Insights into Some Onygenalean Fungi from Freshwater Sediments in Spain and Description of Novel Taxa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and Isolates
2.2. Phenotypic Study
2.3. DNA Extraction, Sequencing, and Phylogenetic Analysis
2.4. Phylogeny and Geographical Distribution of the Related Environmental Sequences
3. Results
3.1. Phylogeny



3.2. Distribution of Species in the Rivers and Streams Sampled
3.3. Global Biogeography and Ecology of the Novel Taxa
3.4. Taxonomy
- Malbrancheaceae (Figure 2, Clade I)
- Malbranchea echinulata Torres-Garcia, Cano & Gené, sp. nov. Figure 5.
- Malbranchea irregularis Torres-Garcia, Gené & Dania García, sp. nov. Figure 6.

- Malbranchea sinuata Torres-Garcia, Gené & Cano, sp. nov. Figure 7.

- Neoarthropsidaceae Torres-Garcia & Gené, fam. Nov. (Figure 2, Clade IV).

- Albidomyces Torres-Garcia, Dania García, Cano & Gené, gen. nov.
- Albidomyces albicans (Apinis) Torres-Garcia, Dania García, Cano & Gené, comb. nov.
- Neoarthropsis Torres-Garcia, Cano & Gené, gen. nov.
- Neoarthropsis hispanica (Ulfig, Gené & Guarro) Torres-Garcia, Cano & Gené, comb. nov.
- Neoarthropsis sexualis Torres-Garcia, Cano & Gené, sp. Nov. Figure 8
- Onygenales, Incertae sedis (Figure 2, Clade X).
- Pseudoamaurascopsis Torres-Garcia, Dania García & Gené, gen. nov.

- Pseudoamaurascopsis spiralis Torres-Garcia, Dania García & Gené, sp. nov. Figure 9.

4. Discussion
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grossart, H.P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 2019, 17, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Lepère, C.; Domaizon, I.; Humbert, J.F.; Jardillier, L.; Hugoni, M.; Debroas, D. Diversity, spatial distribution and activity of fungi in freshwater ecosystems. PeerJ 2019, 7, e6247. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, J.; Gao, G.; Bartlam, M.G.; Wang, Y. Distribution and diversity of fungi in freshwater sediments on a river catchment scale. Front. Microbiol. 2015, 6, 329. [Google Scholar] [CrossRef]
- Debeljak, P.; Baltar, F. Fungal diversity and community composition across ecosystems. J. Fungi 2023, 9, 510. [Google Scholar] [CrossRef] [PubMed]
- Pascoal, C.; Cassio, F. Contribution of fungi and bacteria to leaf litter decomposition in a polluted river. Appl. Environ. Microbiol. 2004, 70, 5266–5273. [Google Scholar] [CrossRef]
- Gessner, M.O.; Gulis, V.; Kuehn, K.A.; Chauvet, E.; Suberkropp, K. Fungal decomposers of plant litter in aquatic ecosystems. In Environmental and Microbial Relationships; Kubicek, C., Druzhinina, I., Eds.; Springer: Heidelberg/Berlin, Germany, 2007; Volume 4, pp. 301–324. [Google Scholar] [CrossRef]
- Gulis, V.; Su, R.; Kuehn, K. Fungal decomposers in freshwater environments. In The Structure and Function of Aquatic Microbial Communities. Advances in Environmental Microbiology; Hurst, C., Ed.; Springer International Publishing: Cham, Switzerland, 2019; Volume 7, pp. 121–155. [Google Scholar] [CrossRef]
- Kandemir, H.; Dukik, K.; de Melo Teixeira, M.; Stielow, J.B.; Delma, F.Z.; Al-Hatmi, A.; Ahmed, S.A.; Ilkit, M.; De Hoog, G.S. Phylogenetic and ecological reevaluation of the order Onygenales. Fungal Divers. 2022, 115, 1–72. [Google Scholar] [CrossRef]
- Currah, R.S. Taxonomy of the Onygenales: Arthrodermataceae, Gymnoascaceae, Myxotrichaceae and Onygenaceae. Mycotaxon 1985, 24, 1–216. [Google Scholar]
- Currah, R.S. Peridial morphology and evolution in the prototunicate ascomycetes. In Ascomycete Systematics: Problems and Perspectives in the Nineties; Hawksworth, D.L., Ed.; Plenum Press: New York, NY, USA, 1994; Volume 269, pp. 281–293. [Google Scholar]
- Hubálek, Z. Keratinophilic fungi associated with free-living mammals and birds. Rev. Iberoam. Micol. 2000, 17, 93–103. [Google Scholar]
- Doveri, F.; Pecchia, S.; Vergara, M.; Sarrocco, S.; Vannacci, G. A comparative study of Neogymnomyces virgineus, a new keratinolytic species from dung, and its relationships with the Onygenales. Fungal Divers. 2012, 52, 13–34. [Google Scholar] [CrossRef]
- Hubka, V.; Dobiasova, S.; Lyskova, P.; Mallátová, N.; Chlebkova, J.; Skorepova, M.; Kubatova, A.; Dobias, R.; Chudickova, M.; Kolarik, M. Auxarthron ostraviense sp. nov., and A. umbrinum associated with non-dermatophytic onychomycosis. Med. Mycol. J. 2013, 51, 614–624. [Google Scholar] [CrossRef]
