Re-Evaluating Botryosphaeriales: Ancestral State Reconstructions of Selected Characters and Evolution of Nutritional Modes
Abstract
1. Introduction
1.1. Botryosphaeriales
1.2. Previous Revisions for the Families in Botryosphaeriales
1.3. Morphologies of Botryosphaerialean Taxa
1.4. Ancestral State Reconstructions for Fungi
1.5. Objectives of the Current Study
2. Materials and Methods
2.1. Data Collection and Analyses
2.2. Molecular Clock Analysis
2.3. Ancestral State Reconstructions
3. Results and Discussion
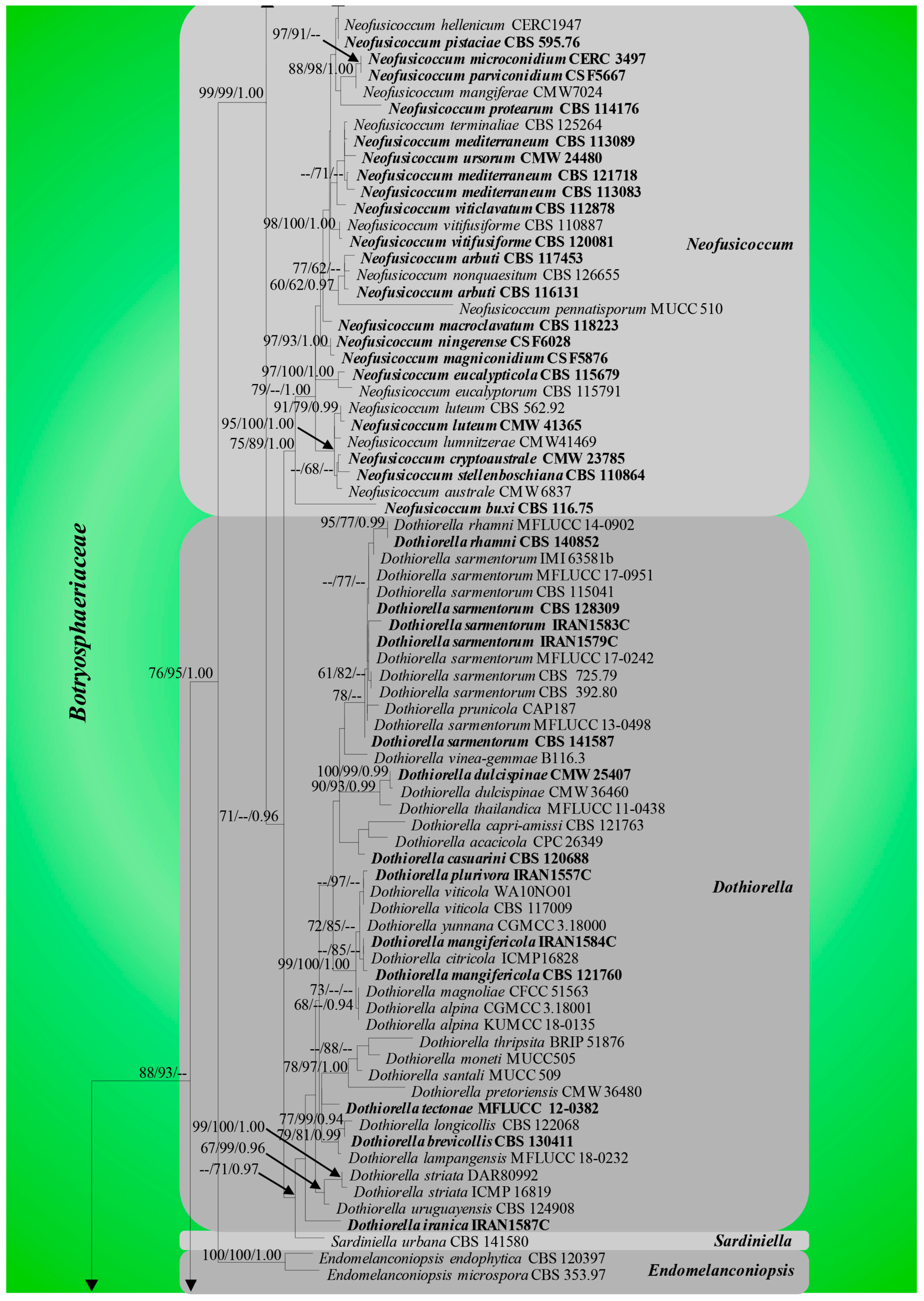
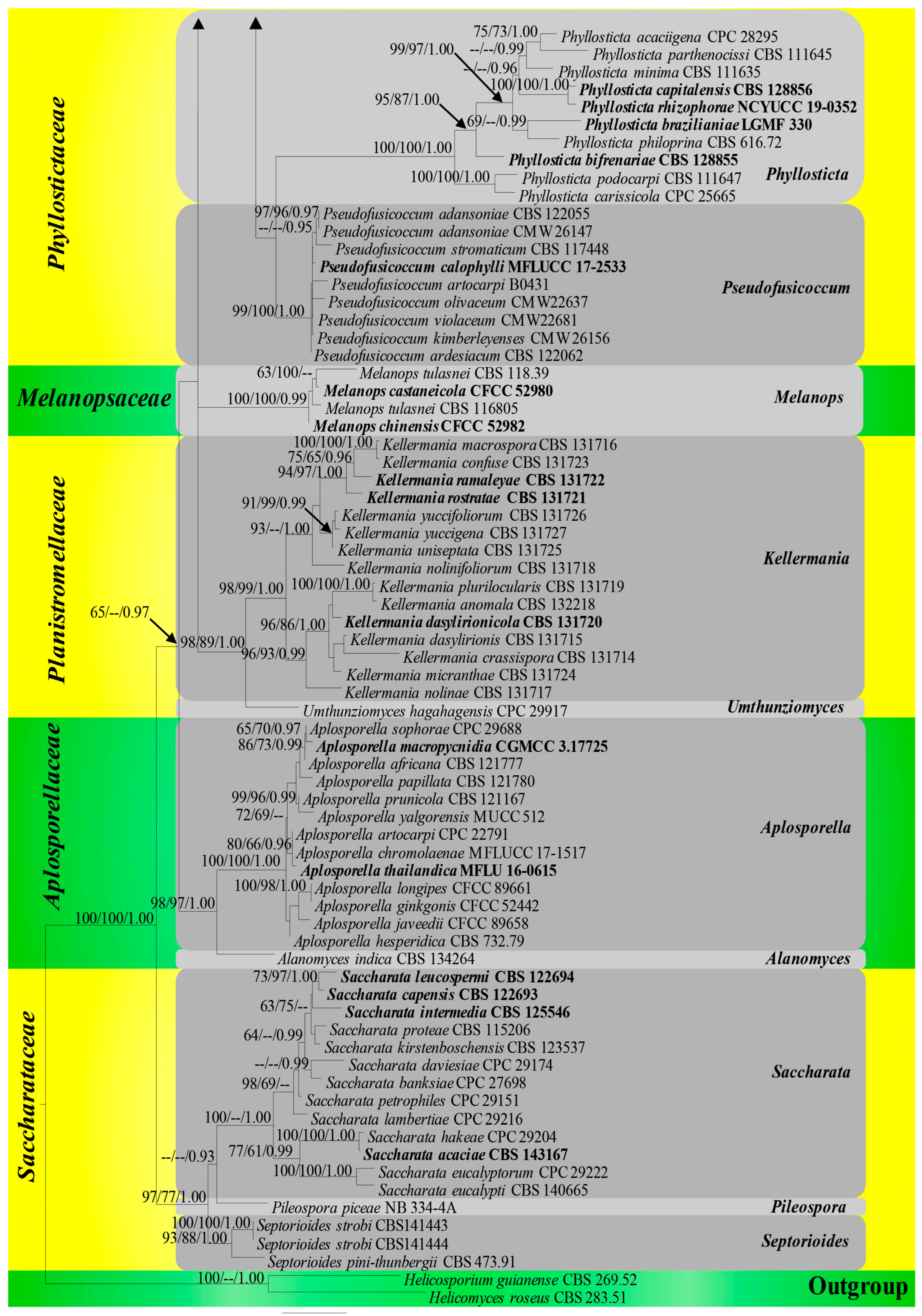
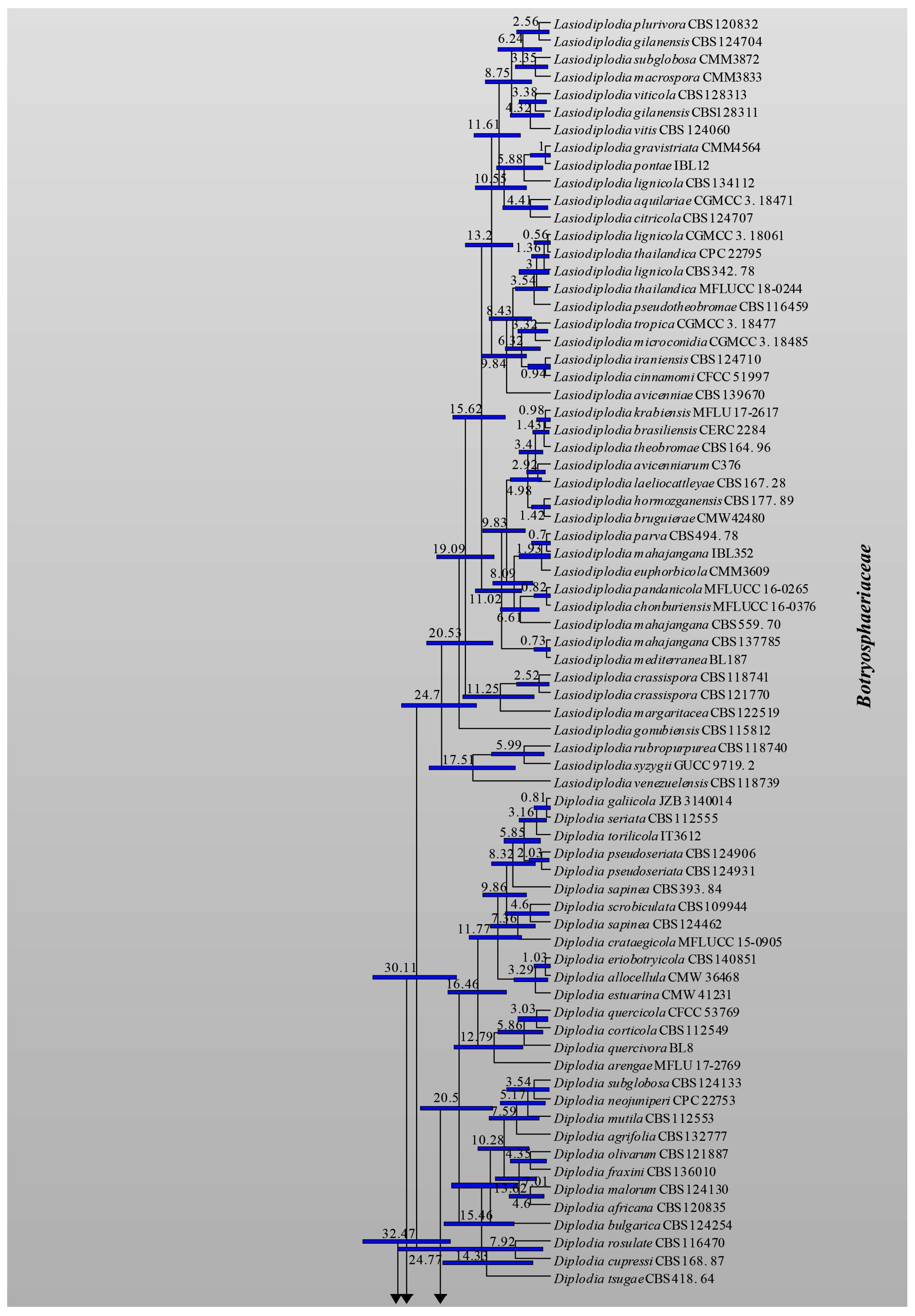
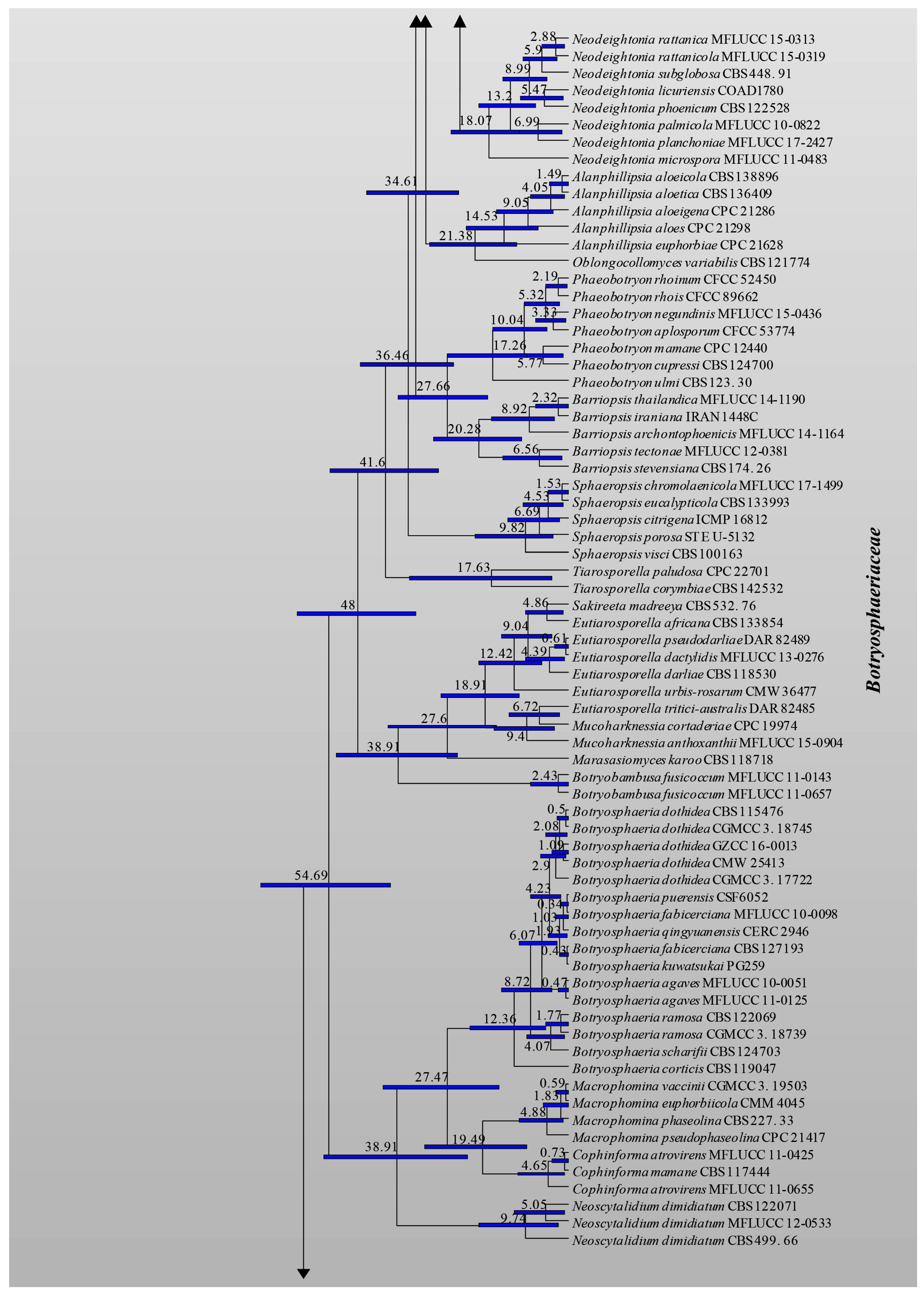
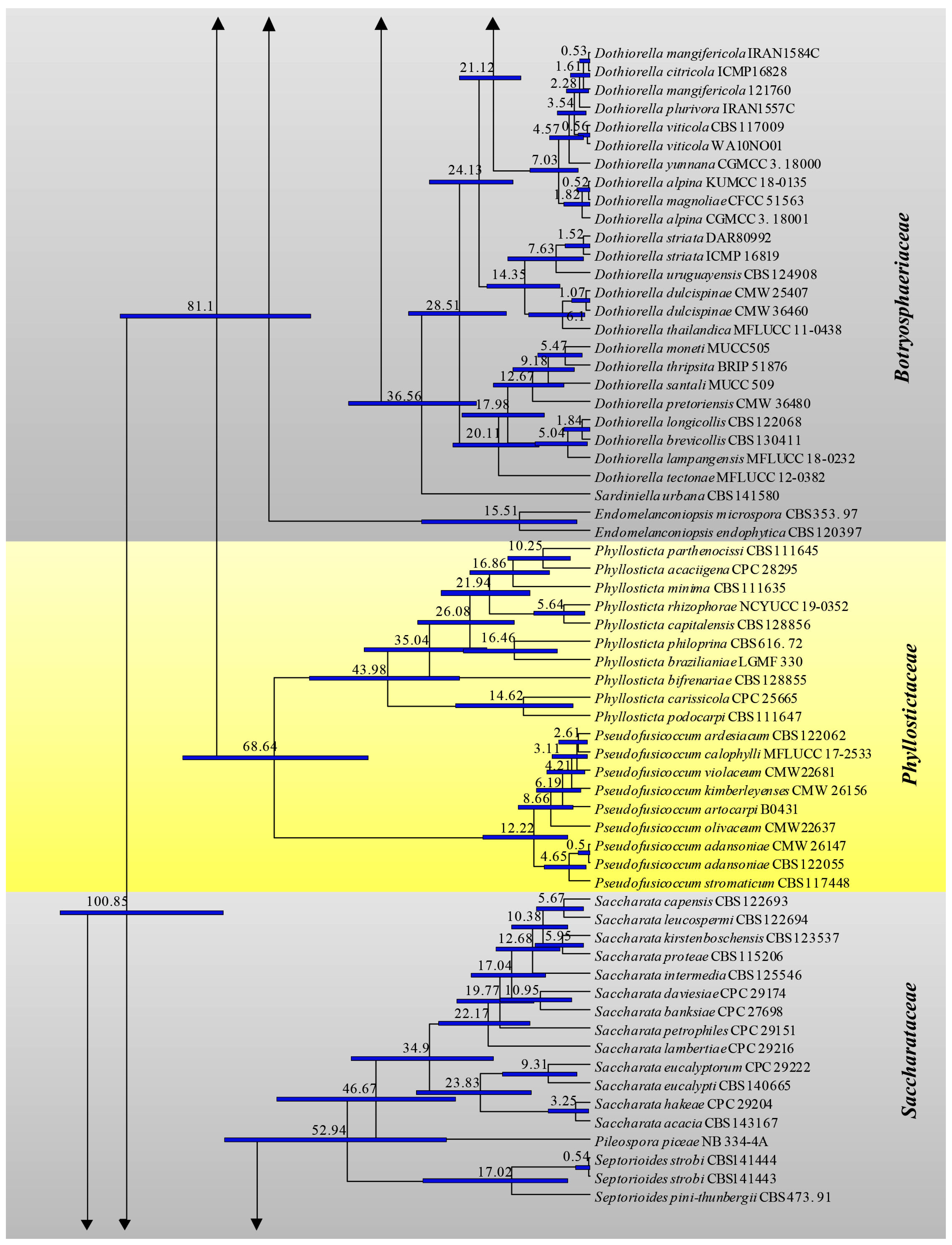
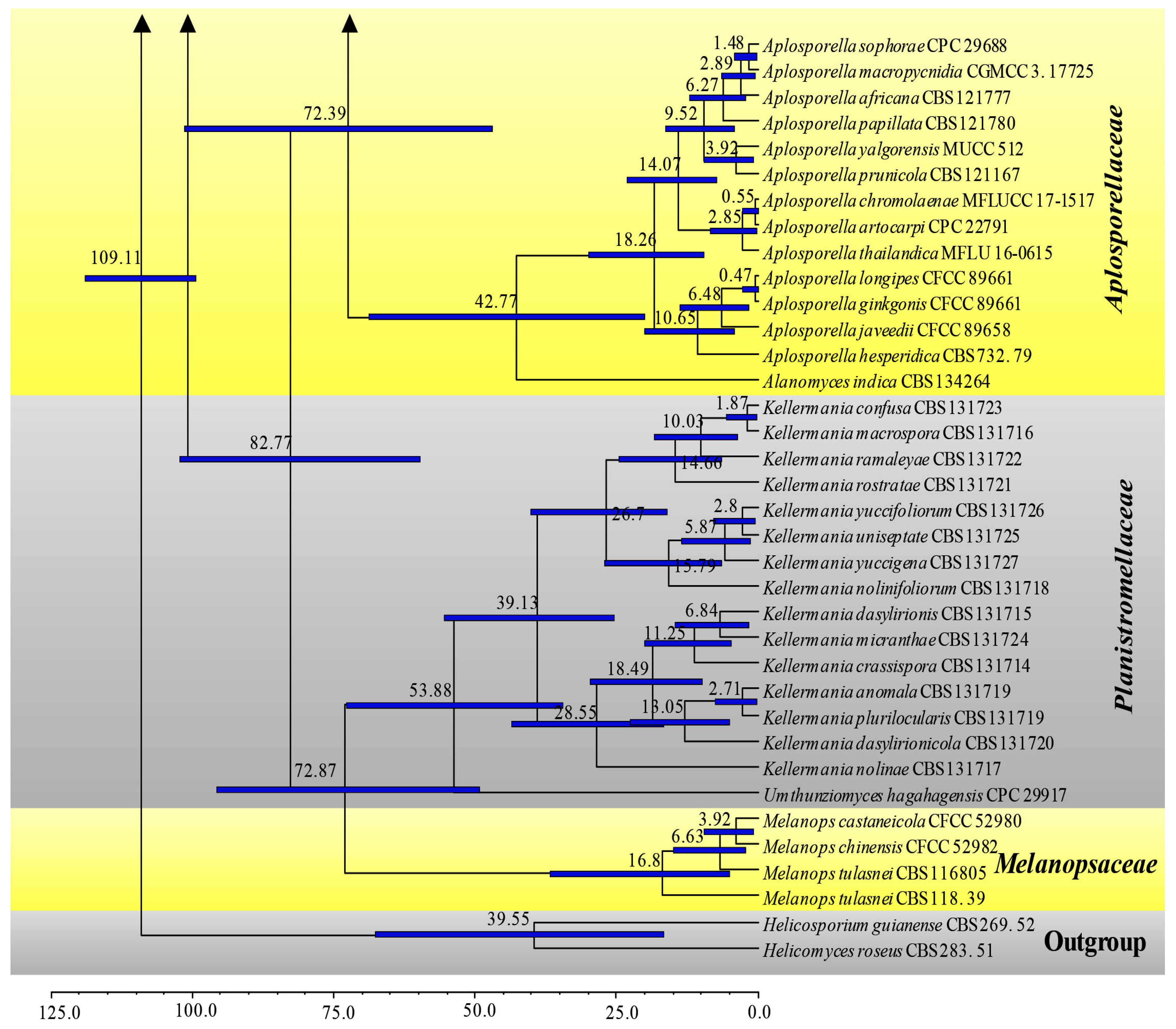
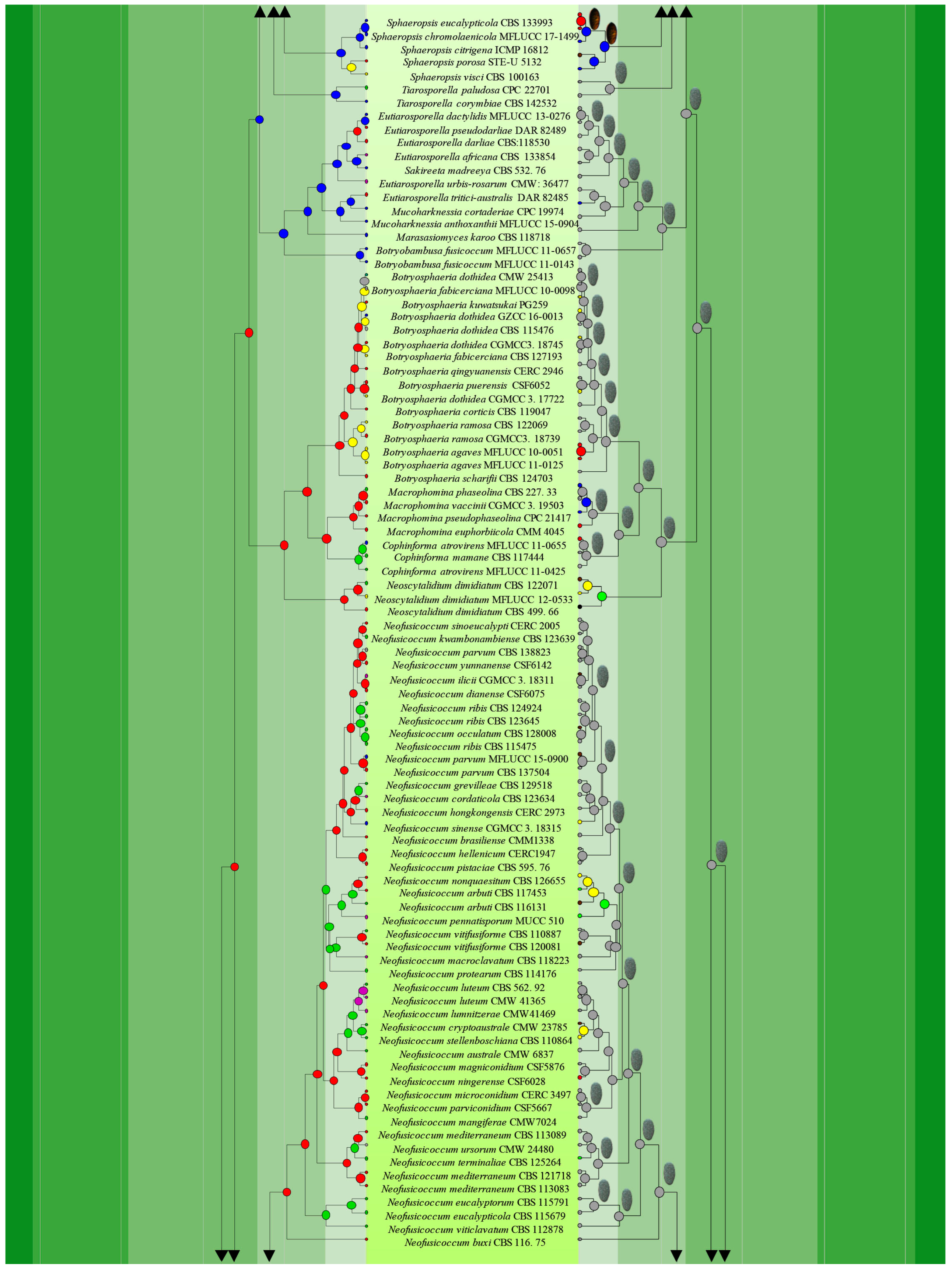
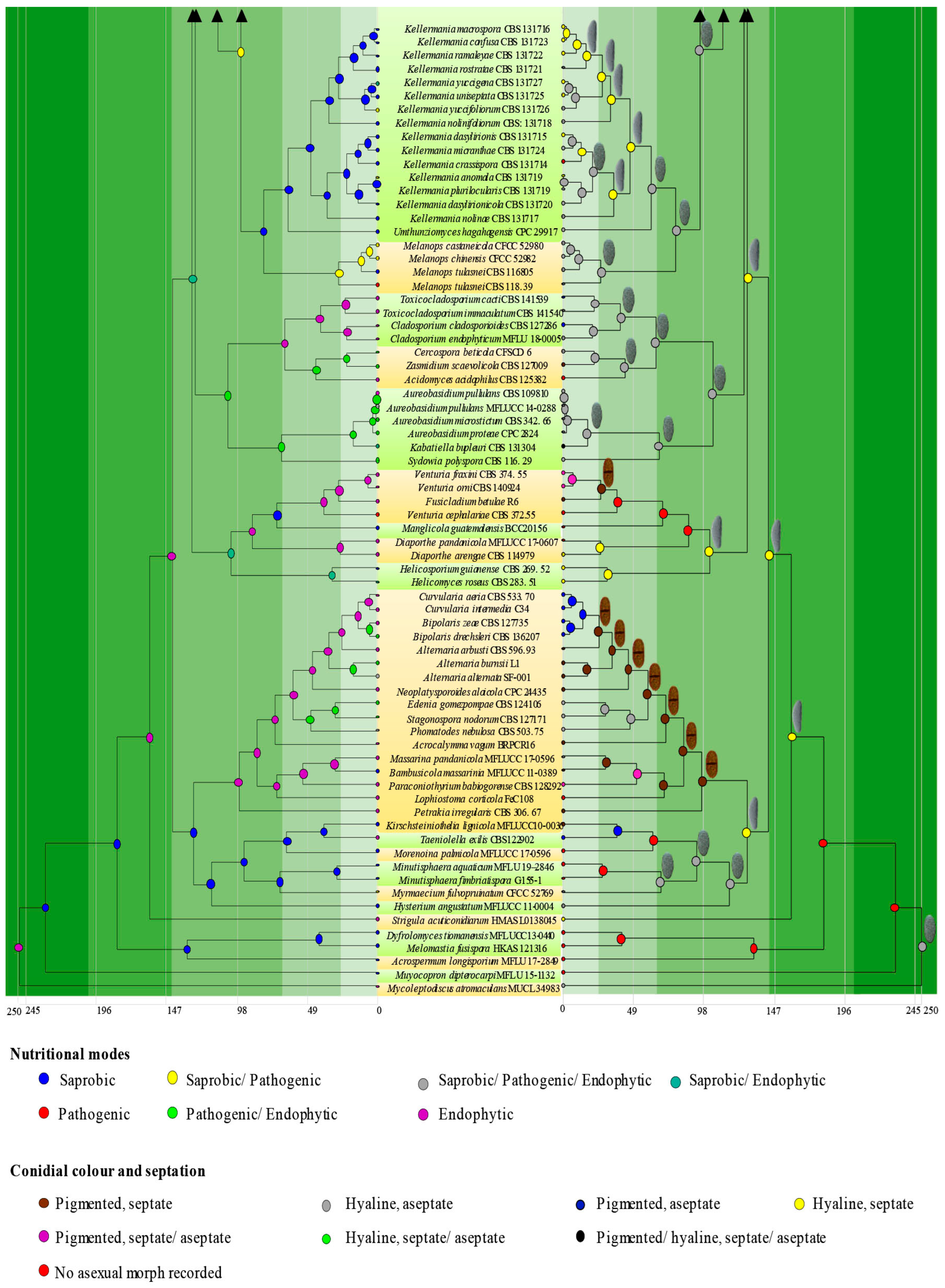
3.1. Phylogenetic Analyses
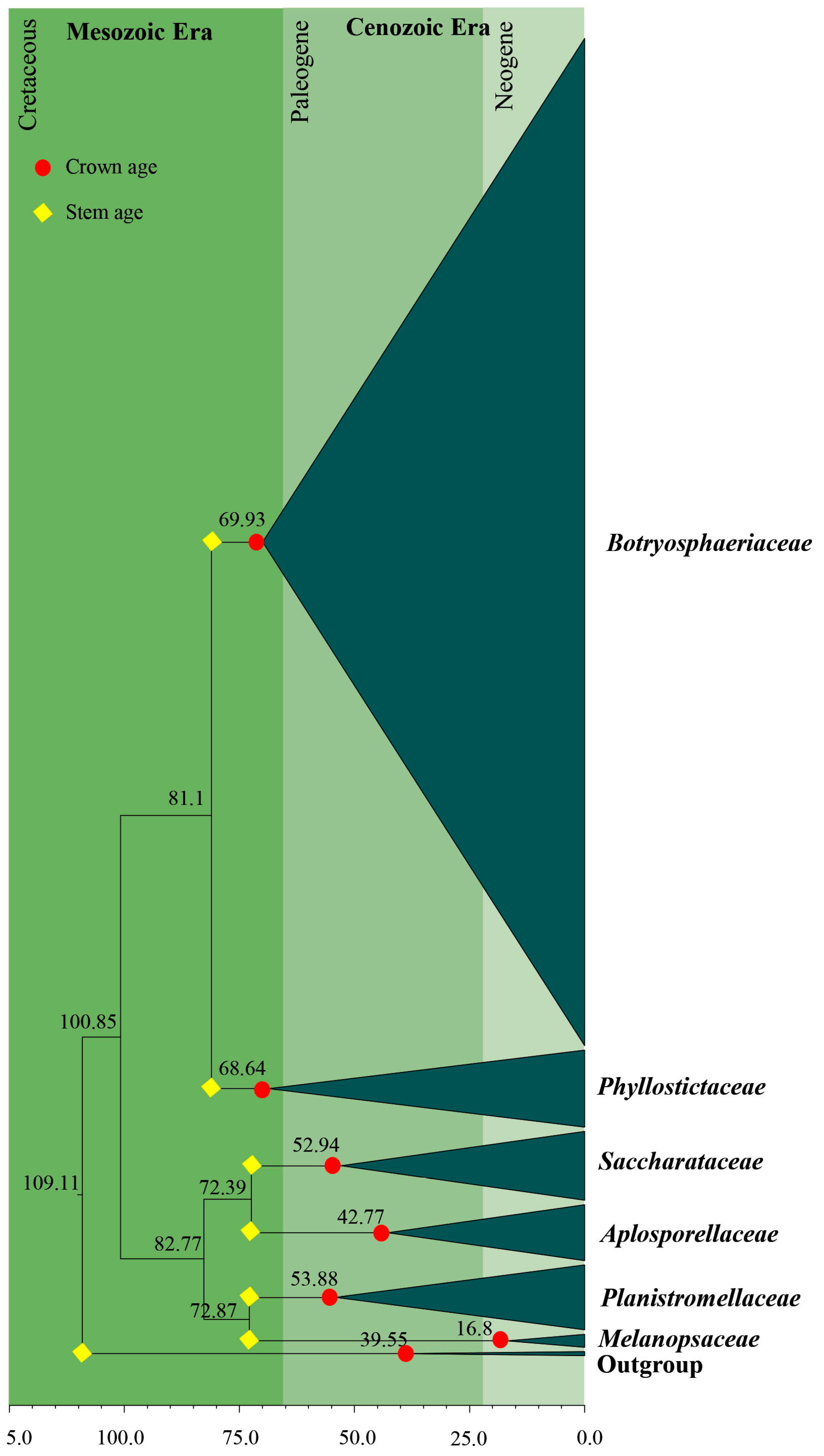
3.2. Divergence Times
3.3. Ancestral State Reconstructions
3.3.1. Ancestral State Reconstructions on Nutritional Modes of Botryosphaeriales Taxa
