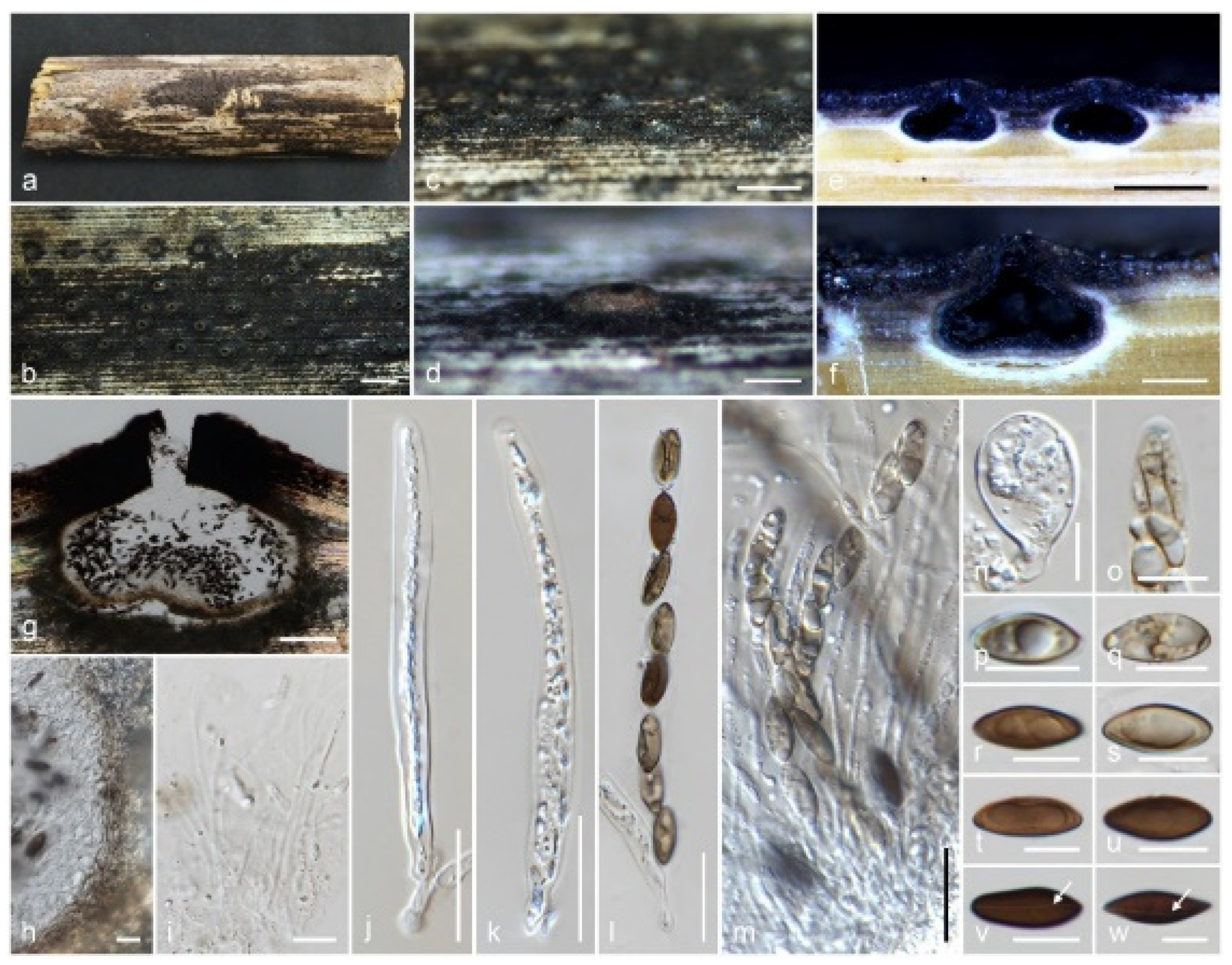
3.2. Taxonomy
3.2.1. Gyrotrichaceae Hern.-Restr. & Crous, in Hernández-Restrepo et al., Persoonia 49: 111 (2022) [35]
3.2.2. Xylariaceae Tul. & C. Tul. [as ‘Xylariei’], Select. fung. carpol. (Paris) 2: 3 (1863)
3.2.3. Xylariales, Genera Incertae Sedis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Novak, L.A.; Kohn, L.M. Electrophoretic and immunological comparisons of developmentally regulated proteins in members of the Sclerotiniaceae and other sclerotial fungi. Appl. Environ. Microbiol. 1991, 57, 525–534. [Google Scholar] [CrossRef]
- Læssøe, T.; Spooner, B.M. Rosellinia & Astrocystis (Xylariaceae): New species and generic concepts. Kew Bull. 1993, 49, 1–70. [Google Scholar] [CrossRef]
- Koroch, A.; Juliani, H.; Bischoff, J.; Lewis, E.; Bills, G.; Simon, J.; White JR, J. Examination of plant biotrophy in the scale insect parasitizing fungus Dussiella tubericornis. Symbiosis 2004, 37, 267–280. [Google Scholar]
- Lembicz, M.; Miszalski, Z.; Kornaś, A.; Turnau, K. Cooling effect of fungal stromata in the Dactylis-Epichloë-Botanophila symbiosis. Commun. Integr. Biol. 2021, 14, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Orton, C.R. Studies in the morphology of the Ascomycetes I. The stroma and the compound fructification of the Dothideaceae and other groups. Mycologia 1924, 16, 49–95. [Google Scholar] [CrossRef]
- Eriksson, O.E.; Winka, K. Supraordinal taxa of Ascomycota. Myconet 1997, 1, 1–16. [Google Scholar]
- Daranagama, D.A.; Jones, E.B.G.; Liu, X.Z.; To-anun, C.; Stadler, M.; Hyde, K.D. Mycosphere Essays 13—Do xylariaceous macromycetes make up most of the Xylariomycetidae? Mycosphere 2016, 7, 582–601. [Google Scholar] [CrossRef]