- Dukik, K.; Muñoz, J.F.; Jiang, Y.; Feng, P.; Sigler, L.; Stielow, J.B.; Freeke, J.; Jamalian, A.; Van den Ende, B.G.; McEwen, J.G.; et al. Novel taxa of thermally dimorphic systemic pathogens in the Ajellomycetaceae (Onygenales). Mycoses 2017, 60, 296–309. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Shouche, Y.S. Diversity of onygenalean fungi in keratin-rich habitats of Maharashtra (India) and description of three novel taxa. Mycopathologia 2020, 185, 67–85. [Google Scholar] [CrossRef] [PubMed]
- Wynns, A.A. Convergent evolution of highly reduced fruiting bodies in Pezizomycotina suggests key adaptations to the bee habitat. BMC Evol. Biol. 2015, 15, 145. [Google Scholar] [CrossRef] [PubMed]
- De Hoog, G.S.; Guarro, J.; Gené, J.; Ahmed, S.; Al-Hatmi, A.M.S.; Figueras, M.J.; Vitale, R.G. Atlas of Clinical Fungi: The Ultimate Bench Tool for Diagnostics. In Part α: Introductions, Lower Fungi, Basidiomycetes, Yeasts, Filamentous Ascomycetes (A-B)/Part β: Filamentous Ascomycetes (C-Z), 4th ed.; Foundation Atlas: Hilversum, The Netherlands, 2020; p. 776. [Google Scholar]
- Hainsworth, S.; Kučerová, I.; Sharma, R.; Cañete-Gibas, C.F.; Hubka, V. Three-gene phylogeny of the genus Arthroderma: Basis for future taxonomic studies. Med. Mycol. 2021, 59, 355–365. [Google Scholar] [CrossRef]
- Simordová, M.; Hejtmanek, M. 1969. Standard methods for Examination of Water and Wastewater. Ceskoslovenská Hyg. 1969, 14, 89. [Google Scholar]
- Bertoldi, M. Pathogenic Fungi Associated with Land Applicantion of Sludge; WHO, Regional Office for Europe: Copenhagen, Denmark, 1981. [Google Scholar]
- Ulfig, K.; Korcz, M. Isolation of keratinophilic fungi from sewage sludge. J. Med. Vet. Mycol. 1983, 21, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Mangiarotti, A.M.; Caretta, G. Keratinophilic fungi isolated from a small pool. Mycopathologia 1984, 85, 9–11. [Google Scholar] [CrossRef]
- Abdel-Hafez, A.I.I.; Bagy, M.M.K.; Shoreit, A.A.M. Keratinophilic fungi in mud of Ibrahimia canal, Egypt. Cryptogam. Mycol. 1989, 10, 275–282. [Google Scholar]
- Ulfig, K.; Ulfig, A. Keratinophilic fungi in bottom sediments of surface waters. J. Med. Vet. Mycol. 1990, 28, 419–422. [Google Scholar] [CrossRef]
- Kornillowicz, T. Occurrence of geophilic keratinophilic fungi in bottom sediments of various trophicity. Acta Mycol. 1993, 28, 171–184. [Google Scholar] [CrossRef]
- Ulfig, K.; Korcz, M. Keratinolytic fungi in sewage sludge applied to devastated urban soil. A preliminary experiment. Int. J. Environ. Health Res. 1994, 4, 244–253. [Google Scholar] [CrossRef]
- Ulfig, K.; Guarro, J.; Cano, J.; Gene, J.; Vidal, P.; Figueras, M.J. General assessment of the occurrence of keratinolytic fungi in river and marine beach sediments of Catalonian waters (Spain). Water Air Soil Pollut. 1997, 94, 275–287. [Google Scholar] [CrossRef]
- Ulfig, K.; Guarro, J.; Cano, J.; Gené, J.; Vidal, P.; Figueras, M.J.; Łukasik, W. The occurrence of keratinophylic fungi in sediments of the river Tordera (Spain). FEMS Microbiol. Ecol. 1997, 22, 111–117. [Google Scholar] [CrossRef]
- Ulfig, K.; Łukasik, W.; Guarro, J.; Cano, J.; Gené, J.; Vidal, P.; Figueras, M.J. The seasonal changes of keratinolytic fungi in sediments of Catalonian rivers (Spain). Water Air Soil Pollut. 1997, 96, 269–290. [Google Scholar] [CrossRef]
- Ulfig, K.; Guarro, J.; Cano, J.; Gené, J.; Vidal, P.; Figueras, M.J.; Łukasik, W. A preliminary study of the occurrence of actidione-resistant fungi in sediments of Catalonian river mouths (Spain). I. Keratinolytic fungi and related Onygenales. Mycopathologia 1998, 141, 143–151. [Google Scholar] [CrossRef]
- Vidal, P. Taxonomía Molecular de Chrysosporium. Ph.D. Thesis, Universitat Rovira i Virgili, Tarragona, Spain, 2000. [Google Scholar]
- Ulfig, K.; Gené, J.; Guarro, J. Studies on keratinophilic fungi. VI. A. new Arthropsis (Fungi Imperfecti) from marine sediments. Mycotaxon 1995, 54, 281–286. [Google Scholar]
- Vidal, P.; Sanchez-Puelles, J.M.; Milan, D.; Guarro, J. Chrysosporium fluviale, a new keratinophilic species from river sediments. Mycol. Res. 2000, 104, 244–250. [Google Scholar] [CrossRef]
- Vidal, P.; Valmaseda, M.; Vinuesa, M.Á.; Guarro, J. Two new species of Chrysosporium. Stud. Mycol. 2002, 47, 199–210. [Google Scholar]
- Gené, J.; Guarro, J.; Ulfig, K.; Vidal, P.; Cano, J. Studies on keratinophilic fungi. II: Chrysosporium pilosum sp. nov. Mycotaxon 1994, 50, 107–113. [Google Scholar]
- Vidal, P.; Ulfig, K.; Valmaseda, M.; Guarro, J. Studies on keratinophilic fungi. XI. Chrysosporium undulatum sp. nov. Antonie Leeuwenhoek 1999, 75, 171–182. [Google Scholar] [CrossRef]
