Regulation of Fungal Morphogenesis and Pathogenicity of Aspergillus flavus by Hexokinase AfHxk1 through Its Domain Hexokinase_2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Growth Conditions
2.2. Bioinformatics Analysis
2.3. Construction of Mutant Fungal Strains
2.4. Real-Time Quantitative Reverse Transcription PCR
2.5. Phenotypic and Carbon Source Analysis
2.6. Aflatoxin Analysis
2.7. Kernel Colonization Assays
2.8. The Insect Infection Model
2.9. Stress Assays
2.10. Statistical Analysis
3. Result
3.1. The Bioinformatic Analysis of AfHxk1 and the Construction of Fungal Mutants
3.2. AfHxk1 Promotes Fungal Capacity to Utilize Various Carbohydrates
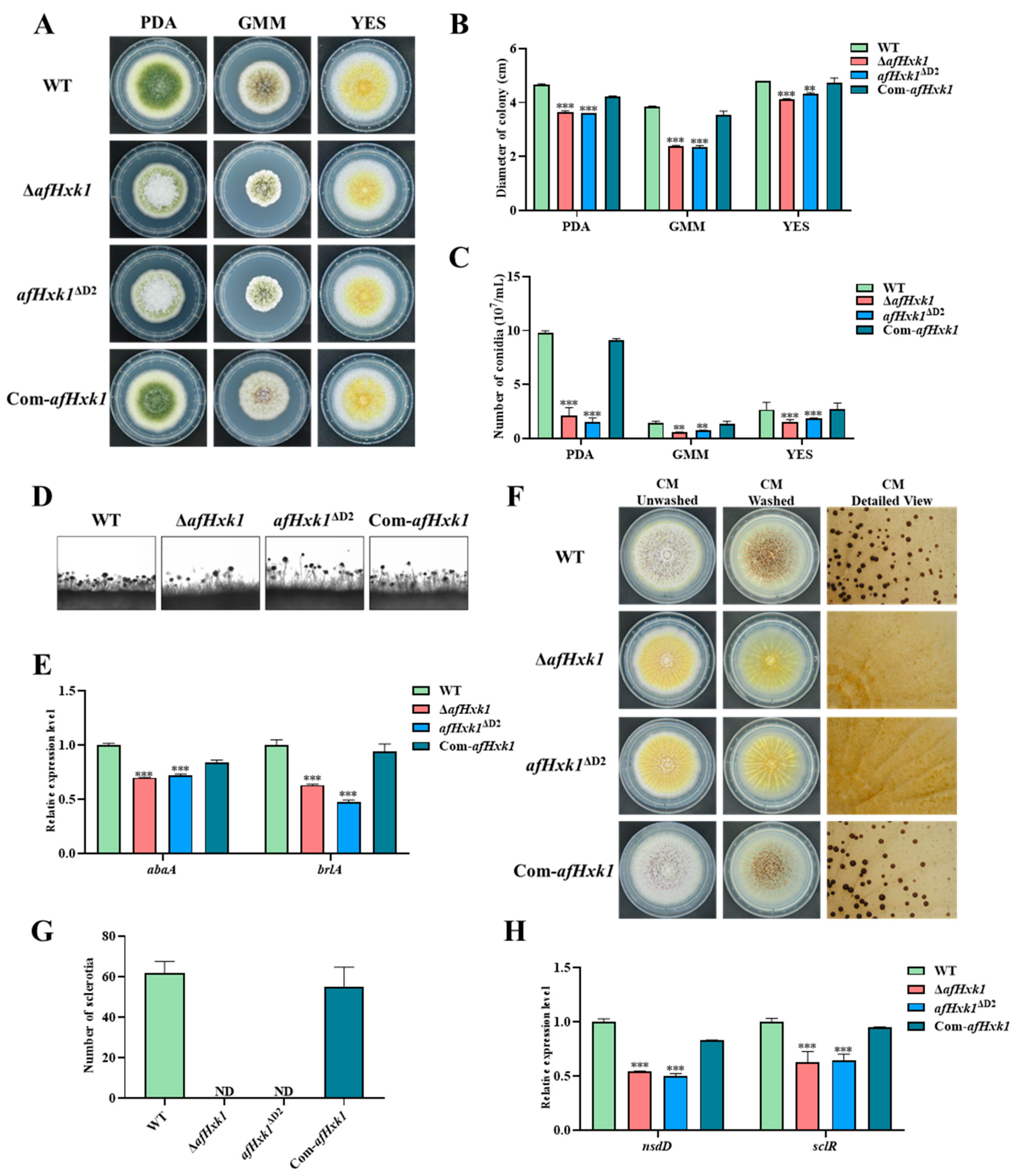
3.3. AfHxk1 Is Deeply Involved in the Morphogenesis of A. flavus
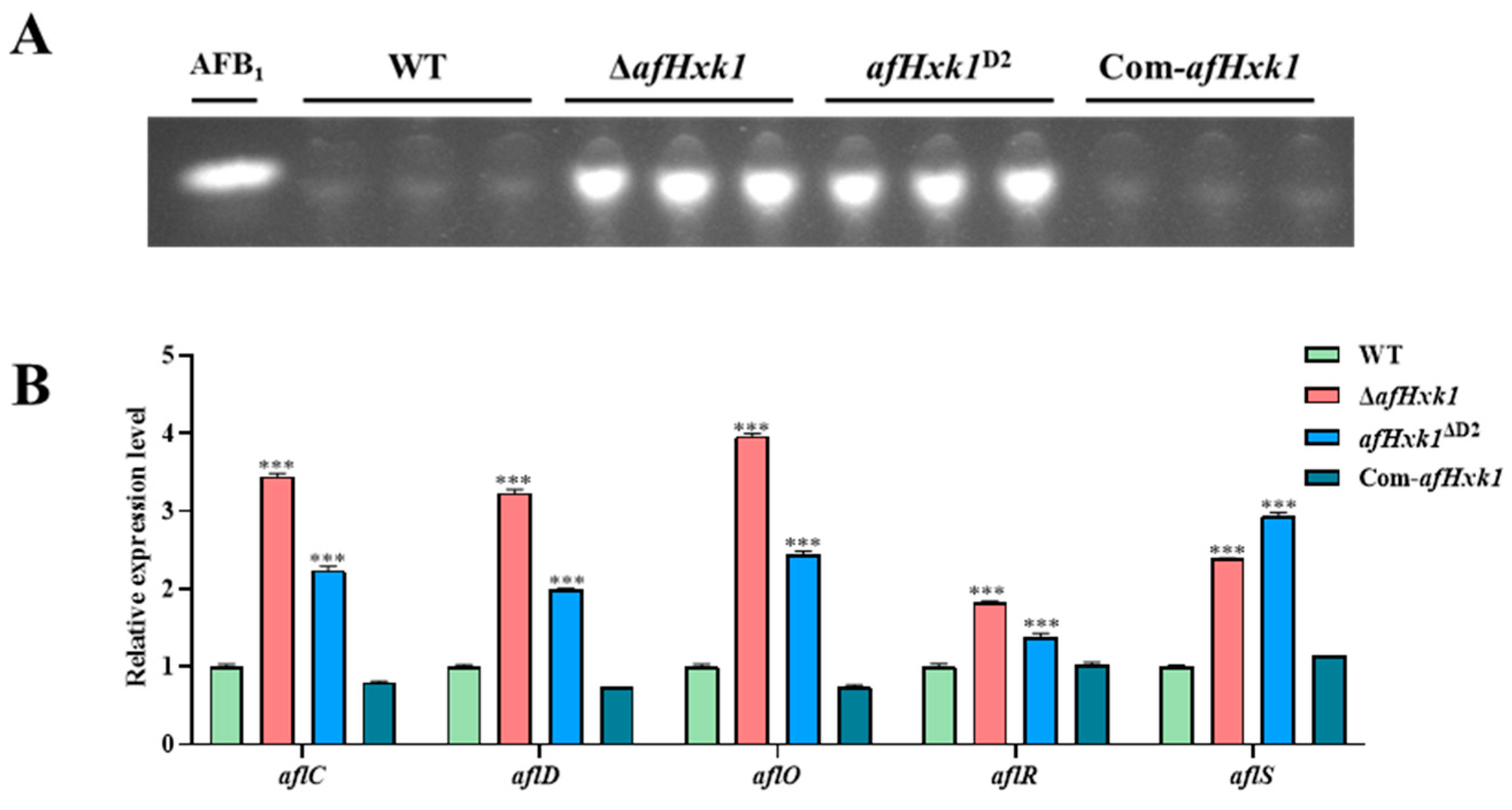
3.4. AfHxk1 Negatively Regulates Aflatoxin Biosynthesis
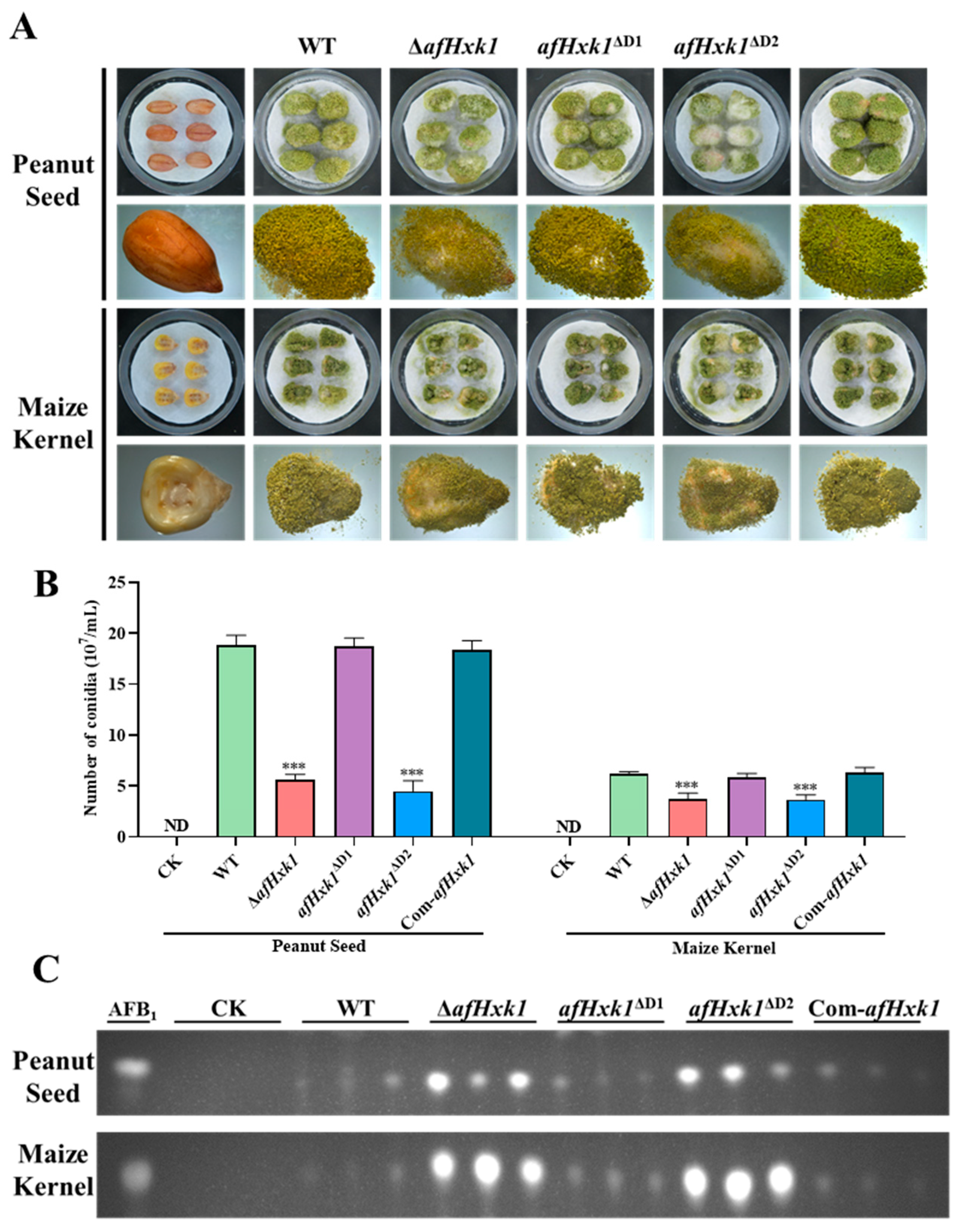
3.5. AfHxk1 Is Involved in the Colonization of A. flavus on Crop Kernels
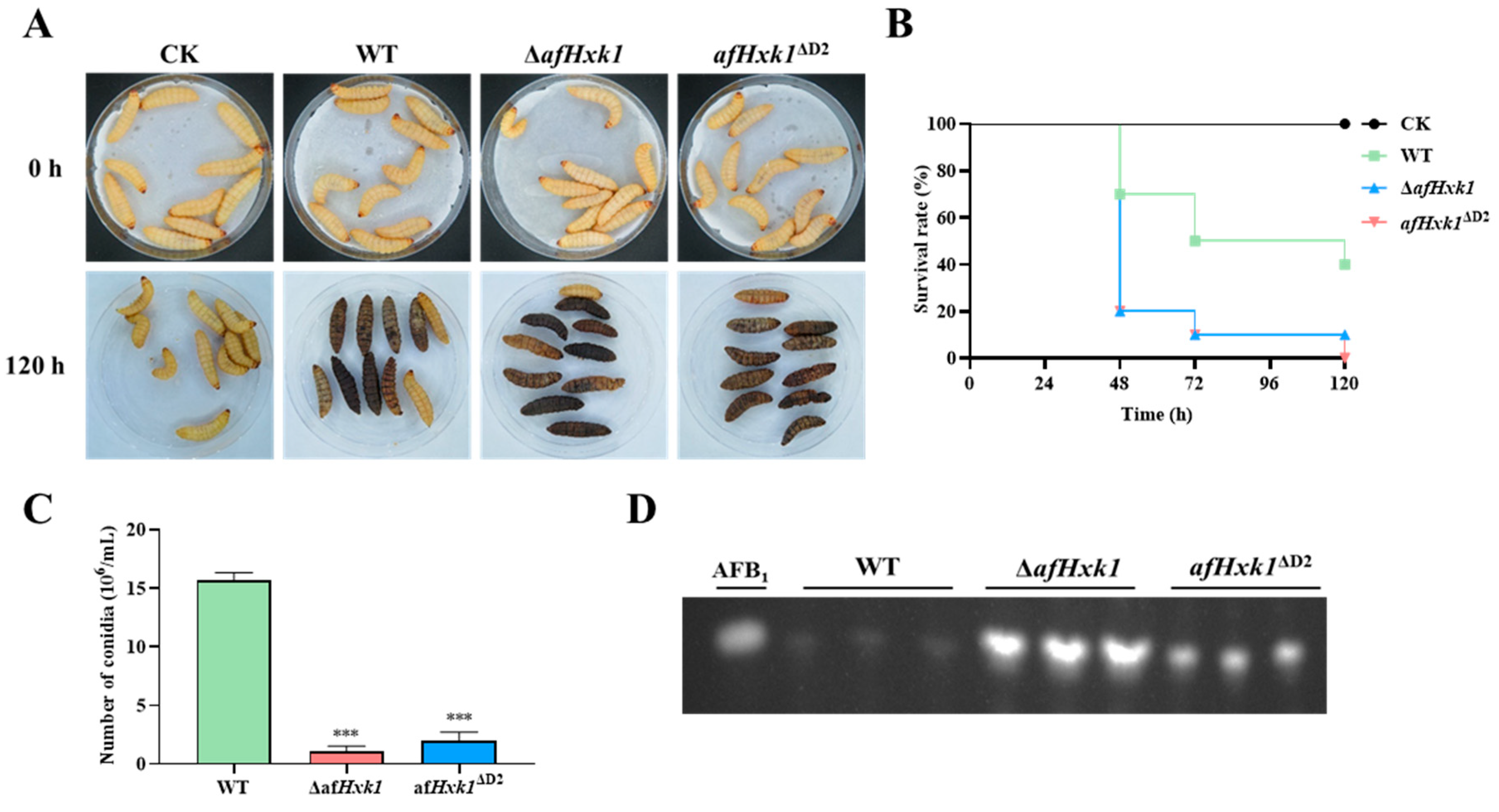
3.6. AfHxk1 Is Involved in Fungal Pathogenicity to Animal
3.7. AfHxk1 Positively Regulates Fungal Spore Germination
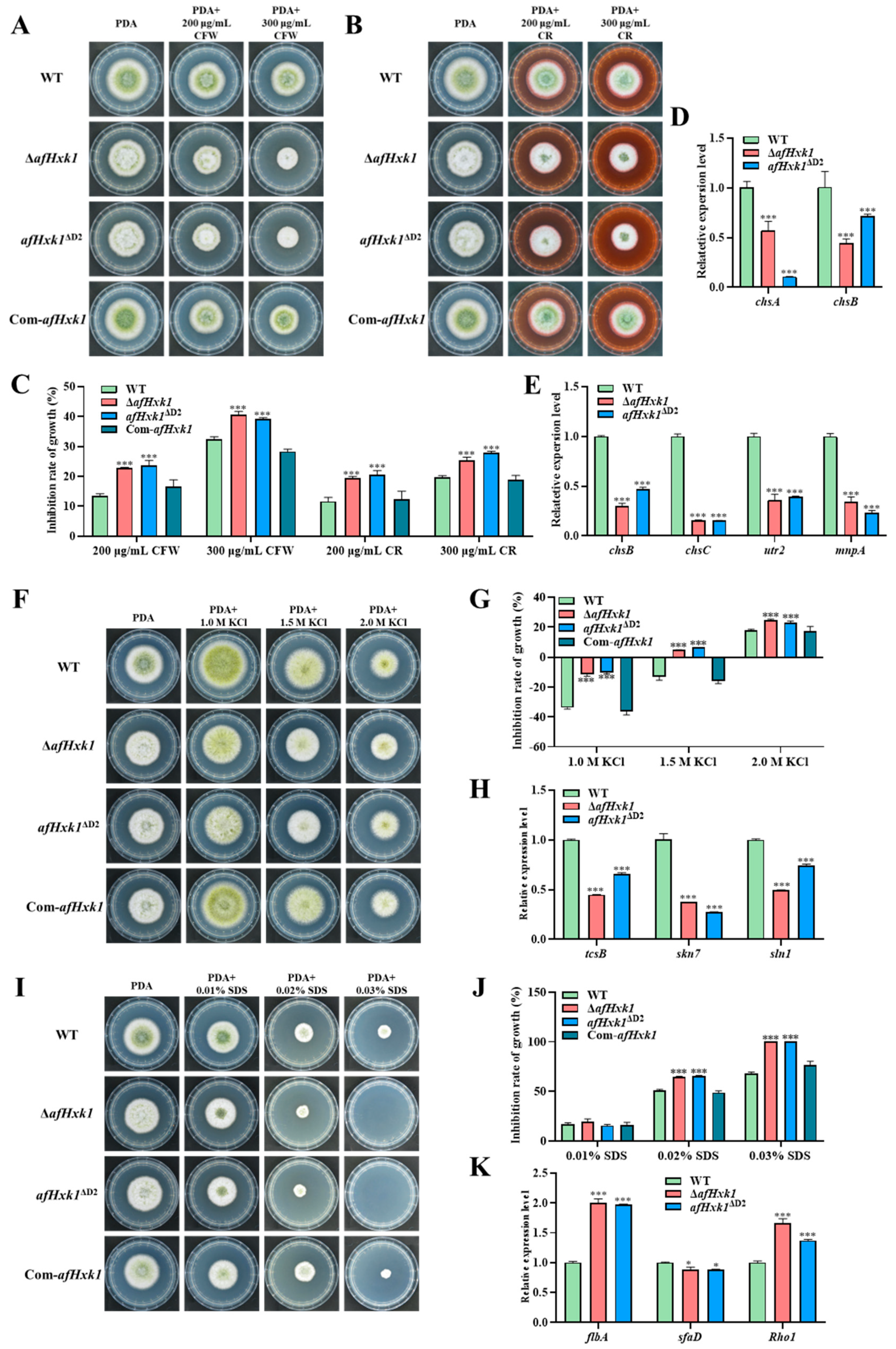
3.8. AfHxk1 Plays Important Role in Fungal Susceptibility Responding to Stresses
4. Discussion
4.1. AfHxk1 Enhances Fungal Colonial Growth and Conidiation
4.2. AfHxk1 Is Indispensable for the Formation of Sclerotia
4.3. AfHxk1 Downregulates the Biosynthesis of AFB1
4.4. AfHxk1 Plays a Vital Role in Fungal Pathogenicity
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cary, J.W.; Gilbert, M.K.; Lebar, M.D.; Majumdar, R.; Calvo, A.M. Secondary metabolites: More than just aflatoxins. Food Saf. 2018, 6, 7–32. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Mahato, D.K.; Kamle, M.; Mohanta, T.K.; Kang, S.G. Aflatoxins: A global concern for food safety, human health and their management. Front. Microbiol. 2016, 7, 2170. [Google Scholar] [CrossRef]