- Samarakoon, M.C.; Hyde, K.D.; Maharachchikumbura, S.S.N.; Stadler, M.; Gareth Jones, E.B.; Promputtha, I.; Suwannarach, N.; Camporesi, E.; Bulgakov, T.S.; Liu, J.-K. Taxonomy, phylogeny, molecular dating and ancestral state reconstruction of Xylariomycetidae (Sordariomycetes). Fungal Divers. 2022, 112, 1–88. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Jones, E.B.G.; Bundhun, D.; Chen, Y.J.; Bao, D.F.; Boonmee, S.; Calabon, M.S.; et al. Refined families of Sordariomycetes. Mycosphere 2020, 11, 305–1059. [Google Scholar] [CrossRef]
- Vasilyeva, L.N.; Stephenson, S.L.; Hyde, K.D.; Bahkali, A.H. Some stromatic pyrenomycetous fungi from northern Thailand—1. Biscogniuxia, Camillea and Hypoxylon (Xylariaceae). Fungal Divers. 2012, 55, 65–76. [Google Scholar] [CrossRef]
- Samarakoon, M.C.; Jack Liu, J.-K.; Hyde, K.D.; Promputtha, I. Two new species of Amphisphaeria (Amphisphaeriaceae) from northern Thailand. Phytotaxa 2019, 391, 207–217. [Google Scholar] [CrossRef]
- Samarakoon, M.C.; Thongbai, B.; Hyde, K.D.; Brönstrup, M.; Beutling, U.; Lambert, C.; Miller, A.N.; Liu, J.-K.; Promputtha, I.; Stadler, M. Elucidation of the life cycle of the endophytic genus Muscodor and its transfer to Induratia in Induratiaceae fam. nov., based on a polyphasic taxonomic approach. Fungal Divers. 2020, 101, 177–210. [Google Scholar] [CrossRef]
- Samarakoon, M.C.; Maharachchikumbura, S.S.N.; Liu, J.-K.; Hyde, K.D.; Promputtha, I.; Stadler, M. Molecular phylogeny and morphology of Amphisphaeria (= Lepteutypa) (Amphisphaeriaceae). J. Fungi 2020, 6, 174. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Elsevier: Amsterdam, The Netherlands, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- O’Donnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef]
- Rehner, S.A.; Samuels, G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994, 98, 625–634. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Zhaxybayeva, O.; Gogarten, J.P. Bootstrap, bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genom. 2002, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v. 1.4.0. Available online: http://tree.bio.ed.ac.uk/%0Asoftware/figtree/ (accessed on 20 August 2023).