- Marin-Félix, Y.; Stchigel, A.M.; Cano-Lira, J.F.; Sanchis, M.; Mayayo, E.; Guarro, J. Emmonsiellopsis, a new genus related to the thermally dimorphic fungi of the family Ajellomycetaceae. Mycoses 2015, 58, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Wingfield, M.J.; Lombard, L.; Roets, F.; Swart, W.J.; Alvarado, P.; Carnegie, A.J.; Moreno, G.; Luangsa-ard, J.; Thangavel, R.; et al. Fungal Planet description sheets: 951–1041. Persoonia Mol. Phylogeny Evol. Fungi 2019, 43, 223–425. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Cowan, D.A.; Maggs-Kölling, G.; Yilmaz, N.; Larsson, E.; Angelini, C.; Brandrud, T.E.; Dearnaley, J.D.W.; Dima, B.; Dovana, F.; et al. Fungal Planet description sheets: 1112–1181. Persoonia Mol. Phylogeny Evol. Fungi 2020, 45, 251–409. [Google Scholar] [CrossRef]
- Crous, P.W.; Boers, J.; Holdom, D.; Osieck, E.R.; Steinrucken, T.V.; Tan, Y.P.; Vitelli, J.S.; Shivas, R.G.; Barrett, M.; Boxshall, A.G.; et al. Fungal Planet description sheets: 1383–1435. Persoonia Mol. Phylogeny Evol. Fungi 2022, 48, 261–371. [Google Scholar] [CrossRef]
- Tan, Y.P.; Bishop-Hurley, S.L.; Shivas, R.G.; Cowan, D.A.; Maggs-Kölling, G.; Maharachchikumbura, S.S.N.; Pinruan, U.; Bransgrove, K.L.; De la Peña-Lastra, S.; Larsson, E.; et al. Fungal Planet description sheets: 1436–1477. Persoonia Mol. Phylogeny Evol. Fungi 2022, 49, 261–350. [Google Scholar] [CrossRef]
- Torres-Garcia, D.; García, D.; Cano-Lira, J.F.; Gené, J. Two novel genera, Neostemphylium and Scleromyces (Pleosporaceae) from freshwater sediments and their global biogeography. J. Fungi 2022, 8, 868. [Google Scholar] [CrossRef]
- Torres-Garcia, D.; Gené, J.; García, D. New and interesting species of Penicillium (Eurotiomycetes, Aspergillaceae) in freshwater sediments from Spain. MycoKeys 2022, 86, 103–145. [Google Scholar] [CrossRef]
- Torres-Garcia, D.; García, D.; Réblová, M.; Jurjević, Ž.; Hubka, V.; Gené, J. Diversity and novel lineages of black yeasts in Chaetothyriales from freshwater sediments in Spain. Persoonia Mol. Phylogeny Evol. Fungi 2023, in press. [CrossRef]
- Gibas, C.F.C.; Sigler, L.; Summerbell, R.C.; Currah, R.S. Phylogeny of the genus Arachnomyces and its anamorphs and the establishment of Arachnomycetales, a new eurotiomycete order in the Ascomycota. Stud. Mycol. 2002, 47, 131–139. [Google Scholar]
- Sun, B.D.; Zhou, Y.G.; Chen, A.J.; Houbraken, J. Phylogeny and a new species of the genus Arachnomyces (Arachnomycetaceae). Phytotaxa 2019, 394, 89–97. [Google Scholar] [CrossRef]
- Labuda, R.; Bernreiter, A.; Hochenauer, D.; Kubatova, A.; Kandemir, H.; Schuller, C. Molecular systematics of Keratinophyton: The inclusion of species formerly referred to Chrysosporium and description of four new species. IMA Fungus 2021, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.W.; Sigler, L. Molecular genetic variation in Emmonsia crescens and Emmonsia parva, etiologic agents of adiaspiromycosis, and their phylogenetic relationship to Blastomyces dermatitidis (Ajellomyces dermatitidis) and other systemic fungal pathogens. J. Clin. Microbiol. 1998, 36, 2918–2925. [Google Scholar] [CrossRef]
- Sugiyama, M.; Summerbell, R.C.; Mikawa, T. Molecular phylogeny of onygenalean fungi based on small subunit (SSU) and large subunity (LSU) ribosomal DNA sequences. Stud. Mycol. 2002, 47, 5–23. [Google Scholar]
- Untereiner, W.A.; Scott, J.A.; Naveau, F.A.; Sigler, L.; Bachewich, J.; Angus, A. The Ajellomycetaceae, a new family of vertebrate-associated Onygenales. Mycologia 2004, 96, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Andrade, E.; Cano-Lira, J.F.; Wiederhold, N.; Perez-Cantero, A.; Guarro, J.; Stchigel, A.M. A revision of malbranchea-like fungi from clinical specimens in the United States of America reveals unexpected novelty. IMA Fungus 2021, 12, 25. [Google Scholar] [CrossRef]
- Větrovský, T.; Morais, D.; Kohout, P.; Lepinay, C.; Algora, C.; Awokunle Hollá, S.; Bahmann, B.D.; Bílohnědá, K.; Brabcová, V.; D’Alò, F.; et al. GlobalFungi, a global database of fungal occurrences from high-throughput-sequencing metabarcoding studies. Sci. Data 2020, 7, 228. [Google Scholar] [CrossRef]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour, 3rd ed.; Methuen: London, UK, 1978; p. 256. [Google Scholar]
