Endophytic Fungal Diversity in Cirsium kawakamii from Taiwan
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Endophytic Fungi
2.2. Morphological and Molecular Identification of Endophytic Fungi
2.3. Alpha and Beta Diversity Analysis of Endophytic Fungi
2.4. Identification of Phytopathogen-like Endophytic Fusarium sp. Isolates
2.5. Pathogenicity Test of Endophytic Fusarium Tricinctum Species Complex (FTSC)-like Isolates toward Apple, Potato, and Wheat
3. Results
3.1. Isolation of Endophytic Fungi from Cirsium kawakamii
3.2. Endophytic Fungal Community Composition in the Three Regions
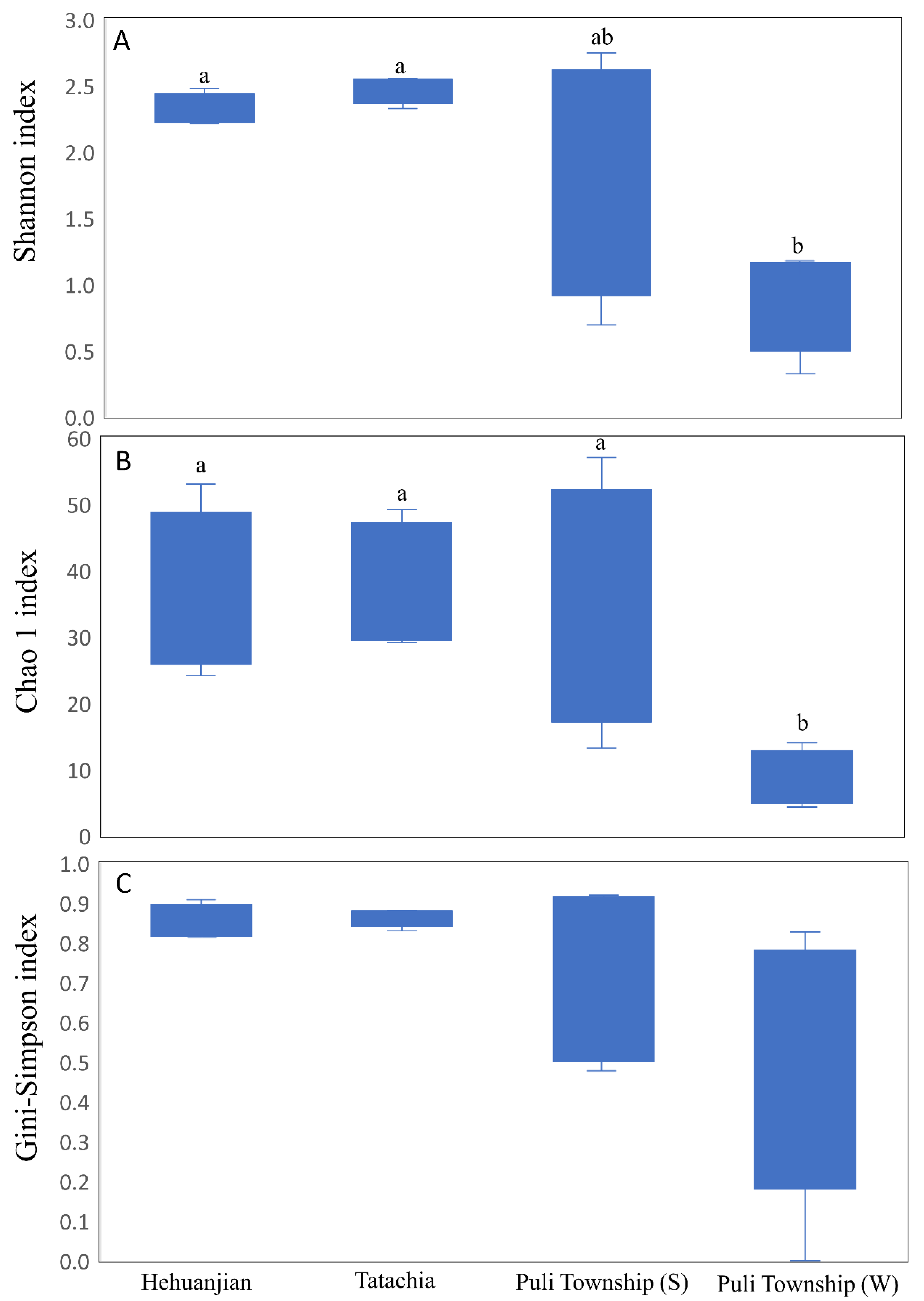
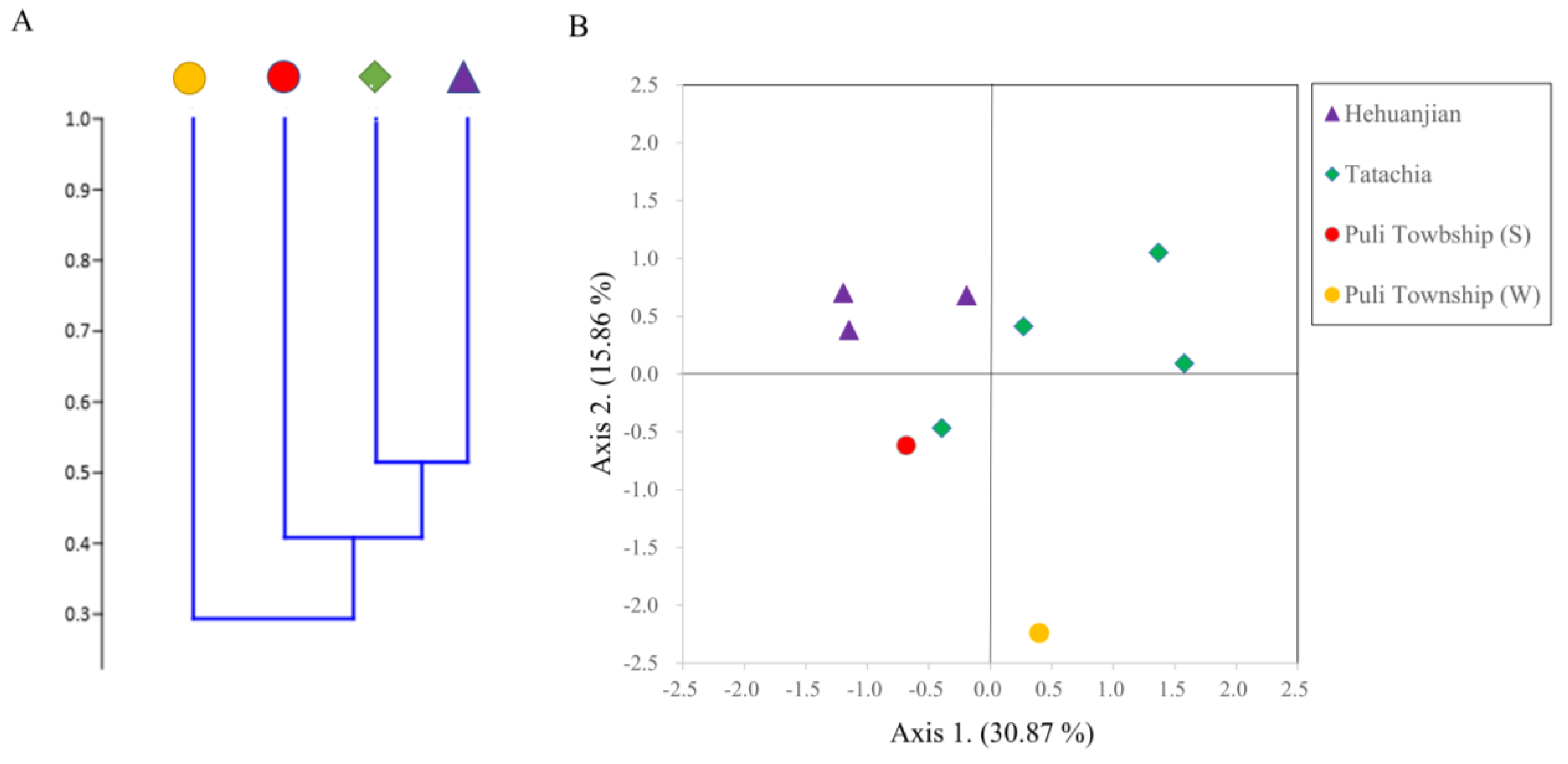
3.3. Endophytic Fungal Alpha and Beta Diversity in Cirsium kawakamii
3.4. Identification of Phytopathogen-like Endophytic Fusarium sp. Isolates
3.5. Pathogenicity Tests of Phytopathogen-like Endophytic Fusarium sp. Isolates on Apple, Potato, and Wheat
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Petrini, O. Fungal endophytes of tree leaves. In Microbial Ecology of Leaves; Andrews, J., Hirano, S., Eds.; Springer: New York, NY, USA, 1991; pp. 179–197. [Google Scholar]
- Wilson, D. Endophyte: The evolution of a term, and clarification of its use and definition. Oikos 1995, 73, 274–276. [Google Scholar] [CrossRef]
- Stone, J.K.; Bacon, C.W.; White, J.F. An overview of endophytic microbes: Endophytism defined. In Microbial Endophytes; Bacon, C.W., White, J.F., Eds.; Marcel Dekker Inc.: New York, NY, USA, 2000; pp. 3–30. [Google Scholar]
- Nair, D.N.; Padmavathy, S. Impact of endophytic microorganisms on plants, environment and humans. Sci. World J. 2014, 2014, 250693. [Google Scholar] [CrossRef] [PubMed]
- Krings, M.; Taylor, T.N.; Hass, H.; Kerp, H.; Dotzler, N.; Hermsen, E.J. Fungal endophytes in a 400-million-yr-old land plant: Infection pathways, spatial distribution, and host responses. New Phytol. 2007, 174, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Brundrett, M.C. Understanding the roles of multifunctional mycorrhizal and endophytic fungi. In Microbial Root Endophytes; Schulz, B.J.E., Boyle, C.J.C., Sieber, T.N., Eds.; Springer: Berlin, Germany, 2006; pp. 281–293. [Google Scholar]
- Rodriguez, R.J.; White, J.F., Jr.; Arnold, A.E.; Redman, R.S. Fungal endophytes: Diversity and functional roles. New Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef] [PubMed]
- Hartley, S.E.; Gange, A.C. Impacts of plant symbiotic fungi on insect herbivores: Mutualism in a multitrophic context. Ann. Rev. Entomol. 2009, 54, 323–342. [Google Scholar] [CrossRef] [PubMed]
