Overexpression of the Transcription Factor Azf1 Reveals Novel Regulatory Functions and Impacts β-Glucosidase Production in Trichoderma reesei
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
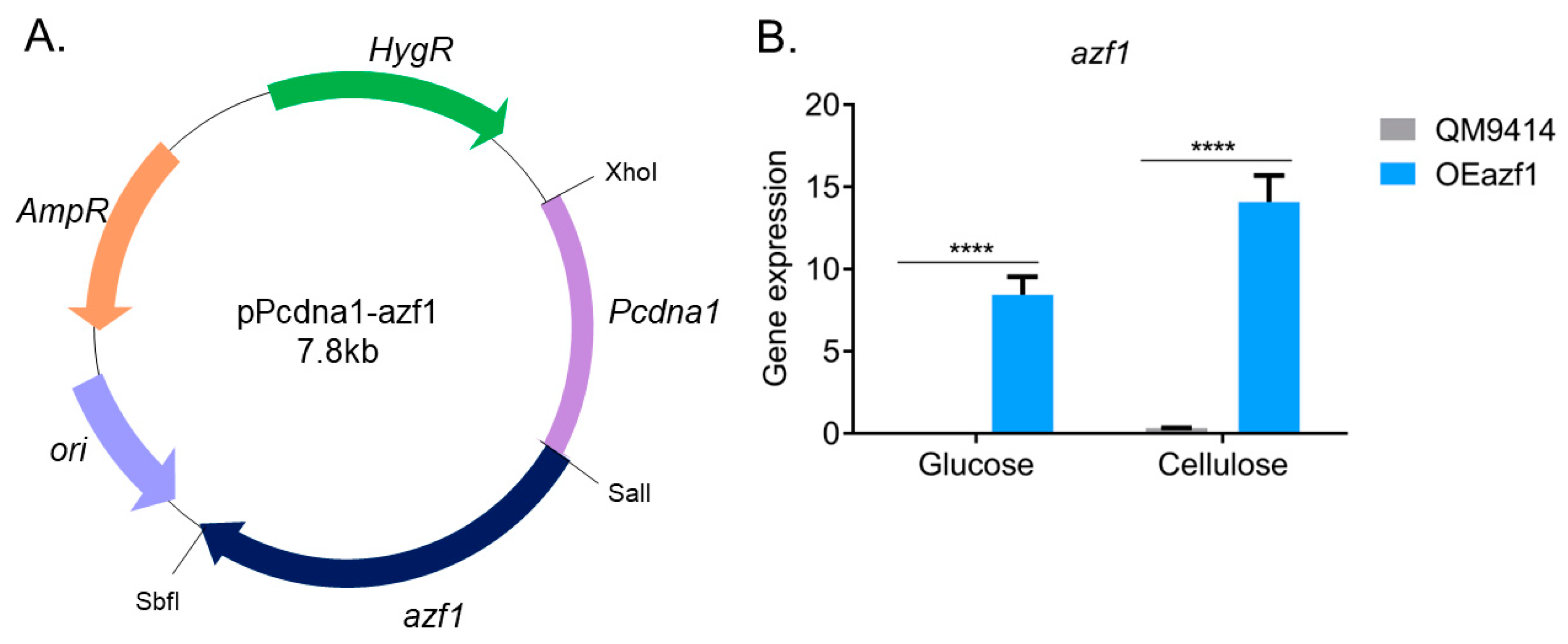
2.2. Vector Construction and Fungal Transformation
2.3. Gene Expression Analysis by RT-qPCR
2.4. Protein and Enzyme Assays
2.5. Intracellular Protein Extraction
2.6. Western Blot
2.7. Cellulase Production in Shake Flasks
2.8. Bioinformatic Analysis
3. Results
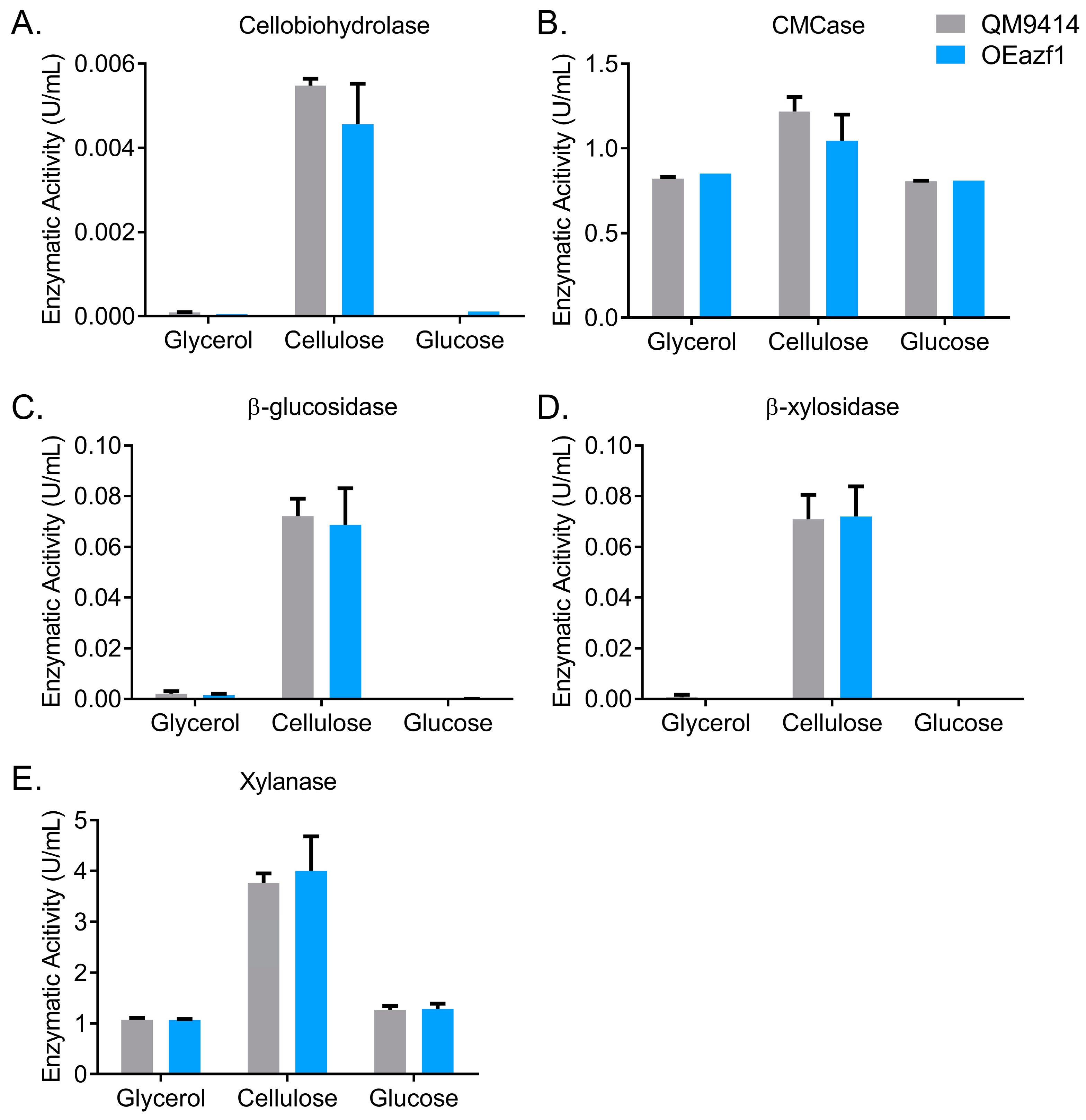
3.1. Azf1-Mediated Cellulase Expression Is Inducer-Dependent
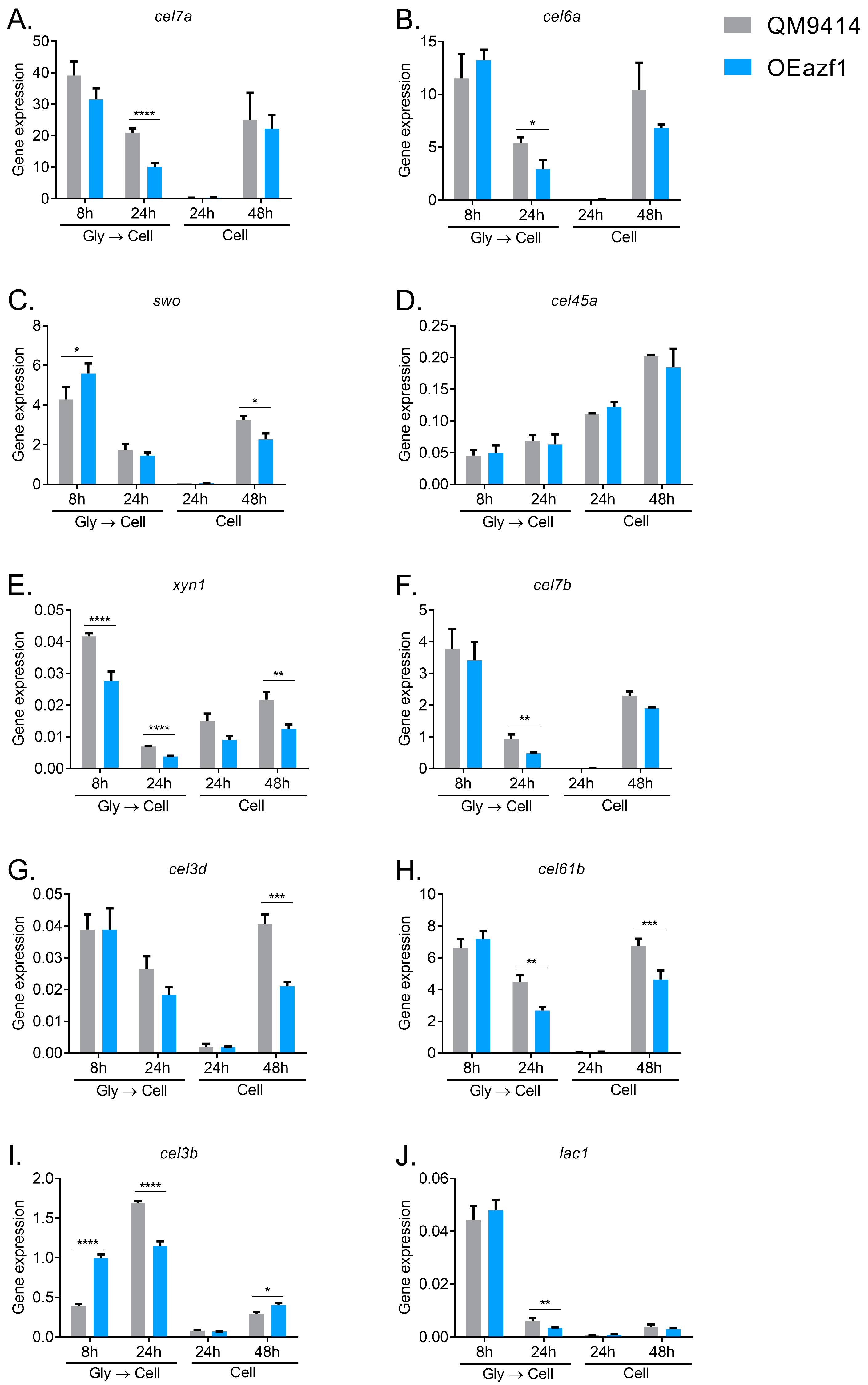
3.2. Azf1 Overexpression Decreases Cellulase Expression in the Presence of Cellulose
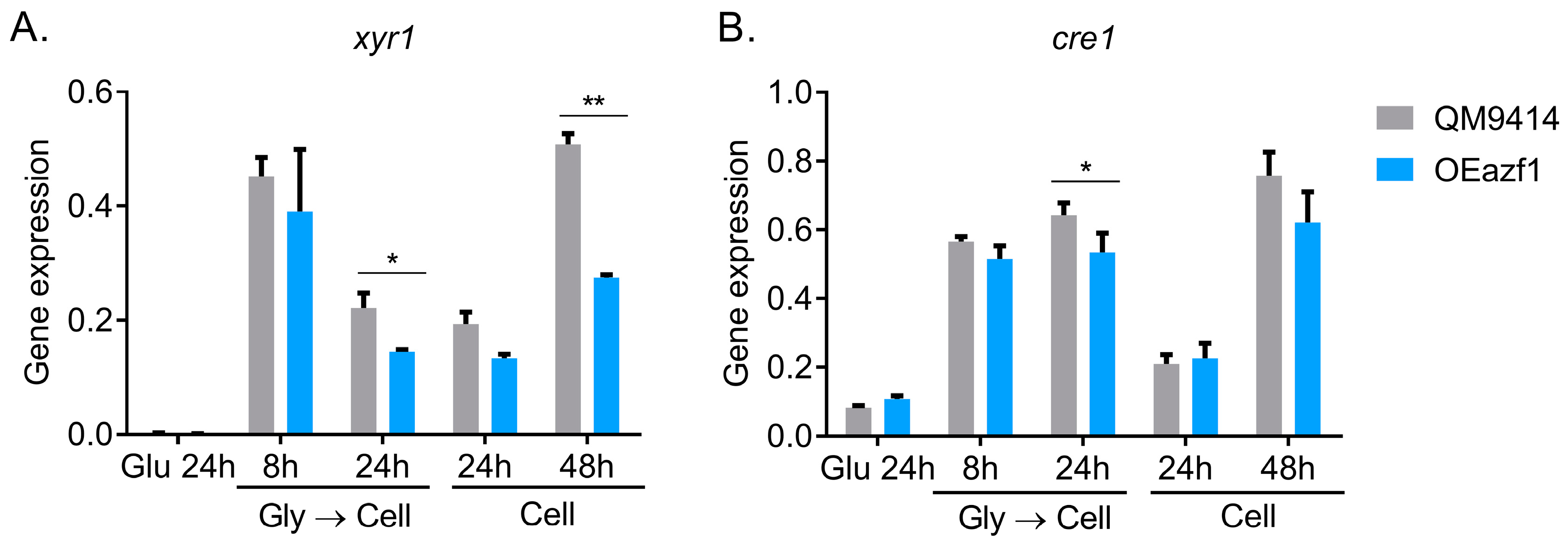
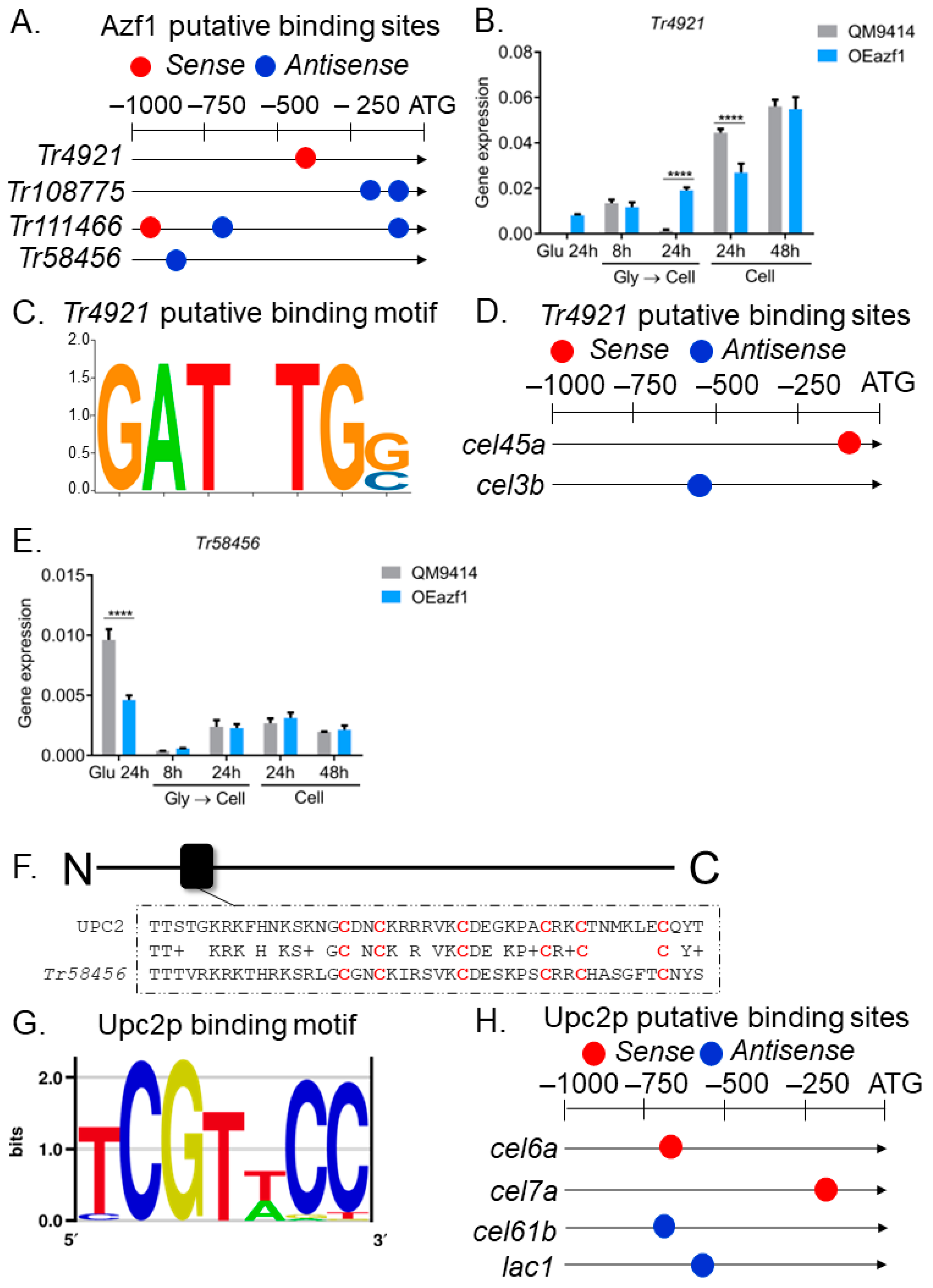
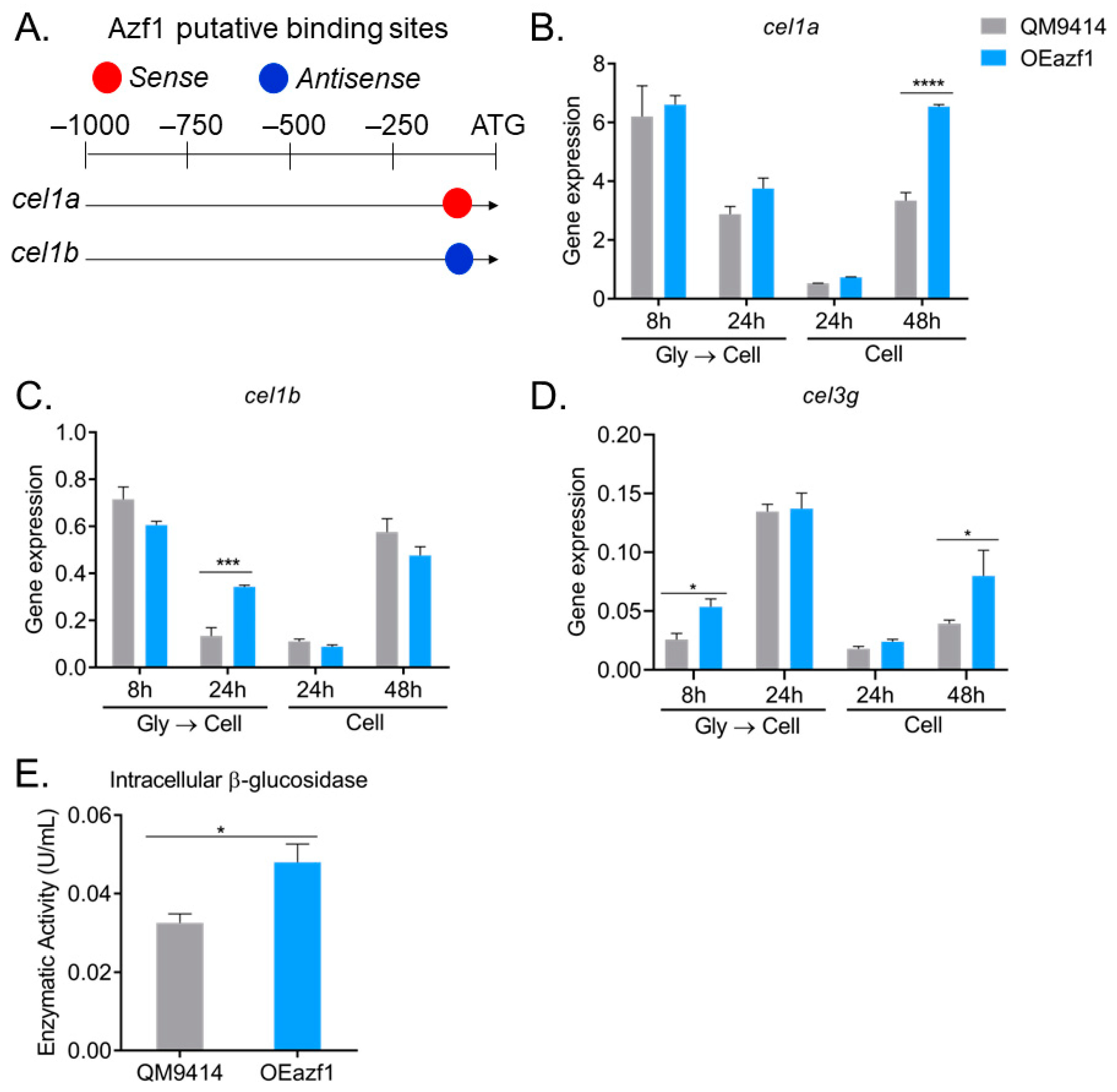
3.3. The Relationship between Azf1 Overexpression and Other Transcription Factors
3.4. Effects of Azf1 Overexpression on Cellulase Production
3.5. Azf1 Overexpression Impacts the Production of Intracellular β-Glucosidases
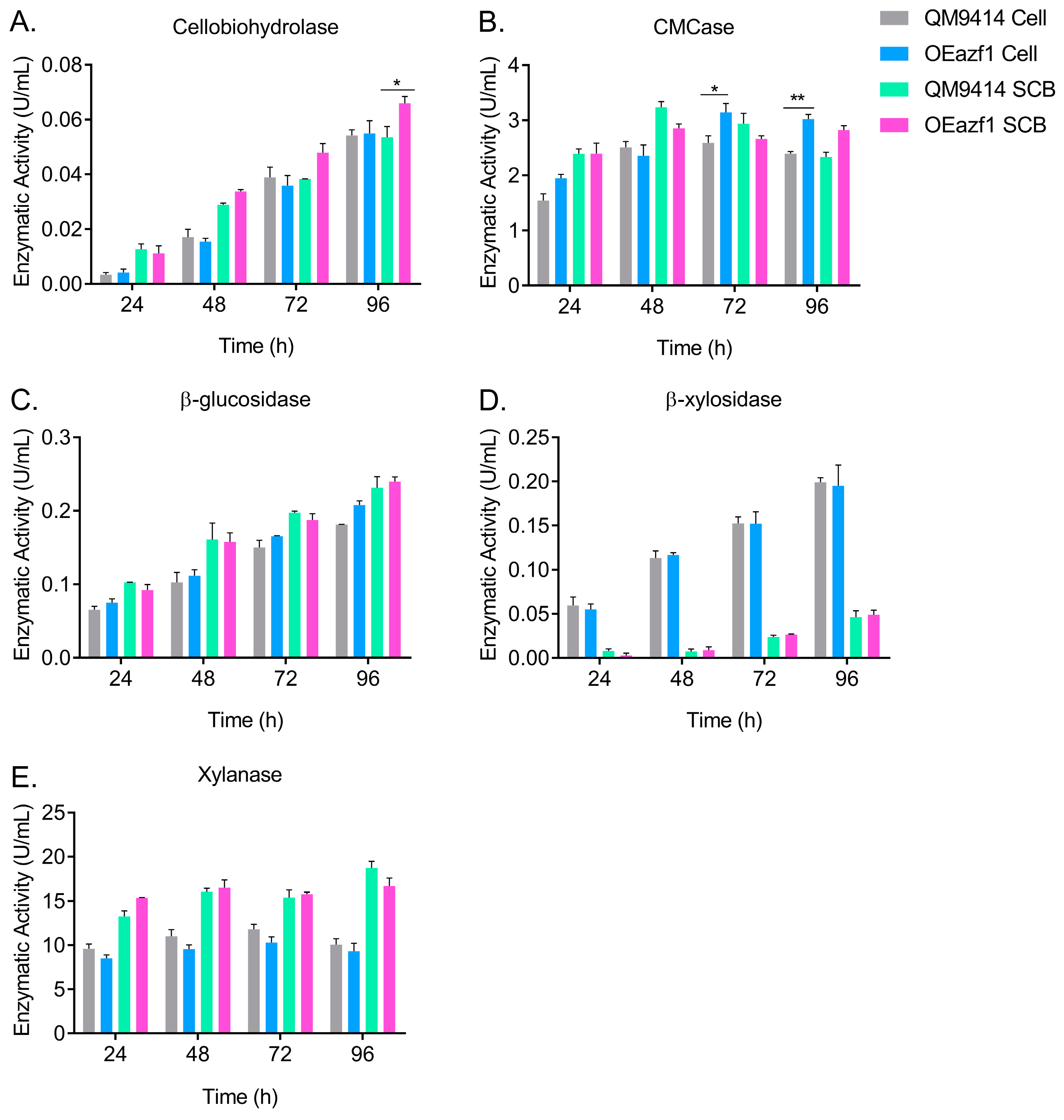
3.6. Azf1 Overexpression Increases Cellulase Production in Long-Term Cultivation
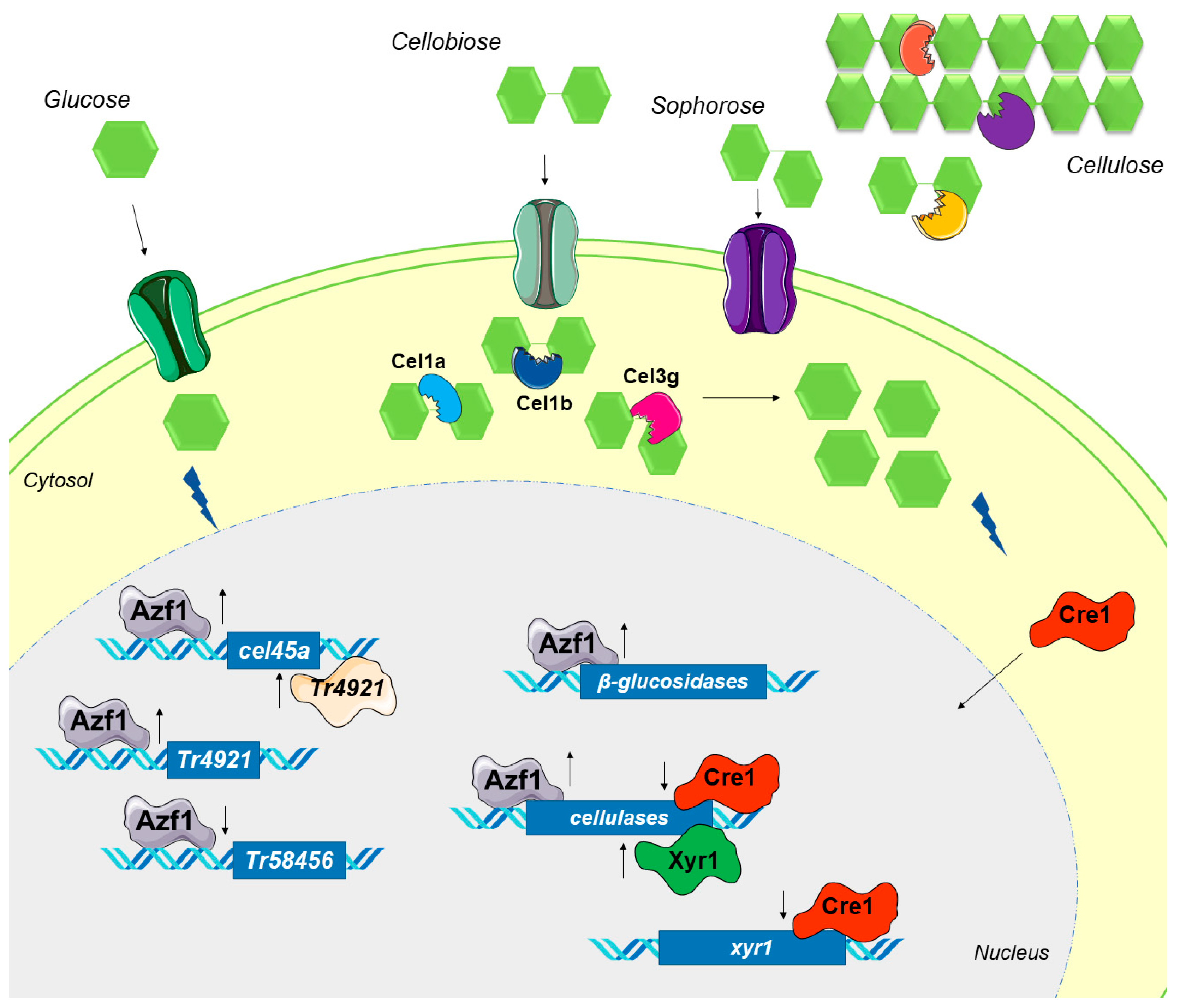
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baruah, J.; Nath, B.K.; Sharma, R.; Kumar, S.; Deka, R.C.; Baruah, D.C.; Kalita, E. Recent Trends in the Pretreatment of Lignocellulosic Biomass for Value-Added Products. Front. Energy Res. 2018, 6, 141. [Google Scholar] [CrossRef]
- Ali, N.; Zhang, Q.; Liu, Z.Y.; Li, F.L.; Lu, M.; Fang, X.C. Emerging Technologies for the Pretreatment of Lignocellulosic Materials for Bio-Based Products. Appl. Microbiol. Biotechnol. 2019, 104, 455–473. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Steindorff, A.S.; de Paula, R.G.; Silva-Rocha, R.; Mach-Aigner, A.R.; Mach, R.L.; Silva, R.N. The Post-Genomic Era of Trichoderma reesei: What’s Next? Trends Biotechnol. 2016, 34, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Häkkinen, M.; Arvas, M.; Oja, M.; Aro, N.; Penttilä, M.; Saloheimo, M.; Pakula, T.M. Re-Annotation of the CAZy Genes of Trichoderma reesei and Transcription in the Presence of Lignocellulosic Substrates. Microb. Cell Fact. 2012, 11, 134. [Google Scholar] [CrossRef] [PubMed]