- Bhatnagar-Mathur, P.; Sunkara, S.; Bhatnagar-Panwar, M.; Waliyar, F.; Sharma, K.K. Biotechnological advances for combating Aspergillus flavus and aflatoxin contamination in crops. Plant Sci. 2015, 234, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, F. Global burden of aflatoxin-induced hepatocellular carcinoma: A risk assessment. Environ. Health Perspect. 2010, 118, 818–824. [Google Scholar] [CrossRef] [PubMed]
- Marchese, S.; Polo, A.; Ariano, A.; Velotto, S.; Costantini, S.; Severino, L. Aflatoxin B1 and M1: Biological properties and their involvement in cancer development. Toxins 2018, 10, 214. [Google Scholar] [CrossRef]
- Pickova, D.; Ostry, V.; Malir, F. A recent overview of producers and important dietary sources of aflatoxins. Toxins 2021, 13, 186. [Google Scholar] [CrossRef]
- Meissonnier, G.M.; Laffitte, J.; Loiseau, N.; Benoit, E.; Raymond, I.; Pinton, P.; Cossalter, A.M.; Bertin, G.; Oswald, I.P.; Galtier, P. Selective impairment of drug-metabolizing enzymes in pig liver during subchronic dietary exposure to aflatoxin B1. Food Chem. Toxicol. 2007, 45, 2145–2154. [Google Scholar] [CrossRef]
- Afshar, P.; Shokrzadeh, M.; Raeisi, S.N.; Ghorbani-HasanSaraei, A.; Nasiraii, L.R. Aflatoxins biodetoxification strategies based on probiotic bacteria. Toxicon 2020, 178, 50–58. [Google Scholar] [CrossRef]
- Intanoo, M.; Kongkeitkajorn, M.B.; Suriyasathaporn, W.; Phasuk, Y.; Bernard, J.K.; Pattarajinda, V. Effect of supplemental kluyveromyces marxianus and pichia kudriavzevii on aflatoxin M1 excretion in Milk of lactating dairy cows. Animals 2020, 10, 709. [Google Scholar] [CrossRef]