- Kharwar, R.N.; Mishra, A.; Gond, S.K.; Stierle, A.; Stierle, D. Anticancer compounds derived from fungal endophytes: Their importance and future challenges. Nat. Prod. Rep. 2011, 28, 1208–1228. [Google Scholar] [CrossRef]
- Preethi, K.; Manon Mani, V.; Lavanya, N. Endophytic Fungi: A potential source of bioactive compounds for commercial and therapeutic applications. In Endophytes; Patil, R.H., Maheshwari, V.L., Eds.; Springer: Singapore, 2021; pp. 247–272. [Google Scholar]
- Zhao, J.; Shan, T.; Mou, Y.; Zhou, L. Plant-derived bioactive compounds produced by endophytic fungi. Mini. Rev. Med. Chem. 2011, 11, 159–168. [Google Scholar] [CrossRef]
- Stierle, A.; Strobel, G.; Stierle, D. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of pacific yew. Science 1993, 260, 214–216. [Google Scholar] [CrossRef]
- Chakravarthi, B.V.S.K.; Das, P.; Surendranath, K.; Karande, A.A.; Jayabaskaran, C. Production of paclitaxel by Fusarium solani isolated from Taxuscelebica. J. Biosci. 2008, 33, 259–267. [Google Scholar] [CrossRef]
- Pongcharoen, W.; Rukachaisirikul, V.; Phongpaichit, S.; Kuehn, T.; Pelzing, M.; Sakayaroj, J.; Taylor, W.C. Metabolites from the endophytic fungus Xylaria sp. PSU-D14. Phytochemistry 2008, 69, 1900–1902. [Google Scholar] [CrossRef]
- Strobel, G.; Daisy, B. Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 2003, 67, 491–502. [Google Scholar] [CrossRef]
- Leuchtmann, A. Systematics, distribution, and host specificity of grass endophytes. Nat. Toxins 1992, 1, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, M.T.; Arnold, A.E. Geographic locality and host identity shape fungal endophyte communities in cupressaceous trees. Mycol. Res. 2008, 112, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Arnold, A.E.; Henk, D.A.; Eells, R.L.; Lutzoni, F.; Vilgalys, R. Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia 2007, 99, 185–206. [Google Scholar] [CrossRef]
- Arnold, A.E.; Lutzoni, F. Diversity and host range of foliar fungal endophytes: Are tropical leaves biodiversity hotspots? Ecology 2007, 88, 541–549. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, M.; Frisullo, S.; Cirulli, M. Endophytic fungi occurring in fennel, lettuce, chicory, and celery-commercial crops in southern Italy. Mycol. Res. 2008, 112, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Gazis, R.; Chaverri, P. Diversity of fungal endophytes in leaves and stems of wild rubber trees (Hevea brasiliensis) in Peru. Fungal Ecol. 2010, 3, 240–254. [Google Scholar] [CrossRef]
- MacArthur, D.; McGee, P. A comparison of the endophytic fungi from leaves of Banksia integrifolia at three sites on the east coast of Australia. Australas. Mycol. 2000, 19, 80–83. [Google Scholar]