- Wang, X.W.; Houbraken, J.; Groenewald, J.Z.; Meijer, M.; Andersen, B.; Nielsen, K.F.; Crous, P.W.; Samson, R.A. Diversity and taxonomy of Chaetomium and chaetomium-like fungi from indoor environments. Stud. Mycol. 2016, 84, 145–224. [Google Scholar] [CrossRef]
- Daranagama, D.A.; Camporesi, E.; Jeewon, R.; Liu, X.; Stadler, M.; Lumyong, S.; Hyde, K.D. Taxonomic rearrangement of Anthostomella (Xylariaceae) based on a multigene phylogeny and morphology. Cryptogam. Mycol. 2016, 37, 509–538. [Google Scholar] [CrossRef]
- Hsieh, H.-M.; Lin, C.-R.; Fang, M.-J.; Rogers, J.D.; Fournier, J.; Lechat, C.; Ju, Y.-M. Phylogenetic status of Xylaria subgenus Pseudoxylaria among taxa of the subfamily Xylarioideae (Xylariaceae) and phylogeny of the taxa involved in the subfamily. Mol. Phylogenetics Evol. 2010, 54, 957–969. [Google Scholar] [CrossRef]
- Daranagama, D.A.; Camporesi, E.; Tian, Q.; Liu, X.; Chamyuang, S.; Stadler, M.; Hyde, K.D. Anthostomella is polyphyletic comprising several genera in Xylariaceae. Fungal Divers. 2015, 73, 203–238. [Google Scholar] [CrossRef]
- Tibpromma, S.; Daranagama, D.A.; Boonmee, S.; Promputtha, I.; Nontachaiyapoom, S.; Hyde, K.D. Anthostomelloides krabiensis gen. et sp. nov. (Xylariaceae) from Pandanus odorifer (Pandanaceae). Turk. J. Bot. 2017, 41, 107–116. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Fournier, J.; Rogers, J.D.; Voglmayr, H. Phylogenetic and taxonomic revision of Lopadostoma. Persoonia—Mol. Phylogeny Evol. Fungi 2014, 32, 52–82. [Google Scholar] [CrossRef]
- Voglmayr, H.; Friebes, G.; Gardiennet, A.; Jaklitsch, W.M. Barrmaelia and Entosordaria in Barrmaeliaceae (Fam. Nov., Xylariales) and critical notes on anthostomella-like genera based on multigene phylogenies. Mycol. Prog. 2018, 17, 155–177. [Google Scholar] [CrossRef]
- Wendt, L.; Sir, E.B.; Kuhnert, E.; Heitkämper, S.; Lambert, C.; Hladki, A.I.; Romero, A.I.; Luangsa-ard, J.J.; Srikitikulchai, P.; Peršoh, D.; et al. Resurrection and emendation of the Hypoxylaceae, recognised from a multigene phylogeny of the Xylariales. Mycol. Prog. 2018, 17, 115–154. [Google Scholar] [CrossRef]
- Wang, X.W.; Lombard, L.; Groenewald, J.Z.; Li, J.; Videira, S.I.R.; Samson, R.A.; Liu, X.Z.; Crous, P.W. Phylogenetic reassessment of the Chaetomium globosum species complex. Persoonia 2016, 36, 83–133. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Restrepo, M.; Decock, C.A.; Costa, M.M.; Crous, P.W. Phylogeny and taxonomy of Circinotrichum, Gyrothrix, Vermiculariopsiella and other setose hyphomycetes. Persoonia 2022, 49, 99–135. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Gardiennet, A.; Voglmayr, H. Resolution of morphology-based taxonomic delusions: Acrocordiella, Basiseptospora, Blogiascospora, Clypeosphaeria, Hymenopleella, Lepteutypa, Pseudapiospora, Requienella, Seiridium and Strickeria. Persoonia—Mol. Phylogeny Evol. Fungi 2016, 37, 82–105. [Google Scholar] [CrossRef]
- Liu, F.; Bonthond, G.; Groenewald, J.Z.; Cai, L.; Crous, P.W. Sporocadaceae, a family of coelomycetous fungi with appendage-bearing conidia. Stud. Mycol. 2019, 92, 287–415. [Google Scholar] [CrossRef]
- Li, Q.R.; Kang, J.C.; Hyde, K.D. Two new species of the genus Collodiscula (Xylariaceae) from China. Mycol Prog. 2015, 14, 52. [Google Scholar] [CrossRef]