- Martinez, D.; Berka, R.M.; Henrissat, B.; Saloheimo, M.; Arvas, M.; Baker, S.E.; Chapman, J.; Chertkov, O.; Coutinho, P.M.; Cullen, D.; et al. Genome Sequencing and Analysis of the Biomass-Degrading Fungus Trichoderma reesei (Syn. Hypocrea jecorina). Nat. Biotechnol. 2008, 26, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Novy, V.; Nielsen, F.; Seiboth, B.; Nidetzky, B. The Influence of Feedstock Characteristics on Enzyme Production in Trichoderma reesei: A Review on Productivity, Gene Regulation and Secretion Profiles. Biotechnol. Biofuels 2019, 12, 238. [Google Scholar] [CrossRef] [PubMed]
- Bazafkan, H.; Tisch, D.; Schmoll, M. Regulation of Glycoside Hydrolase Expression in Trichoderma. In Biotechnology and Biology of Trichoderma; Gupta, V.K., Schmoll, M., Herrera-Estrella, A., Upadhyay, R.S., Druzhinina, I., Tuohy, M.G., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; p. 527. ISBN 978-0-444-59576-8. [Google Scholar]
- Mandels, M.; Parrish, F.W.; Reese, E.T. Sophorose as an Inducer of Cellulase in Trichoderma viride. J. Bacteriol. 1962, 83, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Zheng, F.; Li, C.; Zhang, W.; Chen, G.; Liu, W. Characterization of a Copper Responsive Promoter and Its Mediated Overexpression of the Xylanase Regulator 1 Results in an Induction-Independent Production of Cellulases in Trichoderma reesei. Biotechnol. Biofuels 2015, 8, 67. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Y.; Wu, C.; Liu, P.; Wang, W.; Wei, D. The Transcription Factor ACE3 Controls Cellulase Activities and Lactose Metabolism via Two Additional Regulators in the Fungus Trichoderma reesei. J. Biol. Chem. 2019, 294, 18435–18450. [Google Scholar] [CrossRef]
- Wang, L.; Lv, X.; Cao, Y.; Zheng, F.; Meng, X.; Shen, Y.; Chen, G.; Liu, W.; Zhang, W. A Novel Transcriptional Regulator RXE1 Modulates the Essential Transactivator XYR1 and Cellulase Gene Expression in Trichoderma reesei. Appl. Microbiol. Biotechnol. 2019, 103, 4511–4523. [Google Scholar] [CrossRef]
- Wang, L.; Yang, R.; Cao, Y.; Zheng, F.; Meng, X.; Zhong, Y.; Chen, G.; Zhang, W.; Liu, W. CLP1, a Novel Plant Homeo Domain Protein, Participates in Regulating Cellulase Gene Expression in the Filamentous Fungus Trichoderma reesei. Front. Microbiol. 2019, 10, 100. [Google Scholar] [CrossRef]