- Vega, F.E.; Simpkins, A.; Aime, M.C.; Posada, F.; Peterson, S.W.; Rehner, S.A.; Infante, F.; Castillo, A.; Arnold, A.E. Fungal endophyte diversity in coffee plants from Colombia, Hawai’i, Mexico and Puerto Rico. Fungal Ecol. 2010, 3, 122–138. [Google Scholar] [CrossRef]
- Peng, C.I. Cirsium. In Flora of Taiwan, 2nd ed.; Boufford, D.E., Hsieh, C.F., Kuo, C.S., Ohashi, H., Su, H.J., Eds.; Editorial Committee of the Flora of Taiwan, Department of Botany, National Taiwan University: Taipei, Taiwan, 2003; Volume 4, pp. 903–913. [Google Scholar]
- Lu, Y.S. Investigation of antioxidative components in Cirsium kawakamii Hayata. Master’s Thesis, National Chiayi University, Chiayi City, Taiwan, 2011. [Google Scholar]
- Zhao, Z.W.; Chang, J.C.; Lin, L.W.; Tsai, F.H.; Chang, H.C.; Wu, C.R. Comparison of the hepatoprotective effects of four endemic Cirsium species extracts from Taiwan on ccl4-induced acute liver damage in C57BL/6 Mic. Int. J. Mol. Sci. 2018, 19, 1329. [Google Scholar] [CrossRef]
- Eschen, R.; Hunt, S.; Mykura, C.; Gange, A.C.; Sutton, B.C. The foliar endophytic fungal community composition in Cirsium arvense is affected by mycorrhizal colonization and soil nutrient content. Fungal Biol. 2010, 114, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Bills, G.F.; Redlin, S.; Carris, L. Isolation and analysis of endophytic fungal communities from woody plants. In Endophytic Fungi in Grasses and Woody Plants: Systematics, Ecology, and Evolution; Redlin, S., Carris, L., Eds.; APS Press: Saint Paul, MN, USA, 1996; pp. 31–65. [Google Scholar]
- Goodwin, D.; Lee, S. Microwave miniprep of total genomic DNA from fungi, plants, protists and animals for PCR. Biotechniques 1993, 15, 438–444. [Google Scholar] [PubMed]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Samuels, G.J.; Ismaiel, A. Hypocrea peltata: A mycological Dr Jekyll and Mr Hyde? Mycologia 2011, 103, 616–630. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Laraba, I.; Busman, M.; Geiser, D.M.; O’Donnell, K. Phylogenetic diversity and mycotoxin potential of emergent phytopathogens within the Fusarium tricinctum species complex. Phytopathology 2022, 112, 1284–1298. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Jiang, X.; Sun, M.; Xu, H.L.; Shi, L.; Shan, W.; Feng, Y. Screening of culture conditions for pathogens of potato dry rot. Acta Agric. Scand.-B Soil Plant Sci. 2014, 64, 694–699. [Google Scholar] [CrossRef]
- Bottalico, A.; Perrone, G. Toxigenic Fusarium species and mycotoxins associated with head blight in small-grain cereals in Europe. Eur. J. Plant Pathol. 2002, 108, 611–624. [Google Scholar] [CrossRef]