- Tao, F.; Zhao, K.; Zhao, Q.; Xiang, F.; Han, G. A novel site-specific integration system for genetic modification of Aspergillus flavus. G3 Genes Genomes Genet. 2020, 10, 605–611. [Google Scholar] [CrossRef]
- Karlovsky, P.; Suman, M.; Berthiller, F.; De Meester, J.; Eisenbrand, G.; Perrin, I.; Oswald, I.P.; Speijers, G.; Chiodini, A.; Recker, T.; et al. Impact of food processing and detoxification treatments on mycotoxin contamination. Mycotoxin Res. 2016, 32, 179–205. [Google Scholar] [CrossRef]
- Cho, Y.H.; Yoo, S.D.; Sheen, J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 2006, 127, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Caceres, I.; Khoury, A.A.; Khoury, R.E.; Lorber, S.; Oswald, I.P.; Khoury, A.E.; Atoui, A.; Puel, O.; Bailly, J.D. Aflatoxin biosynthesis and genetic regulation: A review. Toxins 2020, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Amaike, S.; Keller, N.P. Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Eukaryote Cell 2009, 8, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.K.; Scharfenstein, L.L.; Li, P.; Ehrlich, K.C. Aspergillus flavus VelB acts distinctly from VeA in conidiation and may coordinate with FluG to modulate sclerotial production. Fungal Genet. Biol. 2013, 58–59, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Bazán, V.; Jaimes-Arroyo, R.; Sánchez, O.; Lara-Rojas, F.; Aguirre, J. SakA and MpkC stress MAPKs show opposite and common functions during stress responses and development in Aspergillus nidulans. Front. Microbiol. 2018, 9, 2518. [Google Scholar] [CrossRef]
- Eom, T.J.; Moon, H.; Yu, J.H.; Park, H.S. Characterization of the velvet regulators in Aspergillus flavus. J. Microbiol. 2018, 56, 893–901. [Google Scholar] [CrossRef] [PubMed]
- Leiter, É.; Emri, T.; Pákozdi, K.; Hornok, L.; Pócsi, I. The impact of bZIP Atf1 ortholog global regulators in fungi. Appl. Microbiol. Biotechnol. 2021, 105, 5769–5783. [Google Scholar] [CrossRef]