- Antoniêto, A.C.C.; dos Santos Castro, L.; Silva-Rocha, R.; Persinoti, G.F.; Silva, R.N. Defining the Genome-Wide Role of CRE1 during Carbon Catabolite Repression in Trichoderma reesei Using RNA-Seq Analysis. Fungal Genet. Biol. 2014, 73, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zheng, F.; Wang, L.; Zhao, G.; Chen, G.; Zhang, W.; Liu, W. Rce1, a Novel Transcriptional Repressor, Regulates Cellulase Gene Expression by Antagonizing the Transactivator Xyr1 in Trichoderma reesei. Mol. Microbiol. 2017, 105, 65–83. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, W.; Liu, P.; Lin, A.; Fan, X.; Wu, C.; Li, N.; Wei, L.; Wei, D. The Novel Repressor Rce2 Competes with Ace3 to Regulate Cellulase Gene Expression in the Filamentous Fungus Trichoderma reesei. Mol. Microbiol. 2021, 116, 1298–1314. [Google Scholar] [CrossRef] [PubMed]
- Bröhl, S.; Lisowsky, T.; Riemen, G.; Michaelis, G. A New Nuclear Suppressor System for a Mitochondrial RNA Polymerase Mutant Identifies an Unusual Zinc-finger Protein and a Polyglutamine Domain Protein in Saccharomyces cerevisiae. Yeast 1994, 10, 719–731. [Google Scholar] [CrossRef]
- Stein, T.; Kricke, J.; Becher, D.; Lisowsky, T. Azf1p Is a Nuclear-Localized Zinc-Finger Protein That Is Preferentially Expressed under Non-Fermentative Growth Conditions in Saccharomyces cerevisiae. Curr. Genet. 1998, 34, 287–296. [Google Scholar] [CrossRef]
- Newcomb, L.L.; Hall, D.D.; Heideman, W. AZF1 Is a Glucose-Dependent Positive Regulator of CLN3 Transcription in Saccharomyces Serevisiae. Mol. Cell. Biol. 2002, 22, 1607–1614. [Google Scholar] [CrossRef]
- Slattery, M.G.; Liko, D.; Heideman, W. The Function and Properties of the Azf1 Transcriptional Regulator Change with Growth Conditions in Saccharomyces Serevisiae. Eukaryot. Cell 2006, 5, 313–320. [Google Scholar] [CrossRef]
- Antonieto, A.C.C.; Nogueira, K.M.V.; de Paula, R.G.; Nora, L.C.; Cassiano, M.H.A.; Guazzaroni, M.; Almeida, F.; da Silva, T.A.; Ries, L.N.A.; de Assis, L.J.; et al. A Novel Cys2His2 Zinc Finger Homolog of AZF1 Modulates Holocellulase Expression in Trichoderma reesei. mSystems 2019, 4, e00161-19. [Google Scholar] [CrossRef]