- Gao, L.L.; Zhang, Q.; Sun, X.Y.; Jiang, L.; Zhang, R.; Sun, G.Y.; Zha, Y.L.; Biggs, A.R. Etiology of moldy core, core browning, and core rot of Fuji apple in China. Plant Dis. 2013, 97, 510–516. [Google Scholar] [CrossRef]
- Gachango, E.; Hanson, L.; Rojas, A.; Hao, J.; Kirk, W. Fusarium spp. causing dry rot of seed potato tubers in Michigan and their sensitivity to fungicides. Plant Dis. 2012, 96, 1767–1774. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.B.; Li, H.P.; Zhao, C.S.; Liao, Y.C. Comparative pathogenicity of Fusarium graminearum isolates from China revealed by wheat coleoptile and floret inoculations. Mycopathologia 2005, 160, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Sever, Z.; Ivic, D.; Kos, T.; Milicevic, T. Identification of Fusarium species isolated from stored apple fruit in Croatia. Arh. Hig. Rada. Toksikol. 2012, 63, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Booth, C. The Genus Fusarium; Commonwealth Mycological Institute: Surrey, UK, 1971; p. 237. [Google Scholar]
- Araújo, W.L.; Maccheroni, W., Jr.; Aguilar-Vildoso, C.I.; Barroso, P.A.; Saridakis, H.O.; Azevedo, J.L. Variability and interactions between endophytic bacteria and fungi isolated from leaf tissues of citrus rootstocks. Can. J. Microbiol. 2001, 47, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.; Soytong, K. The fungal endophyte dilemma. Fungal Div. 2008, 33, 163–173. [Google Scholar]
- Arnold, A.E. Understanding the diversity of foliar endophytic fungi: Progress, challenges, and frontiers. Fungal Biol. Rev. 2007, 21, 51–66. [Google Scholar] [CrossRef]
- Gange, A.C.; Dey, S.; Currie, A.F.; Sutton, B.C. Site-and species-specific differences in endophyte occurrence in two herbaceous plants. J. Ecol. 2007, 95, 614–622. [Google Scholar] [CrossRef]
- Selim, K.; El-Beih, A.; Abdel-Rahman, T.; El-Diwany, A. Biology of endophytic fungi. Curr. Res. Environ. Appl. Mycol. 2012, 2, 31–82. [Google Scholar] [CrossRef]
- Arnold, A.; Maynard, Z.; Gilbert, G.; Coley, P.; Kursar, T. Are tropical fungal endophytes hyperdiverse? Ecol. Lett. 2000, 3, 267–274. [Google Scholar] [CrossRef]
- Fröhlich, J.; Hyde, K.D. Biodiversity of palm fungi in the tropics: Are global fungal diversity estimates realistic? Biodivers. Conserv. 1999, 8, 977–1004. [Google Scholar] [CrossRef]
- Wu, S.Y.; Chen, Y.J.; Chung, W.C.; Chung, W.H. Diversity of fungal endophytes in Cinnamomum kanehirae in Taiwan and control assessment on crop diseases. J. Plant Med. 2017, 59, 13–26. (In Chinese) [Google Scholar]