- Xie, R.; Yang, K.; Tumukunde, E.; Guo, Z.; Zhang, B.; Liu, Y.; Zhang, Z.; Yuan, J.; Wang, S. Regulator of G protein signaling contributes to the development and aflatoxin biosynthesis in Aspergillus flavus through the regulation of Gα activity. Appl. Environ. Microbiol. 2022, 88, e0024422. [Google Scholar] [CrossRef]
- Wu, M.Y.; Mead, M.E.; Kim, S.C.; Rokas, A.; Yu, J.H. WetA bridges cellular and chemical development in Aspergillus flavus. PLoS ONE 2017, 12, e0179571. [Google Scholar] [CrossRef]
- Wilson, J.E. Isozymes of mammalian hexokinase: Structure, subcellular localization and metabolic function. J. Exp. Biol. 2003, 206, 2049–2057. [Google Scholar] [CrossRef]
- Ciscato, F.; Ferrone, L.; Masgras, I.; Laquatra, C.; Rasola, A. Hexokinase 2 in cancer: A prima donna playing multiple characters. Int. J. Mol. Sci. 2021, 22, 4716. [Google Scholar] [CrossRef]
- Fleck, C.B.; Brock, M. Aspergillus fumigatus catalytic glucokinase and hexokinase: Expression analysis and importance for germination, growth, and conidiation. Eukaryote Cell 2010, 9, 1120–1135. [Google Scholar] [CrossRef]
- Su, Y.; Chen, Y.; Che, Y.; Liu, G. The function of a hexokinase encoding gene Achka in Acremonium chrysogenum. Mycosystema 2017, 36, 323–331. [Google Scholar]
- Rui, O.; Hahn, M. The Botrytis cinerea hexokinase, Hxk1, but not the glucokinase, Glk1, is required for normal growth and sugar metabolism, and for pathogenicity on fruits. Microbiol. Read. 2007, 153 Pt 8, 2791–2802. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, B.; Zhang, Y.; Jia, X.; Zhou, M. Hexokinase plays a critical role in deoxynivalenol (DON) production and fungal development in Fusarium graminearum. Mol. Plant Pathol. 2016, 17, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Pan, X.; Zhang, M.; Liu, Y.; Huang, C.; Li, Y.; Hao, L.; Wang, S. Set2 Family Regulates Mycotoxin Metabolism and Virulence via H3K36 Methylation in Pathogenic Fungus Aspergillus Flavus. Virulence 2022, 13, 1358–1378. [Google Scholar] [CrossRef]