- Schmoll, M.; Schuster, A.; Silva, R.D.N.; Kubicek, C.P. The G-Alpha Protein GNA3 of Hypocrea jecorina (Anamorph Trichoderma reesei) Regulates Cellulase Gene Expression in the Presence of Light. Eukaryot. Cell 2009, 8, 410–420. [Google Scholar] [CrossRef]
- Nakari-Setala, T.; Penttila, M. Production of Trichoderma reesei Cellulases Glucose-Containing Media. Appl. Environ. Microbiol. 1995, 61, 3650–3655. [Google Scholar] [CrossRef] [PubMed]
- Nakari, T.; Alatalo, E.; Penttila, M.E. Isolation of Trichoderma reesei Genes Highly Expressed on Glucose- Containing Media: Characterization of the Tef1 Gene Encoding Translation Elongation Factor 1 Alpha. Gene 1993, 136, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Uzbas, F.; Sezerman, U.; Hartl, L.; Kubicek, C.P.; Seiboth, B. A Homologous Production System for Trichoderma reesei Secreted Proteins in a Cellulase-Free Background. Appl. Microbiol. Biotechnol. 2012, 93, 1601–1608. [Google Scholar] [CrossRef] [PubMed]
- Gruber, F.; Visser, J.; Kubicek, C.P.; de Graaff, L.H. The Development of a Heterologous Transformation System for the Cellulolytic Fungus Trichoderma reesei Based on a PyrG-Negative Mutant Strain. Curr. Genet. 1990, 18, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Steiger, M.G.; Mach, R.L.; Mach-Aigner, A.R. An Accurate Normalization Strategy for RT-QPCR in Hypocrea jecorina (Trichoderma reesei). J. Biotechnol. 2010, 145, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Persikov, A.V.; Singh, M. De Novo Prediction of DNA-Binding Specificities for Cys2His 2 Zinc Finger Proteins. Nucleic Acids Res. 2014, 42, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for Occurrences of a given Motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- De Boer, C.G.; Hughes, T.R. YeTFaSCo: A Database of Evaluated Yeast Transcription Factor Sequence Specificities. Nucleic Acids Res. 2012, 40, 169–179. [Google Scholar] [CrossRef]
- Gordân, R.; Murphy, K.F.; McCord, R.P.; Zhu, C.; Vedenko, A.; Bulyk, M.L. Curated Collection of Yeast Transcription Factor DNA Binding Specificity Data Reveals Novel Structural and Gene Regulatory Insights. Genome Biol. 2011, 12, R125. [Google Scholar] [CrossRef]
- Zou, G.; Jiang, Y.; Liu, R.; Zhu, Z.; Zhou, Z. The Putative β-Glucosidase BGL3I Regulates Cellulase Induction in Trichoderma reesei. Biotechnol. Biofuels 2018, 11, 314. [Google Scholar] [CrossRef]