- Müller, F.M.; Werner, K.E.; Kasai, M.; Francesconi, A.; Mu, F.C.; Chanock, S.J.; Walsh, T.J. Rapid extraction of genomic DNA from medically important yeasts and filamentous fungi by high-speed cell disruption. J. Clin. Microbiol. 1998, 36, 1625–1629. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications, 1st ed.; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Rehner, S.A.; Samuels, G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994, 98, 625–634. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G. Development of primer sets designed for use with PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Sung, G.H.; Sung, J.M.; Hywel-Jones, N.L.; Spatafora, J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogen. Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Whelen, S.; Hall, B.D. 1999. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Farris, J.S.; Kallersjo, M.; Kluge, A.G.; Bult, C. Testing significance of incongruence. Cladistics 1994, 10, 315–319. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES science gateway: Enabling high-impact science for phylogenetics researchers with limited resources. In Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the Extreme to the Campus and Beyond, Chicago, IL, USA, 16–20 July 2012; Association for Computing Machinery: New York, NY, USA, 2012; pp. 1–8. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Hespanhol, L.; Vallio, C.S.; Costa, L.M.; Saragiotto, B.T. Understanding and interpreting confidence and credible intervals around effect estimates. Braz. J. Phys. Ther. 2019, 23, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Réblová, M.; Kolařík, M.; Nekvindová, J.; Miller, A.N.; Hernández-Restrepo, M. Phylogeny, global biogeography and pleomorphism of Zanclospora. Microorganisms 2021, 9, 706. [Google Scholar] [CrossRef] [PubMed]
- Réblová, M.; Kolařík, M.; Nekvindová, J.; Réblová, K.; Sklenář, F.; Miller, A.N.; Hernández-Restrepo, M. Phylogenetic reassessment, taxonomy, and biogeography of Codinaea and similar fungi. J. Fungi 2021, 7, 1097. [Google Scholar] [CrossRef]
- Guarro, J.; Gené, J.; Stchigel, M.A.; Figueras, M.J. Atlas of Soil Ascomycetes; CBS Biodiversity Series No. 10; CBS-KNAW Fungal Biodiversity Centre: Utrecht, The Netherlands, 2012; p. 486. [Google Scholar]
- Von Arx, J.A. On Arachniotus and related genera of the Gymnoascaceae. Persoonia Mol. Phylogeny Evol. Fungi 1971, 6, 371–380. [Google Scholar]
- Gené, J.; Guillamón, J.M.; Ulfig, K.; Guarro, J. Studies on keratinophilic fungi. X. Arthrographis alba sp. nov. Can. J. Microbiol. 1996, 42, 1185–1189. [Google Scholar] [CrossRef]
- Orr, G.F.; Kuehn, H.H.; Plunkett, O.A. A new genus of the Gymnoascaceae with swollen peridial septa. Can. J. Bot. 1963, 41, 1439–1456. [Google Scholar] [CrossRef]
- Sigler, L.; Carmichael, J.W. Taxonomy of Malbranchea and some other Hyphomycetes with arthroconidia. Mycotaxon 1976, 4, 349–488. [Google Scholar]
- Van Oorschot, C.A.N. A revision of Chrysosporium and allied genera. Stud. Mycol. 1980, 20, 1–89. [Google Scholar]
- Cano, J.; Ulfig, K.; Guillamon, J.; Vidal, P.; Guarro, J. Studies on keratinophilic fungi. IX: Neoarachnotheca gen. nov. and a new species of Nannizziopsis. Antonie Leeuwenhoek 1997, 72, 149–158. [Google Scholar] [CrossRef]
- Giraldo, A.; Sutton, D.A.; Gené, J.; Fothergill, A.W.; Cano, J.; Guarro, J. Rare arthroconidial fungi in clinical samples: Scytalidium cuboideum and Arthropsis hispanica. Mycopathologia 2013, 175, 115–121. [Google Scholar] [CrossRef]
- Guarro, J.; Cano, J.; de Vroey, C. Nannizziopsis (Ascomycotina) and related genera. Mycotaxon 1991, 42, 193–200. [Google Scholar]
- Zhang, Z.F.; Zhou, S.Y.; Eurwilaichitr, L.; Ingsriswang, S.; Raza, M.; Chen, Q.; Zhao, P.; Liu, F.; Cai, L. Culturable mycobiota from Karst caves in China II, with descriptions of 33 new species. Fungal Divers. 2021, 106, 29–136. [Google Scholar] [CrossRef]
- Apinis, A.E. A Revision of British Gymnoascaceae. Mycol. Pap. 1964, 96, 1–54. [Google Scholar]
- Von Arx, J.A. The Genera of Fungi Sporulating in Pure Culture, 2nd ed.; J. Cramer: Vaduz, Liechtenstein, 1974; p. 288. [Google Scholar]
- Sigler, L.; Dunn, M.T.; Carmichael, J.W. Arthrocristula and Arthropsis, two new Hyphomycetes with dematiaceous arthroconidia. Mycotaxon 1982, 15, 409–419. [Google Scholar]
- Giraldo, A.; Gené, J.; Sutton, D.A.; Madrid, H.; Cano, J.; Crous, P.W.; Guarro, J. Phylogenetic circumscription of Arthrographis (Eremomycetaceae, Dothideomycetes). Persoonia Mol. Phylogeny Evol. Fungi 2014, 32, 102–114. [Google Scholar] [CrossRef]
- Guarro, J.; Gené, J.; De Vroey, C.H. Amaurascopsis, a new genus of Eurotiales. Mycotaxon 1992, 45, 171–178. [Google Scholar]
- Scott, J.A.; Malloch, D.W.; Gloer, J.B. Polytolypa, an undescribed genus in the Onygenales. Mycologia 1993, 85, 503–508. [Google Scholar] [CrossRef]
- Coleine, C.; Selbmann, L.; Guirado, E.; Singh, B.K.; Delgado-Baquerizo, M. Humidity and low pH boost occurrence of Onygenales fungi in soil at global scale. Soil Biol. Biochem. 2022, 167, 108617. [Google Scholar] [CrossRef]