- Szewczyk, E.; Nayak, T.; Oakley, C.E.; Edgerton, H.; Xiong, Y.; Taheri-Talesh, N.; Osmani, S.A.; Oakley, B.R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2006, 1, 3111–3120. [Google Scholar] [CrossRef]
- Yang, K.; Liang, L.; Ran, F.; Liu, Y.; Li, Z.; Lan, H.; Gao, P.; Zhuang, Z.; Zhang, F.; Nie, X.; et al. The DmtA methyltransferase contributes to Aspergillus flavus conidiation, sclerotial production, aflatoxin biosynthesis and virulence. Sci. Rep. 2016, 6, 23259. [Google Scholar] [CrossRef]
- Nie, X.; Yu, S.; Qiu, M.; Wang, X.; Wang, Y.; Bai, Y.; Zhang, F.; Wang, S. Aspergillus flavus SUMO contributes to fungal virulence and toxin attributes. J. Agric. Food Chem. 2016, 64, 6772–6782. [Google Scholar] [CrossRef]
- Lan, H.; Wu, L.; Fan, K.; Sun, R.; Yang, G.; Zhang, F.; Yang, K.; Lin, X.; Chen, Y.; Tian, J.; et al. Set3 is required for asexual development, aflatoxin niosynthesis, and fungal birulence in. Front. Microbiol. 2019, 10, 530. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.E.; Romano, A.H. Evidence against necessary phosphorylation during hexose transport in Aspergillus nidulans. J. Bacteriol. 1969, 100, 1198–1203. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, G.; Zhang, D.; Liu, Y.; Li, Y.; Lin, G.; Guo, Z.; Wang, S.; Zhuang, Z. The PHD Transcription Factor Rum1 Regulates Morphogenesis and Aflatoxin Biosynthesis in. Toxins 2018, 10, 301. [Google Scholar] [CrossRef] [PubMed]