3.3.2. Ancestral State Reconstructions for Conidial Colour and Septation in Botryosphaeriales Taxa
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schoch, C.L.; Shoemaker, R.A.; Seifert, K.A.; Hambleton, S.; Spatafora, J.W.; Crous, P.W. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 2006, 98, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Minnis, A.M.; Kennedy, A.H.; Grenier, D.B.; Palm, M.E.; Rossman, A.Y. Phylogeny and taxonomic revision of the Planistromellaceae including its coelomycetous anamorphs: Contributions towards a monograph of the genus Kellermania. Pers. Mol. Phylogeny Evol. Fungi 2012, 29, 11–28. [Google Scholar] [CrossRef] [PubMed]
- Wikee, S.; Lombard, L.; Nakashima, C.; Motohashi, K.; Chukeatirote, E.; Cheewangkoon, R.; McKenzie, E.H.C.; Hyde, K.D.; Crous, P.W. A phylogenetic re-evaluation of Phyllosticta (Botryosphaeriales). Stud. Mycol. 2013, 76, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Slippers, B.; Boissin, E.; Phillips, A.J.L.; Groenewald, J.Z.; Lombard, L.; Wingfield, M.J.; Postma, A.; Burgess, T.; Crous, P.W. Phylogenetic lineages in the Botryosphaeriales: A systematic and evolutionary framework. Stud. Mycol. 2013, 76, 31–49. [Google Scholar] [CrossRef]
- Wyka, S.A.; Broders, K.D. The new family Septorioideaceae, within the Botryosphaeriales and Septorioides strobi as a new species associated with needle defoliation of Pinus strobus in the United States. Fungal Biol. 2016, 120, 1030–1040. [Google Scholar] [CrossRef]
- Yang, T.; Groenewald, J.; Cheewangkoon, R.; Jami, F.; Abdollahzadeh, J.; Lombard, L.; Crous, P.W. Families, genera, and species of Botryosphaeriales. Fungal Biol. 2016, 121, 322–346. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.L.; Hyde, K.D.; Alves, A.; Liu, J.K. (Jack) Families in Botryosphaeriales: A phylogenetic, morphological and evolutionary perspective. Fungal Divers. 2019, 94, 1–22. [Google Scholar] [CrossRef]