- Nitta, M.; Furukawa, T.; Shida, Y.; Mori, K.; Kuhara, S.; Morikawa, Y.; Ogasawara, W. A New Zn(II) 2Cys 6-Type Transcription Factor BglR Regulates β-Glucosidase Expression in Trichoderma reesei. Fungal Genet. Biol. 2012, 49, 388–397. [Google Scholar] [CrossRef] [PubMed]
- Coradetti, S.T.; Xiong, Y.; Glass, N.L. Analysis of a Conserved Cellulase Transcriptional Regulator Reveals Inducer-Independent Production of Cellulolytic Enzymes in Neurospora crassa. Microbiologyopen 2013, 2, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yao, G.; Wu, R.; Gao, L.; Kan, Q.; Liu, M.; Yang, P.; Liu, G.; Qin, Y.; Song, X.; et al. Synergistic and Dose-Controlled Regulation of Cellulase Gene Expression in Penicillium oxalicum. PLoS Genet. 2015, 11, e1005509. [Google Scholar] [CrossRef] [PubMed]
- Mello-de-Sousa, T.M.; Rassinger, A.; Pucher, M.E.; dos Santos Castro, L.; Persinoti, G.F.; Silva-Rocha, R.; Poças-Fonseca, M.J.; Mach, R.L.; Nascimento Silva, R.; Mach-Aigner, A.R. The Impact of Chromatin Remodelling on Cellulase Expression in Trichoderma reesei. BMC Genom. 2015, 16, 588. [Google Scholar] [CrossRef] [PubMed]
- Ries, L.; Belshaw, N.J.; Ilmén, M.; Penttilä, M.E.; Alapuranen, M.; Archer, D.B. The Role of CRE1 in Nucleosome Positioning within the Cbh1 Promoter and Coding Regions of Trichoderma reesei. Appl. Microbiol. Biotechnol. 2014, 98, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Zeilinger, S.; Schmoll, M.; Pail, M.; Mach, R.L.; Kubicek, C.P. Nucleosome Transactions on the Hypocrea jecorina (Trichoderma reesei) Cellulase Promoter Cbh2 Associated with Cellulase Induction. Mol. Genet. Genom. 2003, 270, 46–55. [Google Scholar] [CrossRef]
- Mello-De-Sousa, T.M.; Rassinger, A.; Derntl, C.; Poças-Fonseca, M.J.; Mach, R.L.; Mach-Aigner, A.R. The Relation Between Promoter Chromatin Status, Xyr1 and Cellulase Expression in Trichoderma reesei. Curr. Genom. 2016, 17, 145–152. [Google Scholar] [CrossRef]
- Cao, Y.; Zheng, F.; Zhang, W.; Meng, X.; Liu, W. Trichoderma reesei XYR1 Recruits SWI/SNF to Facilitate Cellulase Gene Expression. Mol. Microbiol. 2019, 112, 1145–1162. [Google Scholar] [CrossRef]