- Ichinomiya, M.; Horiuchi, H.; Ohta, A. Different functions of the class I and class II chitin synthase genes, chsC and chsA, are revealed by repression of chsB expression in Aspergillus nidulans. Curr. Genet. 2002, 42, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Klis, F.M.; Sosinska, G.J.; de Groot, P.W.; Brul, S. Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res. 2009, 9, 1013–1028. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.Y.; Kim, H.; Han, D.M.; Jahng, K.Y.; Chae, K.S. Expression of the mnpA gene that encodes the mannoprotein of Aspergillus nidulans is dependent on fadA and flbA as well as veA. Fungal. Genet. Biol. 2003, 38, 228–236. [Google Scholar] [CrossRef]
- Furukawa, K.; Katsuno, Y.; Urao, T.; Yabe, T.; Yamada-Okabe, T.; Yamada-Okabe, H.; Yamagata, Y.; Abe, K.; Nakajima, T. Isolation and functional analysis of a gene, tcsB, encoding a transmembrane hybrid-type histidine kinase from Aspergillus nidulans. Appl. Env. Microbiol. 2002, 68, 5304–5310. [Google Scholar] [CrossRef]
- Li, S.; Ault, A.; Malone, C.L.; Raitt, D.; Dean, S.; Johnston, L.H.; Deschenes, R.J.; Fassler, J.S. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 1998, 17, 6952–6962. [Google Scholar] [CrossRef]
- Levin, D.E. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005, 69, 262–291. [Google Scholar] [CrossRef]
- Park, H.S.; Kim, M.J.; Yu, J.H.; Shin, K.S. Heterotrimeric G-protein signalers and RGSs in Aspergillus fumigatus. Pathogens 2020, 9, 902. [Google Scholar] [CrossRef]
- Coca, M.A.; Damsz, B.; Yun, D.-J.; Hasegawa, P.M.; Bressan, R.A.; Narasimhan, M.L. Heterotrimeric G-proteins of a filamentous fungus regulate cell wall composition and susceptibility to a plant PR-5 protein. Plant J. 2000, 22, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.; De La Cera, T.; Herrero, P.; Moreno, F. The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem. J. 2001, 355 Pt 3, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Vega, M.; Riera, A.; Fernández-Cid, A.; Herrero, P.; Moreno, F. Hexokinase 2 is an intracellular glucose sensor of yeast cells that maintains the structure and activity of Mig1 protein repressor complex. J. Biol. Chem. 2016, 291, 7267–7285. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.H.; Paul, S.; Natarajan, K.; Ghosh, S. N-Acetylglucosamine kinase, Hxk1 is a multifaceted metabolic enzyme in model pathogenic yeast Candida Albicans. Microbiol. Res. 2022, 263, 127146. [Google Scholar] [CrossRef]
- Kim, H.-M.; Kwon, H.; Kim, K.; Lee, S.-E. Antifungal and antiaflatoxigenic activities of 1,8-Cineole and T-Cinnamaldehyde on Aspergillus Flavus. Appl. Sci. 2018, 8, 1655. [Google Scholar] [CrossRef]
- Wijnants, S.; Riedelberger, M.; Penninger, P.; Kuchler, K.; Van Dijck, P. Sugar Phosphorylation Controls Carbon Source Utilization and Virulence of. Front. Microbiol. 2020, 11, 1274. [Google Scholar] [CrossRef]
- Zhang, A.X.; Mouhoumed, A.Z.; Tong, S.M.; Ying, S.H.; Feng, M.G. BrlA and AbaA govern virulence-required dimorphic wwitch, conidiation, and pathogenicity in a fungal insect pathogen. mSystems 2019, 4, e00140-19. [Google Scholar] [CrossRef]
- Calvo, A.M.; Cary, J.W. Association of fungal secondary metabolism and sclerotial biology. Front. Microbiol. 2015, 6, 62. [Google Scholar] [CrossRef]
- Wang, P.; Xu, J.; Chang, P.K.; Liu, Z.; Kong, Q. New insights of transcriptional regulator AflR in Aspergillus flavus physiology. Microbiol. Spectr. 2022, 10, e0079121. [Google Scholar] [CrossRef]
- Abdel-Hadi, A.; Schmidt-Heydt, M.; Parra, R.; Geisen, R.; Magan, N. A systems approach to model the relationship between aflatoxin gene cluster expression, environmental factors, growth and toxin production by Aspergillus flavus. J. R. Soc. Interface 2012, 9, 757–767. [Google Scholar] [CrossRef]
- Yu, J. Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins 2012, 4, 1024–1057. [Google Scholar] [CrossRef]