- Asgari, B.; Zare, R. Contribution to the taxonomy of the genus Coniocessia (Xylariales). Mycol. Prog. 2011, 10, 189–206. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom Fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Voglmayr, H.; Tello, S.; Jaklitsch, W.M.; Friebes, G.; Baral, H.-O.; Fournier, J. About spirals and pores: Xylariaceae with remarkable germ loci. Persoonia—Mol. Phylogeny Evol. Fungi 2022, 49, 58–98. [Google Scholar] [CrossRef]
- Duong, L.M.; Lumyong, S.; Hyde, K.D.; Jeewon, R. Emarcea castanopsidicola gen. et sp. nov. from Thailand, a new xylariaceous taxon based on morphology and DNA sequences. Stud. Mycol. 2004, 50, 253–260. [Google Scholar]
- Crous, P.W.; Wingfield, M.J.; Guarro, J.; Hernández-Restrepo, M.; Sutton, D.A.; Acharya, K.; Barber, P.A.; Boekhout, T.; Dimitrov, R.A.; Dueñas, M.; et al. Fungal Planet Description Sheets: 320–370. Persoonia—Mol. Phylogeny Evol. Fungi 2015, 34, 167–266. [Google Scholar] [CrossRef] [PubMed]
- Johnston, P.R.; Rogers, J.D.; Park, D.; Martin, N.A. Entalbostroma Erumpens gen. et sp. nov. (Xylariaceae) from Phormium in New Zealand. Mycotaxon 2016, 131, 765–771. [Google Scholar] [CrossRef]
- Zhang, N.; Castlebury, L.A.; Miller, A.N.; Huhndorf, S.M.; Schoch, C.L.; Seifert, K.A.; Rossman, A.Y.; Rogers, J.D.; Kohlmeyer, J.; Volkmann-Kohlmeyer, B.; et al. An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia 2006, 98, 1076–1087. [Google Scholar] [CrossRef]
- Stadler, M.; Læssøe, T.; Fournier, J.; Decock, C.; Schmieschek, B.; Tichy, H.-V.; Peršoh, D. A polyphasic taxonomy of Daldinia (Xylariaceae)1. Stud. Mycol. 2014, 77, 1–143. [Google Scholar] [CrossRef]
- Koukol, O.; Kelnarová, I.; Černý, K. Recent observations of sooty bark disease of sycamore maple in Prague (Czech Republic) and the phylogenetic placement of Cryptostroma corticale. For. Pathol. 2015, 45, 21–27. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Chooi, Y.-H.; Gilchrist, C.L.M.; Lacey, E.; Pitt, J.I.; Roets, F.; Swart, W.J.; Cano-Lira, J.F.; Valenzuela-Lopez, N.; et al. Fungal Planet Description Sheets: 1042–1111. Persoonia—Mol. Phylogeny Evol. Fungi 2020, 44, 301–459. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Lombard, L.; Roets, F.; Swart, W.J.; Alvarado, P.; Carnegie, A.J.; Moreno, G.; Luangsa-Ard, J.; Thangavel, R.; et al. Fungal Planet Description Sheets: 951–1041. Persoonia 2019, 43, 223–425. [Google Scholar] [CrossRef]
- Crous, P.W.; Schumacher, R.K.; Akulov, A.; Thangavel, R.; Hernández-Restrepo, M.; Carnegie, A.J.; Cheewangkoon, R.; Wingfield, M.J.; Summerell, B.A.; Quaedvlieg, W.; et al. New and Interesting Fungi. 2. Fungal Syst. Evol. 2019, 3, 57–134. [Google Scholar] [CrossRef]
- U’Ren, J.M.; Miadlikowska, J.; Zimmerman, N.B.; Lutzoni, F.; Stajich, J.E.; Arnold, A.E. Contributions of North American endophytes to the phylogeny, ecology, and taxonomy of Xylariaceae (Sordariomycetes, Ascomycota). Mol. Phylogenet. Evol. 2016, 98, 210–232. [Google Scholar] [CrossRef]
- Cedeño-Sanchez, M.; Schiefelbein, R.; Stadler, M.; Voglmayr, H.; Bensch, K.; Lambert, C. Redisposition of apiosporous genera Induratia and Muscodor in the Xylariales, following the discovery of an authentic strain of Induratia apiospora. Bot. Stud. 2023, 64, 8. [Google Scholar] [CrossRef]
- Stadler, M.; Kuhnert, E.; Peršoh, D.; Fournier, J. The Xylariaceae as model example for a unified nomenclature following the “One Fungus-One Name” (1F1N) concept. Mycol. Int. J. Fungal Biol. 2013, 4, 5–21. [Google Scholar]