- Seiboth, B.; Karimi, R.A.; Phatale, P.A.; Linke, R.; Hartl, L.; Sauer, D.G.; Smith, K.M.; Baker, S.E.; Freitag, M.; Kubicek, C.P. The Putative Protein Methyltransferase LAE1 Controls Cellulase Gene Expression in Trichoderma reesei. Mol. Microbiol. 2012, 84, 1150–1164. [Google Scholar] [CrossRef]
- Cao, Y.; Yang, R.; Zheng, F.; Meng, X.; Zhang, W.; Liu, W. Dual Regulatory Role of Chromatin Remodeler ISW1 in Coordinating Cellulase and Secondary Metabolite Biosynthesis in Trichoderma reesei. MBio 2022, 13, e03456-21. [Google Scholar] [CrossRef] [PubMed]
- Cziferszky, A.; Mach, R.L.; Kubicek, C.P. Phosphorylation Positively Regulates DNA Binding of the Carbon Catabolite Repressor Cre1 of Hypocrea jecorina (Trichoderma reesei). J. Biol. Chem. 2002, 277, 14688–14694. [Google Scholar] [CrossRef] [PubMed]
- Stricker, A.R.; Trefflinger, P.; Aro, N.; Penttilä, M.; Mach, R.L. Role of Ace2 (Activator of Cellulases 2) within the Xyn2 Transcriptosome of Hypocrea jecorina. Fungal Genet. Biol. 2008, 45, 436–445. [Google Scholar] [CrossRef]
- Ribeiro, L.F.C.; Chelius, C.; Boppidi, K.R.; Naik, N.S.; Hossain, S.; Ramsey, J.J.J.; Kumar, J.; Ribeiro, L.F.; Ostermeier, M.; Tran, B.; et al. Comprehensive Analysis of Aspergillus nidulans PKA Phosphorylome Identifies a Novel Mode of CreA Regulation. MBio 2019, 10, e02825-18. [Google Scholar] [CrossRef]
- de Assis, L.J.; Silva, L.P.; Bayram, O.; Dowling, P.; Kniemeyer, O.; Krüger, T.; Brakhage, A.A.; Chen, Y.; Dong, L.; Tan, K.; et al. Carbon Catabolite Repression in Filamentous Fungi Is Regulated by Phosphorylation of the Transcription Factor CreA. MBio 2021, 12, e03146-20. [Google Scholar] [CrossRef] [PubMed]
- Stappler, E.; Dattenböck, C.; Tisch, D.; Schmoll, M. Analysis of Light- and Carbon-Specific Transcriptomes Implicates a Class of G-Protein-Coupled Receptors in Cellulose Sensing. mSphere 2017, 2, e00089-17. [Google Scholar] [CrossRef]
- Collier, L.A.; Ghosh, A.; Borkovich, K.A. Heterotrimeric G-Protein Signaling Is Required for Cellulose Degradation in Neurospora crassa. MBio 2020, 11, e02419-20. [Google Scholar] [CrossRef]
- Vik, Å.; Rine, J. Upc2p and Ecm22p, Dual Regulators of Sterol Biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 6395–6405. [Google Scholar] [CrossRef]
- Bien, C.M.; Espenshade, P.J. Sterol Regulatory Element Binding Proteins in Fungi: Hypoxic Transcription Factors Linked to Pathogenesis. Eukaryot. Cell 2010, 9, 352–359. [Google Scholar] [CrossRef]