- Hopke, A.; Brown, A.J.P.; Hall, R.A.; Wheeler, R.T. Dynamic fungal cell wall architecture in stress adaptation and immune evasion. Trends Microbiol. 2018, 26, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Hwan, J.; Chae, K.; Sung, S. Presence of a Mannoprotein, MnpAp, in the Hyphal cell wall of Aspergillus Nidulans. Mycologia 2004, 96, 52–56. [Google Scholar] [CrossRef]
- Schroeder, L.; Ikui, A.E. Tryptophan confers resistance to SDS-associated cell membrane stress in Saccharomyces cerevisiae. PLoS ONE 2019, 14, e0199484. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.; Wu, D.; Yang, S.; Fu, W.; Ma, D.; Yao, Y.; Lin, H.; Yuan, J.; Yang, Y.; Zhuang, Z. Regulation of Fungal Morphogenesis and Pathogenicity of Aspergillus flavus by Hexokinase AfHxk1 through Its Domain Hexokinase_2. J. Fungi 2023, 9, 1077. https://doi.org/10.3390/jof9111077
Huang Z, Wu D, Yang S, Fu W, Ma D, Yao Y, Lin H, Yuan J, Yang Y, Zhuang Z. Regulation of Fungal Morphogenesis and Pathogenicity of Aspergillus flavus by Hexokinase AfHxk1 through Its Domain Hexokinase_2. Journal of Fungi. 2023; 9(11):1077. https://doi.org/10.3390/jof9111077
Chicago/Turabian StyleHuang, Zongting, Dandan Wu, Sile Yang, Wangzhuo Fu, Dongmei Ma, Yanfang Yao, Hong Lin, Jun Yuan, Yanling Yang, and Zhenhong Zhuang. 2023. "Regulation of Fungal Morphogenesis and Pathogenicity of Aspergillus flavus by Hexokinase AfHxk1 through Its Domain Hexokinase_2" Journal of Fungi 9, no. 11: 1077. https://doi.org/10.3390/jof9111077
APA StyleHuang, Z., Wu, D., Yang, S., Fu, W., Ma, D., Yao, Y., Lin, H., Yuan, J., Yang, Y., & Zhuang, Z. (2023). Regulation of Fungal Morphogenesis and Pathogenicity of Aspergillus flavus by Hexokinase AfHxk1 through Its Domain Hexokinase_2. Journal of Fungi, 9(11), 1077. https://doi.org/10.3390/jof9111077