- Voglmayr, H.; Beenken, L. Linosporopsis, a new leaf-inhabiting scolecosporous genus in Xylariaceae. Mycol. Prog. 2020, 19, 205–222. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Dong, Y.; Phookamsak, R.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Liu, N.-G.; Abeywickrama, P.D.; Mapook, A.; Wei, D.; et al. Fungal Diversity Notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020, 100, 5–277. [Google Scholar] [CrossRef]
- Boonmee, S.; Wanasinghe, D.N.; Calabon, M.S.; Huanraluek, N.; Chandrasiri, S.K.U.; Jones, G.E.B.; Rossi, W.; Leonardi, M.; Singh, S.K.; Rana, S.; et al. Fungal Diversity Notes 1387–1511: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2021, 111, 1–335. [Google Scholar] [CrossRef] [PubMed]
- Jaklitsch, W.M.; Voglmayr, H. Phylogenetic relationships of five genera of Xylariales and Rosasphaeria gen. nov. (Hypocreales). Fungal Divers. 2012, 52, 75–98. [Google Scholar] [CrossRef]
- Hernández-Restrepo, M.; Groenewald, J.Z.; Crous, P.W. Taxonomic and phylogenetic re-evaluation of Microdochium, Monographella and Idriella. Persoonia 2016, 36, 57–82. [Google Scholar] [CrossRef]
- Zhang, C.-L.; Wang, G.-P.; Mao, L.-J.; Komon-Zelazowska, M.; Yuan, Z.-L.; Lin, F.-C.; Druzhinina, I.S.; Kubicek, C.P. Muscodor fengyangensis sp. nov. from southeast China: Morphology, physiology and production of volatile compounds. Fungal Biol. 2010, 114, 797–808. [Google Scholar] [CrossRef]
- Worapong, J.; Strobel, G.; Ford, E.; Li, J.; Baird, G.; Hess, W. Muscodor albus anam. gen. et sp. nov., an endophyte from Cinnamomum zeylanicum. Mycotaxon 2001, 79, 67–79. [Google Scholar]
- Pena, L.C.; Jungklaus, G.H.; Savi, D.C.; Ferreira-Maba, L.; Servienski, A.; Maia, B.H.L.N.S.; Annies, V.; Galli-Terasawa, L.V.; Glienke, C.; Kava, V. Muscodor brasiliensis sp. nov. produces volatile organic compounds with activity against Penicillium digitatum. Microbiol. Res. 2019, 221, 28–35. [Google Scholar] [CrossRef]
- Meshram, V. Muscodor camphora, a new endophytic species from Cinnamomum camphora. Mycosphere 2017, 8, 568–582. [Google Scholar] [CrossRef]
- Suwannarach, N.; Bussaban, B.; Hyde, K.D.; Lumyong, S. Muscodor cinnamomi, a new endophytic species from Cinnamomum bejolghota. Mycotaxon 2011, 114, 15–23. [Google Scholar] [CrossRef]
- Hongsanan, S.; Hyde, K.D.; Bahkali, A.H.; Camporesi, E.; Chomnunti, P.; Ekanayaka, H.; Gomes, A.A.M.; Hofstetter, V.; Jones, E.B.G.; Pinho, D.B.; et al. Fungal Biodiversity Profiles 11–20. Cryptogam. Mycol. 2015, 36, 355–380. [Google Scholar] [CrossRef]
- Li, Q.-R.; Kang, J.-C. The sexual morph of Induratia coffeana, a new record from Thailand. MycoAsia 2022, 1, 1–14. [Google Scholar] [CrossRef]
- Mitchell, A.M.; Strobel, G.A.; Moore, E.; Robison, R.; Sears, J. Volatile antimicrobials from Muscodor crispans, a novel endophytic fungus. Microbiology 2010, 156, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Suwannarach, N.; Kumla, J.; Bussaban, B.; Hyde, K.D.; Matsui, K.; Lumyong, S. Molecular and morphological evidence support four new species in the genus Muscodor from northern Thailand. Ann. Microbiol. 2013, 63, 1341–1351. [Google Scholar] [CrossRef]
- Meshram, V.; Gupta, M.; Saxena, S. Muscodor ghoomensis and Muscodor indica: New endophytic species based on morphological features and molecular and volatile organic analysis from northeast India. Sydowia 2015, 67, 133–146. [Google Scholar] [CrossRef]