- Reilly, M.C.; Qin, L.; Craig, J.P.; Starr, T.L.; Glass, N.L. Deletion of Homologs of the SREPB Pathway Results in Hyper-Production of Cellulases in Neurospora crassa and Trichoderma reesei. Biotechnol. Biofuels 2015, 8, 121. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Wu, V.W.; Glass, N.L. Deciphering the Regulatory Network between the SREBP Pathway and Protein Secretion in Neurospora crassa. MBio 2017, 8, e00233-17. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhao, G.; Kou, Y.; Zhang, W.; Zhou, Q.; Chen, G.; Liu, W. Intracellular β-Glucosidases CEL1a and CEL1b Are Essential for Cellulase Induction on Lactose in Trichoderma reesei. Eukaryot. Cell 2014, 13, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Xu, J.; Kou, Y.; Lv, X.; Zhang, X.; Zhao, G.; Zhang, W.; Chen, G.; Liu, W. Differential Involvement of β-Glucosidases from Hypocrea jecorina in Rapid Induction of Cellulase Genes by Cellulose and Cellobiose. Eukaryot. Cell 2012, 11, 1371–1381. [Google Scholar] [CrossRef] [PubMed]
- Lichius, A.; Seidl-Seiboth, V.; Seiboth, B.; Kubicek, C.P. Nucleo-Cytoplasmic Shuttling Dynamics of the Transcriptional Regulators XYR1 and CRE1 under Conditions of Cellulase and Xylanase Gene Expression in Trichoderma reesei. Mol. Microbiol. 2014, 94, 1162–1178. [Google Scholar] [CrossRef]
- Pang, A.; Wang, H.; Luo, Y.; Yang, Z.; Liu, Z.; Wang, Z.; Li, B.; Yang, S.; Zhou, Z.; Lu, X.; et al. Dissecting Cellular Function and Distribution of β-Glucosidases in Trichoderma reesei. MBio 2021, 12, e03671-20. [Google Scholar] [CrossRef]
- Pang, A.-P.; Luo, Y.; Hu, X.; Zhang, F.; Wang, H.; Gao, Y.; Durrani, S.; Li, C.; Shi, X.; Wu, F.-G.; et al. Transmembrane Transport Process and Endoplasmic Reticulum Function Facilitate the Role of Gene Cel1b in Cellulase Production of Trichoderma reesei. Microb. Cell Fact. 2022, 21, 90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Kou, Y.; Xu, J.; Cao, Y.; Zhao, G.; Shao, J.; Wang, H.; Wang, Z.; Bao, X.; Chen, G.; et al. Two Major Facilitator Superfamily Sugar Transporters from Trichoderma reesei and Their Roles in Induction of Cellulase Biosynthesis. J. Biol. Chem. 2013, 288, 32861–32872. [Google Scholar] [CrossRef]