- Zhang, W.; Groenewald, J.Z.; Lombard, L.; Schumacher, R.K.; Phillips, A.J.L.; Crous, P.W. Evaluating species in Botryosphaeriales. Pers. Mol. Phylogeny Evol. Fungi 2021, 46, 63–115. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.A.; Dissanayake, A.J.; Manawasinghe, I.S.; Rathnayaka, A.R.; Liu, J.K.; Phillips, A.J.L.; Promputtha, I.; Hyde, K.D. https://botryosphaeriales.org/, an online platform for up-to-date classification and account of taxa of Botryosphaeriales. Database 2021, 2021, baab061. [Google Scholar] [CrossRef] [PubMed]
- Slippers, B.; Wingfield, M.J. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: Diversity, ecology and impact. Fungal Biol. Rev. 2007, 21, 90–106. [Google Scholar] [CrossRef]
- Liu, J.K.; Phookamsak, R.; Doilom, M.; Wikee, S.; Li, Y.M.; Ariyawansha, H.; Boonmee, S.; Chomnunti, P.; Dai, D.Q.; Bhat, J.D.; et al. Towards a natural classification of Botryosphaeriales. Fungal Divers. 2012, 57, 149–210. [Google Scholar] [CrossRef]
- Barr, M.E. Prodromus to Class Loculoascomycetes; University of Massachusetts: Amherst, MA, USA, 1987. [Google Scholar]