- Meshram, V.; Kapoor, N.; Saxena, S. Muscodor kashayum sp. nov.—A new volatile anti-microbial producing endophytic fungus. Mycol. Int. J. Fungal Biol. 2013, 4, 196–204. [Google Scholar] [CrossRef]
- Worapong, J.; Strobel, G.; Daisy, B.; Castillo, U.; Baird, G.; Hess, W. Muscodor roseus anam. sp. nov., an endophyte from Grevillea pteridifolia. Mycotaxon 2002, 81, 463–475. [Google Scholar]
- Miller, A.N.; Huhndorf, S.M. Multi-gene phylogenies indicate ascomal wall morphology is a better predictor of phylogenetic relationships than ascospore morphology in the Sordariales (Ascomycota, Fungi). Mol. Phylogenet. Evol. 2005, 35, 60–75. [Google Scholar] [CrossRef]
- Meshram, V.; Saxena, S.; Kapoor, N. Muscodor strobelii, a new endophytic species from south India. Mycotaxon 2014, 128, 93–104. [Google Scholar] [CrossRef]
- Kudalkar, P.; Strobel, G.; Riyaz-Ul-Hassan, S.; Geary, B.; Sears, J. Muscodor sutura, a novel endophytic fungus with volatile antibiotic activities. Mycoscience 2012, 53, 319–325. [Google Scholar] [CrossRef]
- Saxena, S.; Meshram, V.; Kapoor, N. Muscodor tigerii sp. nov.-Volatile antibiotic producing endophytic fungus from the northeastern Himalayas. Ann. Microbiol. 2015, 65, 47–57. [Google Scholar] [CrossRef]
- Daisy, B.; Strobel, G.; Ezra, D.; Castillo, U.; Baird, G.; Hess, W. Muscodor vitigentis anam. nov., an endophyte from Paullinia paullinioides. Mycotaxon 2002, 84, 39–50. [Google Scholar]
- González, M.C.; Anaya, A.L.; Glenn, A.E.; Macías-Rubalcava, M.L.; Hernández-Bautista, B.E.; Hanlin, R.T. Muscodor yucatanensis, a new endophytic ascomycete from Mexican Chakah, Bursera simaruba. Mycotaxon 2009, 110, 363–372. [Google Scholar] [CrossRef]
- Chen, J.; Feng, X.; Xia, C.; Kong, D.; Qi, Z.; Liu, F.; Chen, D.; Lin, F.; Zhang, C. Confirming the phylogenetic position of the genus Muscodor and the description of a new Muscodor species. Mycosphere 2019, 10, 187–201. [Google Scholar] [CrossRef]
- Tennakoon, D.S.; Kuo, C.-H.; Maharachchikumbura, S.S.N.; Thambugala, K.M.; Gentekaki, E.; Phillips, A.J.L.; Bhat, D.J.; Wanasinghe, D.N.; De Silva, N.I.; Promputtha, I.; et al. Taxonomic and phylogenetic contributions to Celtis formosana, Ficus ampelas, F. septica, Macaranga tanarius and Morus australis leaf litter inhabiting microfungi. Fungal Divers. 2021, 108, 1–215. [Google Scholar] [CrossRef]
- Dai, D.Q.; Phookamsak, R.; Wijayawardene, N.N.; Li, W.J.; Bhat, D.J.; Xu, J.C.; Taylor, J.E.; Hyde, K.D.; Chukeatirote, E. Bambusicolous Fungi. Fungal Divers. 2017, 82, 1–105. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Phukhamsakda, C.; Hyde, K.D.; Jeewon, R.; Lee, H.B.; Gareth Jones, E.B.; Tibpromma, S.; Tennakoon, D.S.; Dissanayake, A.J.; Jayasiri, S.C.; et al. Fungal Diversity Notes 709–839: Taxonomic and phylogenetic contributions to fungal Taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018, 89, 1–236. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Bhat, J.D.; Jones, E.B.G.; McKenzie, E.H.C.; Dai, D.Q.; Daranagama, D.A.; Dayarathne, M.C.; Goonasekara, I.D.; et al. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers. 2015, 73, 73–144. [Google Scholar] [CrossRef]