- Havukainen, S.; Valkonen, M.; Koivuranta, K.; Landowski, C.P. Studies on Sugar Transporter CRT1 Reveal New Characteristics That Are Critical for Cellulase Induction in Trichoderma reesei. Biotechnol. Biofuels 2020, 13, 158. [Google Scholar] [CrossRef]
- Yao, C.; Yan, M.; Li, K.; Gao, W.; Li, X.; Zhang, J.; Liu, H.; Zhong, Y. The ERAD Pathway Participates in Fungal Growth and Cellulase Secretion in Trichoderma reesei. J. Fungi 2023, 9, 74. [Google Scholar] [CrossRef]
- Shida, Y.; Yamaguchi, K.; Nitta, M.; Nakamura, A.; Takahashi, M.; Kidokoro, S.I.; Mori, K.; Tashiro, K.; Kuhara, S.; Matsuzawa, T.; et al. The Impact of a Single-Nucleotide Mutation of Bgl2 on Cellulase Induction in a Trichoderma reesei Mutant. Biotechnol. Biofuels 2015, 8, 230. [Google Scholar] [CrossRef]
- Znameroski, E.A.; Coradetti, S.T.; Roche, C.M.; Tsai, J.C.; Iavarone, A.T.; Cate, J.H.D.; Glass, N.L. Induction of Lignocellulose-Degrading Enzymes in Neurospora crassa by Cellodextrins. Proc. Natl. Acad. Sci. USA 2012, 109, 6012–6017. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maués, D.B.; Maraschin, J.C.; Duarte, D.Â.; Antoniêto, A.C.C.; Silva, R.N. Overexpression of the Transcription Factor Azf1 Reveals Novel Regulatory Functions and Impacts β-Glucosidase Production in Trichoderma reesei. J. Fungi 2023, 9, 1173. https://doi.org/10.3390/jof9121173
Maués DB, Maraschin JC, Duarte DÂ, Antoniêto ACC, Silva RN. Overexpression of the Transcription Factor Azf1 Reveals Novel Regulatory Functions and Impacts β-Glucosidase Production in Trichoderma reesei. Journal of Fungi. 2023; 9(12):1173. https://doi.org/10.3390/jof9121173
Chicago/Turabian StyleMaués, David Batista, Jhonatan Christian Maraschin, Diego Ângelo Duarte, Amanda Cristina Campos Antoniêto, and Roberto N. Silva. 2023. "Overexpression of the Transcription Factor Azf1 Reveals Novel Regulatory Functions and Impacts β-Glucosidase Production in Trichoderma reesei" Journal of Fungi 9, no. 12: 1173. https://doi.org/10.3390/jof9121173
APA StyleMaués, D. B., Maraschin, J. C., Duarte, D. Â., Antoniêto, A. C. C., & Silva, R. N. (2023). Overexpression of the Transcription Factor Azf1 Reveals Novel Regulatory Functions and Impacts β-Glucosidase Production in Trichoderma reesei. Journal of Fungi, 9(12), 1173. https://doi.org/10.3390/jof9121173