- Von Arx, J.A. Plant pathogenic fungi. Mycologia 1987, 79, 919–920. [Google Scholar] [CrossRef]
- Crous, P.W.; Wood, A.R.; Okada, G.; Groenewald, J.Z. Foliicolous microfungi occurring on Encephalartos. Pers. Mol. Phylogeny Evol. Fungi 2008, 21, 135–146. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Hardy, G.E.S.J.; Crane, C.; Barrett, S.; Le Roux, J.J.; Thangavel, R.; Guarro, J.; Stchigel, A.M.; et al. Fungal Planet description sheets: 469–557. Persoonia 2016, 37, 218–403. [Google Scholar] [CrossRef]
- Prusky, D.; Alkan, N.; Mengiste, T.; Fluhr, R. Quiescent and Necrotrophic lifestyle choice during postharvest disease development. Annu. Rev. Phytopathol. 2013, 51, 155–176. [Google Scholar] [CrossRef]
- Desprez-Loustau, M.L.; Marçais, B.; Nageleisen, L.M.; Piou, D.; Vannini, A. Interactive effects of drought and pathogens in forest trees. Ann. For. Sci. 2006, 63, 597–612. [Google Scholar] [CrossRef]
- Theissen, F.; Sydow, H. Vorentwurfe zu den Pseudosphaeriales. Ann. Mycol. 1918, 16, 1–34. [Google Scholar]