- Salazar-Hamm, P.S.; Montoya, K.N.; Montoya, L.; Cook, K.; Liphardt, S.; Taylor, J.W.; Cook, J.A.; Natvig, D.O. Breathing can be dangerous: Opportunistic fungal pathogens and the diverse community of the small mammal lung mycobiome. Front. Fungal Biol. 2022, 3, 996574. [Google Scholar] [CrossRef]
- Jiang, Y.; Dukik, K.; Muñoz, J.F.; Sigler, L.; Schwartz, I.S.; Govender, N.P.; Kenyon, C.; Feng, P.; Van den Ende, B.G.; Stielow, J.B.; et al. Phylogeny, ecology and taxonomy of systemic pathogens and their relatives in Ajellomycetaceae (Onygenales): Blastomyces, Emergomyces, Emmonsia, Emmonsiellopsis. Fungal Divers. 2018, 90, 245–291. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Han, Y.F.; Chen, W.H.; Liang, Z.Q. Phylogeny and taxonomy of three new Ctenomyces (Arthrodermataceae, Onygenales) species from China. MycoKeys 2019, 47, 1–16. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Ren, Y.L.; Li, X.; Chen, W.H.; Liang, J.D.; Han, Y.F.; Liang, Z.Q. New taxonomic framework for Arthrodermataceae: A comprehensive analysis based on their phylogenetic reconstruction, divergence time estimation, phylogenetic split network, and phylogeography. Antonie Leeuwenhoek 2022, 115, 1319–1333. [Google Scholar] [CrossRef]
- Woodburn, D.B.; Miller, A.N.; Allender, M.C.; Maddox, C.W.; Terio, K.A. Emydomyces testavorans, a new genus and species of onygenalean fungus isolated from shell lesions of freshwater aquatic turtles. J. Clin. Microbiol. 2019, 57, e00628-18. [Google Scholar] [CrossRef]
- Moulíková, Š.; Kolařík, M.; Lorch, J.M.; Kolarczyková, D.; Hubka, V.; Čmoková, A. Wild rodents harbour high diversity of Arthroderma. Persoonia Mol. Phylogeny Evol. Fungi 2023, 50, 27–47. [Google Scholar] [CrossRef]
- Kochhar, S.; Gupta, V.S.; Sethi, H.S.; Capoor, M.R. A rare case of keratomycosis due to Myriodontium keratinophilum. Delhi J. Ophthal. 2018, 29, 54–56. [Google Scholar] [CrossRef]
- Sigler, L.; Hambleton, S.; Paré, J.A. Molecular characterization of reptile pathogens currently known as members of the Chrysosporium anamorph of Nannizziopsis vriesii complex and relationship with some human-associated isolates. J. Clin. Microbiol. 2013, 51, 3338–3357. [Google Scholar] [CrossRef]
- Cano, J.; Guarro, J.; Figueras, M.J. Some keratinophilic fungi from Spain. Mycopathologia 1987, 100, 163–167. [Google Scholar] [CrossRef]
- Stchigel, A.M.; Sutton, D.A.; Cano-Lira, J.F.; Cabañes, F.J.; Abarca, M.L.; Tintelnot, K.; Wickes, B.L.; García, D.; Guarro, J. Phylogeny of chrysosporia infecting reptiles: Proposal of the new family Nannizziopsiaceae and five new species. Persoonia Mol. Phylogeny Evol. Fungi 2013, 31, 86–100. [Google Scholar] [CrossRef]
- Cabañes, F.J.; Sutton, D.A.; Guarro, J. Chrysosporium-related fungi and reptiles: A Fatal Attraction. PLoS Pathog. 2014, 10, e1004367. [Google Scholar] [CrossRef]
- Seena, S.; Baschien, C.; Barros, J.; Sridhar, K.R.; Graça, M.A.; Mykrä, H.; Bundschuh, M. Ecosystem services provided by fungi in freshwaters: A wake-up call. Hydrobiologia 2022, 850, 2779–2794. [Google Scholar] [CrossRef]
- Todaro, V.; D’Oria, M.; Secci, D.; Zanini, A.; Tanda, M.G. Climate change over the Mediterranean region: Local temperature and precipitation variations at five pilot sites. Water 2022, 14, 2499. [Google Scholar] [CrossRef]
- Lumbsch, H.T.; Lindemuth, R.; Schmitt, I. Evolution of filamentous ascomycetes inferred from LSU rDNA sequence data. Plant Biology 2000, 2, 525–529. [Google Scholar] [CrossRef]
- Wynns, A.A.; Jensen, A.B.; Eilenberg, J.; James, R. Ascosphaera subglobosa, a new spore cyst fungus from North America associated with the solitary bee Megachile rotundata. Mycologia 2012, 04, 108–114. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; De Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Vidal, P.; Vinuesa, M.D.L.A.; Sánchez-Puelles, J.M.; Guarro, J. Phylogeny of the anamorphic genus Chrysosporium and related taxa based on rDNA internal transcribed spacer sequences. Rev. Iberoam. Micol. 2000, 17, 22–29. [Google Scholar]
- Solé, M.; Cano, J.; Guarro, J. Molecular taxonomy of Amauroascus, Auxarthron and other related genera of the Onygenales. Mycol. Res. 2002, 106, 388–396. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; White, M.M. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Nat. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Suankratay, C.; Dhissayakamol, O.; Uaprasert, N.; Chindamporn, A. Invasive pulmonary infection caused by Chrysosporium articulatum: The first case report. Mycoses 2015, 58, 1–3. [Google Scholar] [CrossRef]