- Higgins, K.L.; Arnold, A.E.; Miadlikowska, J.; Sarvate, S.D.; Lutzoni, F. Phylogenetic relationships, host affinity, and geographic structure of boreal and arctic endophytes from three major plant lineages. Mol. Phylogenet. Evol. 2007, 42, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, N.B.; Vitousek, P.M. Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proc. Natl. Acad. Sci. USA 2012, 109, 13022–13027. [Google Scholar] [CrossRef] [PubMed]
- Arnold, A.E.; Mejia, L.C.; Kyllo, D.; Rojas, E.I.; Maynard, Z.; Robbins, N.; Herre, E.A. Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. USA 2003, 100, 15649–15654. [Google Scholar] [CrossRef] [PubMed]
- Shipunov, A.; Newcombe, G.; Raghavendra, A.K.; Anderson, C.L. Hidden diversity of endophytic fungi in an invasive plant. Am. J. Bot. 2008, 95, 1096–1108. [Google Scholar] [CrossRef] [PubMed]
- Hashizume, Y.; Fukuda, K.; Sahashi, N. Effects of summer temperature on fungal endophyte assemblages in Japanese beech (Fagus crenata) leaves in pure beech stands. Botany 2010, 88, 266–274. [Google Scholar] [CrossRef]
- Naik, B.S.; Shashikala, J.; Krishnamurthy, Y. Diversity of fungal endophytes in shrubby medicinal plants of Malnad region, Western Ghats, Southern India. Fungal Ecol. 2008, 1, 89–93. [Google Scholar] [CrossRef]
- Mishra, A.; Gond, S.K.; Kumar, A.; Sharma, V.K.; Verma, S.K.; Kharwar, R.N.; Sieber, T.N. Season and tissue type affect fungal endophyte communities of the Indian medicinal plant Tinospora cordifolia more strongly than geographic location. Microb. Ecol. 2012, 64, 388–398. [Google Scholar] [CrossRef]
- Göre, M.; Bucak, C. Geographical and seasonal influences on the distribution of fungal endophytes in Laurus nobilis. For. Pathol. 2007, 37, 281–288. [Google Scholar] [CrossRef]
- Krishnamurthy, Y.L.; Naik, S.B.; Jayaram, S. Fungal communities in herbaceous medicinal plants from the Malnad region, Southern India. Microbes Environ. 2008, 23, 24–28. [Google Scholar] [CrossRef][Green Version]
- Naik, B.S.; Krishnappa, M.; Krishnamurthy, Y. Biodiversity of endophytic fungi from seven herbaceous medicinal plants of Malnad region, Western Ghats, southern India. J. For. Res. 2014, 25, 707–711. [Google Scholar] [CrossRef]
- Ma, L.J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Swett, C.L.; Porter, B.; Fourie, G.; Steenkamp, E.T.; Gordon, T.R.; Wingfield, M.J. Association of the pitch canker pathogen Fusarium circinatum with grass hosts in commercial pine production areas of South Africa. South. For. 2014, 76, 161–166. [Google Scholar] [CrossRef]