- Hernández-Restrepo, M.; Gené, J.; Castañeda-Ruiz, R.F.; Mena-Portales, J.; Crous, P.W.; Guarro, J. Phylogeny of saprobic microfungi from southern Europe. Stud. Mycol. 2017, 86, 53–97. [Google Scholar] [CrossRef]
- Tang, A.M.C.; Jeewon, R.; Hyde, K.D. Phylogenetic utility of protein (RPB2, β-Tubulin) and ribosomal (LSU, SSU) gene sequences in the systematics of Sordariomycetes (Ascomycota, Fungi). Antonie Van Leeuwenhoek 2007, 91, 327–349. [Google Scholar] [CrossRef] [PubMed]
- Konta, S.; Tibpromma, S.; Karunarathna, S.; Samarakoon, M.; Steven, L.; Mapook, A.; Boonmee, S.; Senwanna, C.; Balasuriya, A.; Eungwanichayapant, P.; et al. Morphology and multigene phylogeny reveal ten novel taxa in Ascomycota from terrestrial palm substrates (Arecaceae) in Thailand. Mycosphere 2023, 14, 107–152. [Google Scholar] [CrossRef]
- Peršoh, D.; Melcher, M.; Graf, K.; Fournier, J.; Stadler, M.; Rambold, G. Molecular and morphological evidence for the delimitation of Xylaria hypoxylon. Mycologia 2009, 101, 256–268. [Google Scholar] [CrossRef]
- Lu, B.S.; Hyde, K.D. A World Monograph of Anthostomella; Fungal Diversity Press: Hong Kong, China, 2000. [Google Scholar]
- Hyde, K.D.; Fröhlich, J.; Taylor, J.E. Fungi from palms XXXVI. Reflections on unitunicate ascomycetes with aiospores. Sydowia 1998, 50, 21–80. [Google Scholar]
- Samuels, G.J.; Müller, E.; Petrini, O. Studies in the Amphisphaeriaceae (Sensu Lato) 3. New species of Monographella and Pestalosphaeria, and two new genera. Mycotaxon 1987, 28, 473–499. [Google Scholar]
- Sugita, R.; Hirayama, K.; Shirouzu, T.; Tanaka, K. Spirodecosporaceae fam. nov. (Xylariales, Sordariomycetes) and two new species of Spirodecospora. Fungal Syst. Evol. 2022, 10, 217–229. [Google Scholar] [CrossRef]
- Konta, S.; Hongsanan, S.; Tibpromma, S.; Thongbai, B.; Maharachchikumbura, S.S.N.; Bahkali, A.H.; Hyde, K.D.; Boonmee, S. An advance in the endophyte story: Oxydothidaceae fam. nov. with six new species of Oxydothis. Mycosphere 2016, 7, 1425–1446. [Google Scholar] [CrossRef]
- Stadler, M.; Lambert, C.; Wibberg, D.; Kalinowski, J.; Cox, R.J.; Kolařík, M.; Kuhnert, E. Intragenomic polymorphisms in the ITS region of high-quality genomes of the Hypoxylaceae (Xylariales, Ascomycota). Mycol. Prog. 2020, 19, 235–245. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samarakoon, M.C.; Lumyong, S.; Manawasinghe, I.S.; Suwannarach, N.; Cheewangkoon, R. Addition of Five Novel Fungal Flora to the Xylariomycetidae (Sordariomycetes, Ascomycota) in Northern Thailand. J. Fungi 2023, 9, 1065. https://doi.org/10.3390/jof9111065
Samarakoon MC, Lumyong S, Manawasinghe IS, Suwannarach N, Cheewangkoon R. Addition of Five Novel Fungal Flora to the Xylariomycetidae (Sordariomycetes, Ascomycota) in Northern Thailand. Journal of Fungi. 2023; 9(11):1065. https://doi.org/10.3390/jof9111065
Chicago/Turabian StyleSamarakoon, Milan C., Saisamorn Lumyong, Ishara S. Manawasinghe, Nakarin Suwannarach, and Ratchadawan Cheewangkoon. 2023. "Addition of Five Novel Fungal Flora to the Xylariomycetidae (Sordariomycetes, Ascomycota) in Northern Thailand" Journal of Fungi 9, no. 11: 1065. https://doi.org/10.3390/jof9111065
APA StyleSamarakoon, M. C., Lumyong, S., Manawasinghe, I. S., Suwannarach, N., & Cheewangkoon, R. (2023). Addition of Five Novel Fungal Flora to the Xylariomycetidae (Sordariomycetes, Ascomycota) in Northern Thailand. Journal of Fungi, 9(11), 1065. https://doi.org/10.3390/jof9111065