- Untereiner, W.A.; Scott, J.A.; Naveau, F.A.; Bachewich, J. Phylogeny of Ajellomyces, Polytolypa and Spiromastix (Onygenaceae) inferred from rDNA sequence and non-molecular data. Stud. Mycol. 2002, 47, 25–35. [Google Scholar]
- Zhang, Z.F.; Liu, F.; Zhou, X.; Liu, X.Z.; Liu, S.J.; Cai, L. Culturable mycobiota from Karst caves in China, with descriptions of 20 new species. Persoonia Mol. Phylogeny Evol. Fungi 2017, 39, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Robbertse, B.; Robert, V.; Vu, D.; Cardinali, G.; Irinyi, L.; Meyer, W.; Nilsson, R.H.; Hughes, K.; Miller, A.N.; et al. Finding needles in haystacks: Linking scientific names, reference specimens and molecular data for Fungi. Database 2014, bau061. [Google Scholar] [CrossRef] [PubMed]
- Nováková, A.; Kolařík, M. Chrysosporium speluncarum, a new species resembling Ajellomyces capsulatus, obtained from bat guano in caves of temperate Europe. Mycol. Prog. 2010, 9, 253–260. [Google Scholar] [CrossRef]
- Stielow, J.B.; Lévesque, C.A.; Seifert, K.A.; Meyer, W.; Iriny, L.; Smits, D.; Renfurm, R.; Verkley, G.J.; Groenewald, M.; Chaduli, D.; et al. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia Mol. Phylogeny Evol. Fungi 2015, 35, 242–263. [Google Scholar] [CrossRef]
- Brasch, J.; Gräser, Y. Trichophyton eboreum sp. nov. isolated from human skin. J. Clin. Microbiol. 2005, 43, 5230–5237. [Google Scholar] [CrossRef]
- Sugiyama, M.; Mikawa, T. Phylogenetic analysis of the non-pathogenic genus Spiromastix (Onygenaceae) and related onygenalean taxa based on large subunit ribosomal DNA sequences. Mycoscience 2001, 42, 413–421. [Google Scholar] [CrossRef]
- Gueidan, C.; Villaseñor, C.R.; De Hoog, G.S.; Gorbushina, A.A.; Untereiner, W.A.; Lutzoni, F. A rock-inhabiting ancestor for mutualistic and pathogen-rich fungal lineages. Stud. Mycol. 2008, 61, 111–119. [Google Scholar] [CrossRef]
- Rangel-Grimaldo, M.; Macías-Rubalcava, M.L.; González-Andrade, M.; Raja, H.; Figueroa, M.; Mata, R. α-Glucosidase and protein tyrosine phosphatase 1B inhibitors from Malbranchea circinata. J. Nat. Prod. 2020, 83, 675–683. [Google Scholar] [CrossRef]
- Sigler, L.; Hambleton, S.; Flis, A.L.; Pare, J.A. Auxarthron teleomorphs for Malbranchea filamentosa and Malbranchea albolutea and relationships within Auxarthron. Stud. Mycol. 2002, 47, 111–122. [Google Scholar]
- Crous, P.W.; Wingfield, M.J.; Guarro, J.; Cheewangkoon, R.; Van der Bank, M.; Swart, W.J.; Stchigel, A.M.; Cano-Lira, J.F.; Roux, J.; Madrid, H.; et al. Fungal Planet description sheets: 154–213. Persoonia Mol. Phylogeny Evol. Fungi 2013, 31, 188–296. [Google Scholar] [CrossRef]
- Gräser, Y.; El Fari, M.; Vilgalys, R.; Kuijpers, A.F.A.; De Hoog, G.S.; Presber, W.; Tietz, H.J. Phylogeny and taxonomy of the family Arthrodermataceae (dermatophytes) using sequence analysis of the ribosomal ITS region. Med. Mycol. 1999, 37, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Hardy, G.S.J.; Barber, P.A.; Alvarado, P.; Barnes, C.W.; Buchanan, P.K.; Heykoop, M.; Moreno, G.; et al. Fungal Planet description sheets: 558–624. Persoonia Mol. Phylogeny Evol. Fungi 2017, 38, 240. [Google Scholar] [CrossRef]
- Irinyi, L.; Serena, C.; Garcia-Hermoso, D.; Arabatzis, M.; Desnos-Ollivier, M.; Vu, D.; Cardinali, G.; Arthur, I.A.; Normand, A.C.; Giraldo, A.; et al. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database—the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med. Mycol. 2015, 53, 313–337. [Google Scholar] [CrossRef]
- Hirooka, Y.; Tanney, J.B.; Nguyen, H.D.; Seifert, K.A. Xerotolerant fungi in house dust: Taxonomy of Spiromastix, Pseudospiromastix and Sigleria gen. nov. in Spiromastigaceae (Onygenales, Eurotiomycetes). Mycologia 2016, 108, 135–156. [Google Scholar] [CrossRef] [PubMed]
- Stchigel, A.M.; Sutton, D.A.; Cano-Lira, J.F.; Wiederhold, N.; Guarro, J. New species Spiromastigoides albida from a lung biopsy. Mycopathologia 2017, 182, 967–978. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Sutton, D.A.; Wiederhold, N.P.; Thompson, E.H.; Friedman, R.; Wickes, B.L.; Cano-Lina, J.F.; Stchigel, A.M.; Guarro, J. Isolation and characterisation of the fungus Spiromastix asexualis sp. nov. from discospondylitis in a German Shepherd dog, and review of Spiromastix with the proposal of the new order Spiromastixales (Ascomycota). Mycoses 2014, 57, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Heidemann, S.; Monod, M.; Gräser, Y. Signature polymorphisms in the internal transcribed spacer region relevant for the differentiation of zoophilic and anthropophilic strains of Trichophyton interdigitale and other species of T. mentagrophytes sensu lato. Br. J. Dermatol. 2010, 162, 282–295. [Google Scholar] [CrossRef]