- Promputtha, I.; Lumyong, S.; Dhanasekaran, V.; McKenzie, E.H.C.; Hyde, K.D.; Jeewon, R. A phylogenetic evaluation of whether endophytes become saprotrophs at host senescence. Microb. Ecol. 2007, 53, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Redman, R.S.; Dunigan, D.D.; Rodriguez, R.J. Fungal symbiosis from mutualism to parasitism: Who controls the outcome, host or invader? New Phytol. 2001, 151, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Stergiopoulos, I.; Gordon, T.R. Cryptic fungal infections: The hidden agenda of plant pathogens. Front. Plant Sci. 2014, 5, 506. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.J.; Lin, Y.S.; Pan, H.R.; Chung, W.H. Distribution and multiplication of Ralstonia solanacearum strain race 1 biovar 4 in vegetable sweet potato cuttings. J. Phytopathol. 2020, 168, 36–46. [Google Scholar] [CrossRef]
- Swanson, J.K.; Yao, J.; Tans-Kersten, J.; Allen, C. Behavior of Ralstonia solanacearum race 3 biovar 2 during latent and active infection of geranium. Phytopathology 2005, 95, 136–143. [Google Scholar] [CrossRef]
- Malcolm, G.M.; Kuldau, G.A.; Gugino, B.K.; Jiménez-Gasco, M. Hidden host plant associations of soilborne fungal pathogens: An ecological perspective. Phytopathology 2013, 103, 538–544. [Google Scholar] [CrossRef]
- Gilbert, G.S. Evolutionary ecology of plant diseases in natural ecosystems. Annu. Rev. Phytopathol. 2002, 40, 13–43. [Google Scholar] [CrossRef]
- Bennett, R.; Davis, R.M. Method for rapid production of Fusarium oxysporum f. sp. vasinfectum chlamydospores. J. Cotton Sci. 2013, 17, 52–59. [Google Scholar]
- Seifert, K.A.; Cereal, E.; Cereal, E.; Cereal, E. Fuskey: Fusarium Interactive Key; Agriculture & Agri-Food Canada, Research Branch, Eastern Cereal & Oilseed Research Centre: Ottawa, ON, Canada, 1996. [Google Scholar]

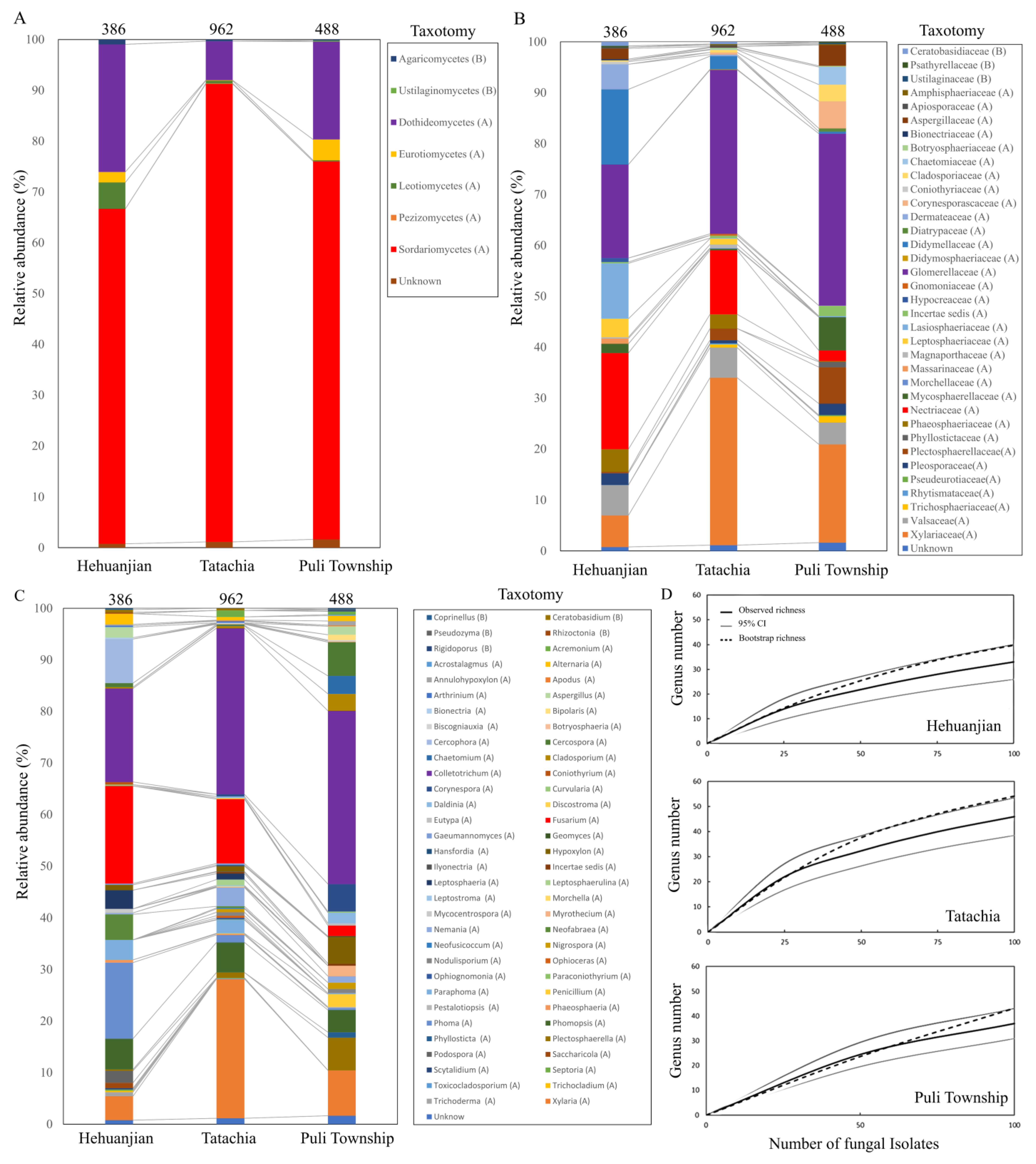
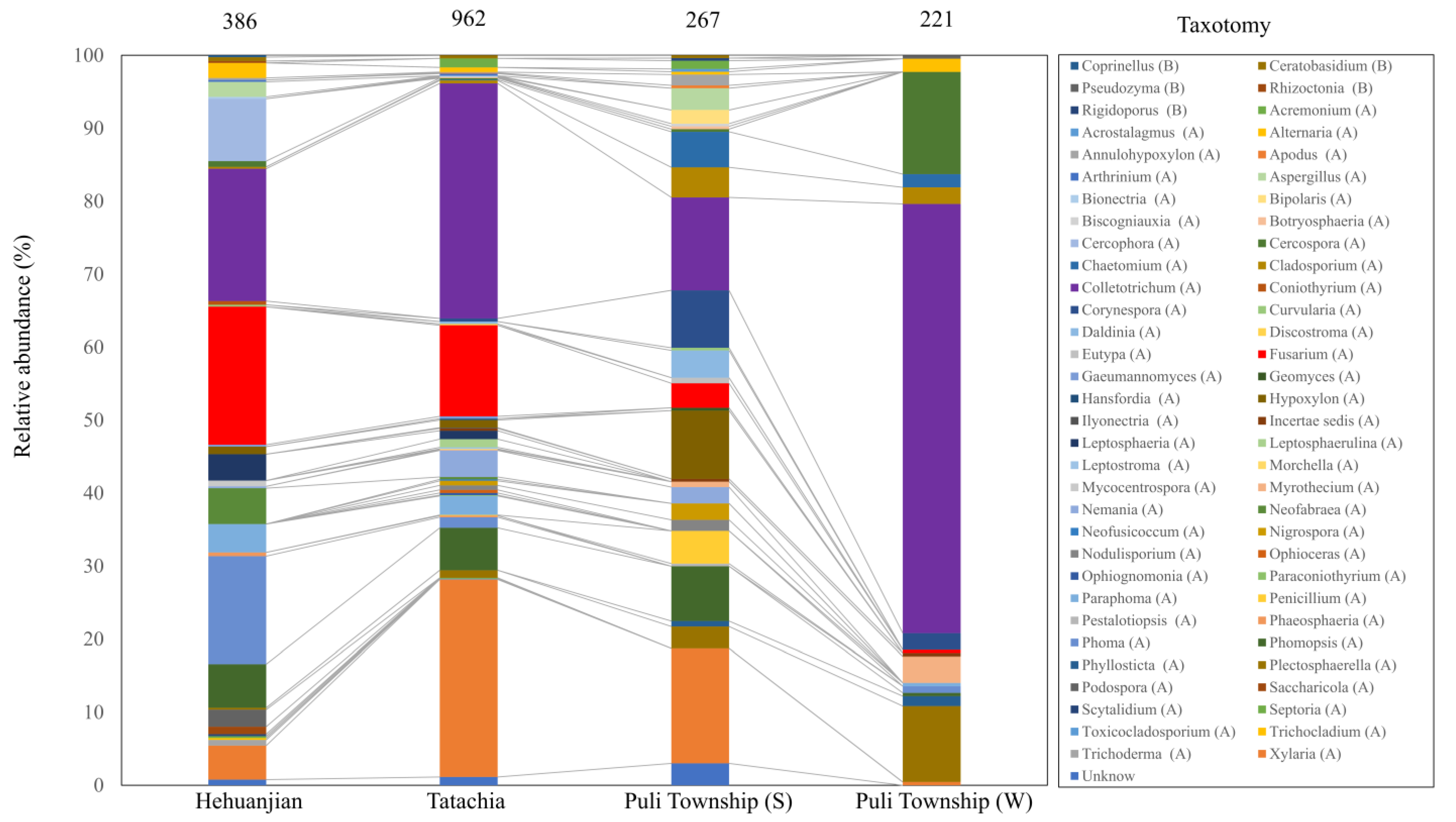
| Fungal Family | Fungal Genus | Number of Isolates (Isolation Frequency (%)) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Hehuanjian | Tatachia | Puli Township | Total | ||||||
| Ascomycota | |||||||||
| Dothideomycetes | |||||||||
| Cladosporiaceae | Cladosporium | 1 | (0.3) | 4 | (0.4) | 16 | (3.3) | 21 | (1.2) |
| Toxicocladosporium | 0 | (0.0) | 1 | (0.1) | 0 | (0.0) | 1 | (0.1) | |
| Coniothyriaceae | Coniothyrium | 2 | (0.5) | 0 | (0.0) | 0 | (0.0) | 2 | (0.1) |
| Corynesporascaceae | Corynespora | 0 | (0.0) | 4 | (0.4) | 26 | (5.4) | 30 | (1.7) |
| Didymellaceae | Leptosphaerulina | 0 | (0.0) | 11 | (1.2) | 0 | (0.0) | 11 | (0.6) |
| Phoma | 57 | (14.9) | 14 | (1.5) | 2 | (0.4) | 73 | (4.0) | |
| Didymosphaeriaceae | Paraconiothyrium | 0 | (0.0) | 1 | (0.1) | 0 | (0.0) | 1 | (0.1) |
| Massarinaceae | Saccharicola | 4 | (1.0) | 0 | (0.0) | 0 | (0.0) | 4 | (0.2) |
| Mycosphaerellaceae | Cercospora | 3 | (0.8) | 2 | (0.2) | 32 | (6.7) | 37 | (2.0) |
| Mycocentrospora | 3 | (0.8) | 0 | (0.0) | 0 | (0.0) | 3 | (0.2) | |
| Septoria | 1 | (0.3) | 1 | (0.1) | 0 | (0.0) | 2 | (0.1) | |