- Phillips, A.J.L.; Alves, A.; Abdollahzadeh, J.; Slippers, B.; Wingfield, M.J.; Groenewald, J.Z.; Crous, P.W. The Botryosphaeriaceae: Genera and species known from culture. Stud. Mycol. 2013, 76, 51–167. [Google Scholar] [CrossRef]
- Smith, H.; Wingfield, M.J.; Crous, P.W.; Coutinho, I.A. Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South Afr. J. Bot. 1996, 62, 86–88. [Google Scholar] [CrossRef]
- Denman, S.; Crous, P.W.; Taylor, J.E.; Kang, J.C.; Pascoe, I.; Wingfield, M.J. An overview of the taxonomic history of Botryosphaeria, and a re-evaluation of its anamorphs based on morphology and ITS rDNa phylogeny. Stud. Mycol. 2000, 45, 129–140. [Google Scholar]
- Phillips, A.J.L.; Oudemans, P.V.; Correia, A.; Alves, A. Characterisation and epitypification of Botryosphaeria corticis, the cause of blueberry cane canker. Fungal Divers. 2006, 21, 141–155. [Google Scholar]
- Huang, W.Y.; Cai, Y.Z.; Hyde, K.D.; Corke, H.; Sun, M. Biodiversity of endophytic fungi associated with 29 traditional Chinese medicinal plants. Fungal Divers. 2009, 33, 61–75. [Google Scholar]