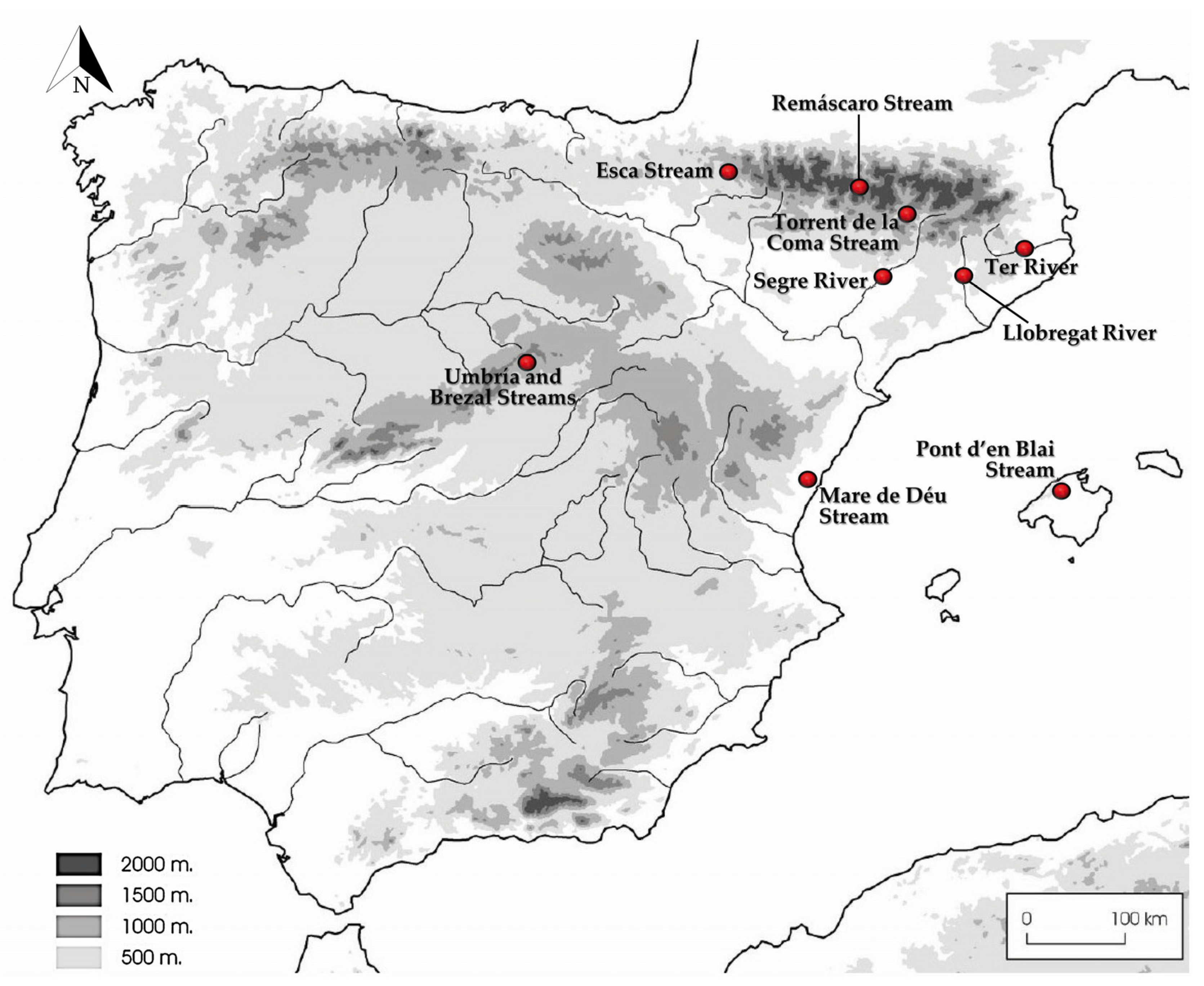

| Species | Strain Numbers 1 | Rivers and Streams in Spain (Coordinates) | GenBank Accession Numbers | |||
|---|---|---|---|---|---|---|
| ITS | LSU | tub2 | rpb2 | |||
| Albidomyces albicans (≡Arachniotus albicans) | FMR 17576 | Remáscaro Stream, Cerler, Huesca (N 42.58698° E 0.54993°) | ON720226 | ON720765 | - | - |
| FMR 18029 | Torrent de la Coma Stream, Lleida (N 42.21889° E 1.65159°) | ON721307 | ON720780 | - | - | |
| FMR 18030 | Torrent de la Coma Stream, Lleida (N 42.21889° E 1.65159°) | ON720223 | ON720762 | - | - | |
| FMR 18031 | Torrent de la Coma Stream, Lleida (N 42.21889° E 1.65159°) | ON720231 | ON720770 | - | - | |
| FMR 18033 | Torrent de la Coma Stream, Lleida (N 42.21889° E 1.65159°) | ON720228 | ON720767 | - | - | |
| FMR 18697 | Ter River, Vilanova de Sau, Barcelona (N 41.97084° E 2.38108°) | OP373730 | OP373734 | - | - | |
| FMR 18698 | Ter River, Llanars, Girona (N 42.32383° E 2.32989°) | ON720224 | ON720763 | - | - | |
| FMR 18699 | Ter River, Sant Joan de les Abadesses, Girona (N 42.24138° E 2.28830°) | OP373731 | OP373735 | - | - | |
| FMR 19026 | Llobregat River, Castellar de n’Hug, Barcelona (N 42.24012° E 1.93742°) | ON720229 | ON720768 | - | - | |
| FMR 19027 | Llobregat River, Castellar de n’Hug, Barcelona (N 42.24012° E 1.93742°) | ON720225 | ON720764 | - | - | |
| FMR 19028 | Llobregat River, Castellar de n’Hug, Barcelona (N 42.24012° E 1.93742°) | ON720227 | ON720766 | - | - | |
| FMR 19029 | Llobregat River, Balsareny, Barcelona (N 41.87818° E 1.88900°) | ON720230 | ON720769 | - | - | |
| Aphanoascus crassitunicatus | FMR 18700 | Ter River, Vilanova de Sau, Barcelona (N 41.97084° E 2.38108°) | ON720203 | ON720742 | - | - |
| FMR 18701 | Ter River, Roda de Ter, Barcelona (N 41.96974° E 2.31211°) | ON720204 | ON720743 | - | - | |
| FMR 18702 | Ter River, Ripoll, Girona (N 42.17258° E 2.19372°) | ON721311 | ON720784 | - | - | |
| Aphanoascus fulvescens | FMR 18235 | Segre River, Aitona, Lleida (N 41.97084° E 2.38108°) | ON720208 | ON720747 | - | - |
| FMR 18236 | Segre River, La Granja d’Escarp, Lleida (N 41.42754° E 0.35020°) | ON720210 | ON720749 | - | - | |
| FMR 18241 | Segre River, La Granja d’Escarp, Lleida (N 41.42754° E 0.35020°) | ON721308 | ON720781 | - | - | |
| FMR 18242 | Segre River, La Granja d’Escarp, Lleida (N 41.42754° E 0.35020°) | ON720211 | ON720750 | - | - | |
| FMR 18243 | Segre River, Camarassa, Lleida (N 41.89834° E 0.88110°) | ON942224 | ON942226 | - | - | |
| FMR 18704 | Ter River, Les Masies de Voltregà, Barcelona (N 42.02951° E 2.25359°) | ON720207 | ON720746 | - | - | |
| FMR 19018 | Mare de Déu Stream, Borriana, Castellón (N 39.87727° W 0.05374°) | ON720212 | ON720751 | - | - | |
| FMR 19019 | Mare de Déu Stream, Borriana, Castellón (N 39.87727° W 0.05374°) | ON720209 | ON720748 | - | - | |
| FMR 19022 | Mare de Déu Stream, Borriana, Castellón (N 39.87727° W 0.05374°) | ON720206 | ON720745 | - | - | |
| FMR 19020 | Llobregat River, Cercs, Barcelona (N 42.10512° E 1.88044°) | ON721312 | ON720785 | - | - | |
| FMR 19021 | Llobregat River, Cercs, Barcelona (N 42.10512° E 1.88044°) | ON720205 | ON720744 | - | - | |
| Aphanoascus reticulisporus | FMR 18004 | Esca Stream, Navarra (N 42.81466° W 0.95221°) | ON720200 | ON720739 | - | - |
| FMR 19012 | Llobregat River, Cercs, Barcelona (N 42.10512° E 1.88044°) | ON720201 | ON720740 | - | - | |
| FMR 19013 | Llobregat River, Cercs, Barcelona (N 42.10512° E 1.88044°) | ON720202 | ON720741 | - | - | |
| FMR 19033 | Llobregat River, La Pobla de Lillet, Barcelona (N 42.24294° E 1.96998°) | OP373732 | OP373736 | - | - | |
| Arthroderma curreyi | FMR 19038 | Llobregat River, Balsareny, Barcelona (N 41.87818° E 1.88900°) | ON720238 | ON720777 | - | - |
| Emmonsiellopsis tuberculata | FMR 17582T | Remáscaro Stream, Cerler, Huesca (N 42.58698° E 0.54993°) | LR598892 | LR598891 | - | - |
| Gymnoascoideus petalosporus | FMR 19036 | Llobregat River, Cercs, Barcelona (N 42.10512° E 1.88044°) | ON720236 | ON720775 | - | - |
| Leucothecium emdenii | FMR 18703 | Ter River, Sant Joan de les Abadesses, Girona (N 42.24138° E 2.28830°) | ON720239 | ON720778 | - | - |
| Malbranchea chinensis | FMR 18267 | Segre River, Camarassa, Lleida (N 41.89834° E 0.88110°) | ON720190 | ON720729 | OP425706 | OP425715 |
| Malbranchea echinulata (=Malbranchea sp. II) | FMR 17906 T = CBS 149936 | Umbría Stream, Rascafría, Madrid (N 40.86011° W 3.90849°) | ON720198 | ON720737 | OP425705 | - |