| Phaeosphaeriaceae | Paraphoma | 15 | (3.9) | 25 | (2.6) | 1 | (0.2) | 41 | (2.3) |
| Phaeosphaeria | 2 | (0.5) | 2 | (0.2) | 0 | (0.0) | 4 | (0.2) | |
| Pleosporaceae | Alternaria | 8 | (2.1) | 6 | (0.6) | 5 | (1.0) | 19 | (1.0) |
| Bipolaris | 0 | (0.0) | 0 | (0.0) | 5 | (1.0) | 5 | (0.3) | |
| Curvularia | 1 | (0.3) | 0 | (0.0) | 1 | (0.2) | 2 | (0.1) | |
| Botryosphaeriaceae | Botryosphaeria | 0 | (0.0) | 1 | (0.1) | 1 | (0.2) | 2 | (0.1) |
| Neofusicoccum | 0 | (0.0) | 2 | (0.2) | 0 | (0.0) | 2 | (0.1) | |
| Phyllostictaceae | Phyllosticta | 0 | (0.0) | 0 | (0.0) | 5 | (1.0) | 5 | (0.3) |
| Eurotiomycetes | |||||||||
| Aspergillaceae | Aspergillus | 8 | (2.1) | 0 | (0.0) | 8 | (1.7) | 16 | (0.9) |
| Penicillium | 0 | (0.0) | 1 | (0.1) | 12 | (2.5) | 13 | (0.7) | |
| Leotiomycetes | |||||||||
| Dermateaceae | Neofabraea | 19 | (5.0) | 3 | (0.3) | 0 | (0.0) | 22 | (1.2) |
| Incertae sedis | Scytalidium | 1 | (0.3) | 0 | (0.0) | 0 | (0.0) | 1 | (0.1) |
| Pseudeurotiaceae | Geomyces | 0 | (0.0) | 0 | (0.0) | 1 | (0.2) | 1 | (0.1) |
| Rhytismataceae | Leptostroma | 0 | (0.0) | 2 | (0.2) | 0 | (0.0) | 2 | (0.1) |
| Pezizomycetes | |||||||||
| Morchellaceae | Morchella | 0 | (0.0) | 1 | (0.1) | 0 | (0.0) | 1 | (0.1) |
| Sordariomycetes | |||||||||
| Amphisphaeriaceae | Discostroma | 0 | (0.0) | 2 | (0.2) | 0 | (0.0) | 2 | (0.1) |
| Pestalotiopsis | 0 | (0.0) | 0 | (0.0) | 1 | (0.2) | 1 | (0.1) | |
| Apiosporaceae | Arthrinium | 1 | (0.3) | 3 | (0.3) | 0 | (0.0) | 4 | (0.2) |
| Bionectriaceae | Bionectria | 1 | (0.3) | 1 | (0.1) | 0 | (0.0) | 2 | (0.1) |
| Chaetomiaceae | Chaetomium | 0 | (0.0) | 1 | (0.1) | 17 | (3.5) | 18 | (1.0) |
| Diatrypaceae | Eutypa | 0 | (0.0) | 0 | (0.0) | 2 | (0.4) | 2 | (0.1) |
| Incertae sedis | 0 | (0.0) | 0 | (0.0) | 1 | (0.2) | 1 | (0.1) | |
| Glomerellaceae | Colletotrichum | 70 | (18.3) | 310 | (32.6) | 164 | (34.2) | 544 | (30.0) |
| Gnomoniaceae | Ophiognomonia | 0 | (0.0) | 3 | (0.3) | 0 | (0.0) | 3 | (0.2) |
| Hypocreaceae | Trichoderma | 3 | (0.8) | 0 | (0.0) | 0 | (0.0) | 3 | (0.2) |
| Incertae sedis | Hansfordia | 0 | (0.0) | 2 | (0.2) | 0 | (0.0) | 2 | (0.1) |
| Incertae sedis | 0 | (0.0) | 2 | (0.2) | 0 | (0.0) | 2 | (0.1) | |
| Myrothecium | 0 | (0.0) | 1 | (0.1) | 10 | (2.1) | 11 | (0.6) | |
| Lasiosphaeriaceae | Apodus | 0 | (0.0) | 1 | (0.1) | 1 | (0.2) | 2 | (0.1) |
| Cercophora | 33 | (8.6) | 0 | (0.0) | 0 | (0.0) | 33 | (1.8) | |
| Podospora | 9 | (2.3) | 0 | (0.0) | 0 | (0.0) | 9 | (0.5) | |
| Leptosphaeriaceae | Leptosphaeria | 14 | (3.7) | 11 | (1.2) | 0 | (0.0) | 25 | (1.4) |
| Magnaporthaceae | Gaeumannomyces | 1 | (0.3) | 3 | (0.3) | 0 | (0.0) | 4 | (0.2) |
| Ophioceras | 0 | (0.0) | 4 | (0.4) | 0 | (0.0) | 4 | (0.2) | |
| Nectriaceae | Fusarium | 73 | (19.1) | 120 | (12.6) | 10 | (2.1) | 203 | (11.2) |
| Ilyonectria | 0 | (0.0) | 1 | (0.1) | 0 | (0.0) | 1 | (0.1) | |
| Plectosphaerellaceae | Acremonium | 0 | (0.0) | 12 | (1.3) | 3 | (0.6) | 15 | (0.8) |
| Acrostalagmus | 0 | (0.0) | 0 | (0.0) | 1 | (0.2) | 1 | (0.1) | |
| Plectosphaerella | 1 | (0.3) | 10 | (1.1) | 31 | (6.5) | 42 | (2.3) | |
| Trichosphaeriaceae | Nigrospora | 0 | (0.0) | 6 | (0.6) | 6 | (1.3) | 12 | (0.7) |
| Valsaceae | Incertae sedis | 0 | (0.0) | 1 | (0.1) | 0 | (0.0) | 1 | (0.1) |
| Phomopsis | 23 | (6.0) | 56 | (5.9) | 21 | (4.4) | 100 | (5.5) | |
| Xylariaceae | Annulohypoxylon | 1 | (0.3) | 1 | (0.1) | 4 | (0.8) | 6 | (0.3) |
| Biscogniauxia | 0 | (0.0) | 1 | (0.1) | 1 | (0.2) | 2 | (0.1) | |
| Daldinia | 0 | (0.0) | 3 | (0.3) | 10 | (2.1) | 13 | (0.7) | |
| Hypoxylon | 4 | (1.0) | 10 | (1.1) | 25 | (5.2) | 39 | (2.1) | |
| Incertae sedis | 0 | (0.0) | 0 | (0.0) | 1 | (0.2) | 1 | (0.1) | |
| Nemania | 1 | (0.3) | 35 | (3.7) | 6 | (1.3) | 42 | (2.3) | |
| Nodulisporium | 0 | (0.0) | 6 | (0.6) | 4 | (0.8) | 10 | (0.6) | |
| Xylaria | 18 | (4.7) | 260 | (27.3) | 43 | (9.0) | 321 | (17.7) | |
| Incertae sedis | |||||||||
| Incertae sedis | Trichocladium | 1 | (0.3) | 0 | (0.0) | 0 | (0.0) | 1 | (0.1) |
| Basidiomycota | |||||||||
| Agaricomycetes | |||||||||
| Ceratobasidiaceae | Ceratobasidium | 2 | (0.5) | 4 | (0.4) | 0 | (0.0) | 6 | (0.3) |
| Rhizoctonia | 1 | (0.3) | 0 | (0.0) | 0 | (0.0) | 1 | (0.1) | |
| Incertae sedis | Rigidoporus | 0 | (0.0) | 0 | (0.0) | 1 | (0.2) | 1 | (0.1) |
| Psathyrellaceae | Coprinellus | 1 | (0.3) | 0 | (0.0) | 1 | (0.2) | 2 | (0.1) |
| Ustilaginomycetes | |||||||||
| Ustilaginaceae | Pseudozyma | 0 | (0.0) | 0 | (0.0) | 1 | (0.2) | 1 | (0.1) |
| Unknown | 3 | 11 | 8 | 22 | |||||
| Total | 386 | 962 | 488 | 1836 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-J.; Chen, H.-J.; Chung, W.-H. Endophytic Fungal Diversity in Cirsium kawakamii from Taiwan. J. Fungi 2023, 9, 1076. https://doi.org/10.3390/jof9111076
Chen Y-J, Chen H-J, Chung W-H. Endophytic Fungal Diversity in Cirsium kawakamii from Taiwan. Journal of Fungi. 2023; 9(11):1076. https://doi.org/10.3390/jof9111076
Chicago/Turabian StyleChen, Yi-Jeng, Hui-Juan Chen, and Wen-Hsin Chung. 2023. "Endophytic Fungal Diversity in Cirsium kawakamii from Taiwan" Journal of Fungi 9, no. 11: 1076. https://doi.org/10.3390/jof9111076
APA StyleChen, Y.-J., Chen, H.-J., & Chung, W.-H. (2023). Endophytic Fungal Diversity in Cirsium kawakamii from Taiwan. Journal of Fungi, 9(11), 1076. https://doi.org/10.3390/jof9111076