| Malbranchea irregularis (=Malbranchea sp. III) | FMR 19016 T = CBS 149937 | Llobregat River, Castellar de n’Hug, Barcelona (N 42.24012° E 1.93742°) | ON720191 | ON720730 | OP425710 | OP425719 |
| FMR 19017 | Llobregat River, Castellar de n’Hug, Barcelona (N 42.24012° E 1.93742°) | ON720192 | ON720731 | OP425713 | OP425722 | |
| FMR 19030 | Llobregat River, Castellar de n’Hug, Barcelona (N 42.24012° E 1.93742°) | ON720193 | ON720732 | OP425712 | OP425721 | |
| FMR 19015 | Llobregat River, Balsareny, Barcelona (N 41.87818° E 1.88900°) | ON720194 | ON720733 | OP425711 | OP425720 | |
| Malbranchea ostraviensis | FMR 18693 | Ter River, Roda de Ter, Barcelona (N 41.96974° E 2.31211°) | ON720199 | ON720738 | OP425707 | OP425716 |
| Malbranchea reticulata | FMR 18696 | Ter River, Roda de Ter, Barcelona (N 41.96974° E 2.31211°) | ON721310 | ON720783 | - | - |
| Malbranchea sinuata (=Malbranchea sp. I) | FMR 18266 T = CBS 149938 | Segre River, La Granja d’Escarp, Lleida (N 41.42754° E 0.35020°) | ON720195 | ON720734 | OP425704 | OP425714 |
| Malbranchea umbrina | FMR 17899 | Ter River, Sant Joan de les Abadesses, Girona (N 42.24138° E 2.28830°) | ON721306 | ON720779 | - | - |
| FMR 18695 | Ter River, Sant Joan de les Abadesses, Girona (N 42.24138° E 2.28830°) | ON720196 | ON720735 | OP425709 | OP425718 | |
| FMR 18766 | Brezal Stream, Rascafría, Madrid (N 40.86011° W 3.90849°) | ON720197 | ON720736 | OP425708 | OP425717 | |
| Myriodontium keratinophilum | FMR 17624 | Pont d’en Blai Stream, Mallorca, Balears Islands (N 39.76907° E 2.88418°) | ON720220 | ON720759 | - | - |
| FMR 18244 | Segre River, Aitona, Lleida (N 41.48444° E 0.46446°) | ON720222 | ON720761 | - | - | |
| FMR 18247 | Segre River, Aitona, Lleida (N 41.48444° E 0.46446°) | ON721309 | ON720782 | - | - | |
| FMR 18248 | Segre River, Aitona, Lleida (N 41.48444° E 0.46446°) | ON720215 | ON720754 | - | - | |
| FMR 18245 | Segre River, La Granja d’Escarp, Lleida (N 41.42754° E 0.35020°) | ON720213 | ON720752 | - | - | |
| FMR 18246 | Segre River, La Granja d’Escarp, Lleida (N 41.42754° E 0.35020°) | ON720221 | ON720760 | - | - | |
| FMR 18251 | Segre River, La Granja d’Escarp, Lleida (N 41.42754° E 0.35020°) | ON720217 | ON720756 | - | - | |
| FMR 18258 | Segre River, La Granja d’Escarp, Lleida (N 41.42754° E 0.35020°) | ON720219 | ON720758 | - | - | |
| FMR 18249 | Segre River, Camarassa, Lleida (N 41.89834° E 0.88110°) | ON720216 | ON720755 | - | - | |
| FMR 18250 | Segre River, Camarassa, Lleida (N 41.89834° E 0.88110°) | ON720218 | ON720757 | - | - | |
| FMR 18257 | Segre River, Camarassa, Lleida (N 41.89834° E 0.88110°) | ON720214 | ON720753 | - | - | |
| FMR 19031 | Llobregat River, Guardiola del Berguedà, Barcelona (N 42.24012°E 1.93742°) | OP070725 | OP077087 | - | - | |
| Neoarthropsis hispanica (≡Arthropsis hispanica) | FMR 18694 | Ter River, Les Masies de Voltregà, Barcelona (N 42.02951° E 2.25359°) | ON720234 | ON720773 | - | - |
| FMR 19034 | Llobregat River, Balsareny, Barcelona (N 41.87818° E 1.88900°) | ON720232 | ON720771 | - | - | |
| FMR 19035 | Llobregat River, Balsareny, Barcelona (N 41.87818° E 1.88900°) | ON720233 | ON720772 | - | - | |
| Neoarthropsis sexualis | FMR 19025T = CBS 149940 | Llobregat River, Balsareny, Barcelona (N 41.87818° E 1.88900°) | ON720235 | ON720774 | - | - |
| Pseudoamaurascopsis spiralis | FMR 19014T = CBS 149941 | Llobregat River, Balsareny, Barcelona (N 41.87818° E 1.88900°) | ON720237 | ON720776 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres-Garcia, D.; Gené, J.; García, D.; Cano-Lira, J.F. Insights into Some Onygenalean Fungi from Freshwater Sediments in Spain and Description of Novel Taxa. J. Fungi 2023, 9, 1129. https://doi.org/10.3390/jof9121129
Torres-Garcia D, Gené J, García D, Cano-Lira JF. Insights into Some Onygenalean Fungi from Freshwater Sediments in Spain and Description of Novel Taxa. Journal of Fungi. 2023; 9(12):1129. https://doi.org/10.3390/jof9121129
Chicago/Turabian StyleTorres-Garcia, Daniel, Josepa Gené, Dania García, and Jose F. Cano-Lira. 2023. "Insights into Some Onygenalean Fungi from Freshwater Sediments in Spain and Description of Novel Taxa" Journal of Fungi 9, no. 12: 1129. https://doi.org/10.3390/jof9121129
APA StyleTorres-Garcia, D., Gené, J., García, D., & Cano-Lira, J. F. (2023). Insights into Some Onygenalean Fungi from Freshwater Sediments in Spain and Description of Novel Taxa. Journal of Fungi, 9(12), 1129. https://doi.org/10.3390/jof9121129