- Pérez, C.A.; Wingfield, M.J.; Slippers, B.; Altier, N.A.; Blanchette, R.A. Endophytic and canker-associated Botryosphaeriaceae occurring on non-native Eucalyptus and native Myrtaceae trees in Uruguay. Fungal Divers. 2010, 41, 53–69. [Google Scholar] [CrossRef]
- Ghimire, S.R.; Charlton, N.D.; Bell, J.D.; Krishnamurthy, Y.L.; Craven, K.D. Biodiversity of fungal endophyte communities inhabiting switchgrass (Panicum virgatum L.) growing in the native tallgrass prairie of northern Oklahoma. Fungal Divers. 2010, 47, 19–27. [Google Scholar] [CrossRef]
- González, V.; Tello, M.L. The endophytic mycota associated with Vitis vinifera in central Spain. Fungal Divers. 2011, 47, 29–42. [Google Scholar] [CrossRef]
- Laurent, B.; Marchand, M.; Chancerel, E.; Saint-Jean, G.; Capdevielle, X.; Poeydebat, C.; Bellée, A.; Comont, G.; Villate, L.; Desprez-Loustau, M.L. A richer community of Botryosphaeriaceae within a less diverse community of fungal endophytes in grapevines than in adjacent forest trees revealed by a mixed metabarcoding strategy. Phytobiomes J. 2020, 4, 252–267. [Google Scholar] [CrossRef]
- Mohali, S.R.; Slippers, B.; Wingfield, M.J. Identification of Botryosphaeriaceae from Eucalyptus, Acacia and Pinus in Venezuela. Fungal Divers. 2007, 25, 103–125. [Google Scholar]
- Lazzizera, C.; Frisullo, S.; Alves, A.; Lopes, J.; Phillips, A.J.L. Phylogeny and morphology of Diplodia species on olives in Southern Italy and description of Diplodia olivarum sp. nov. Fungal Divers. 2008, 31, 63–71. [Google Scholar]
- Marincowitz, S.; Groenewald, J.Z.; Wingfield, M.J.; Crous, P.W. Species of Botryosphaeriaceae occurring on Proteaceae. Pers. Mol. Phylogeny Evol. Fungi 2008, 21, 111–118. [Google Scholar] [CrossRef]
- Burgess, T.I.; Tan, Y.P.; Garnas, J.; Edwards, J.; Scarlett, K.A.; Shuttleworth, L.A.; Daniel, R.; Dann, E.K.; Parkinson, L.E.; Dinh, Q.; et al. Current status of the Botryosphaeriaceae in Australia. Australas. Plant Pathol. 2019, 48, 35–44. [Google Scholar] [CrossRef]
- Garcia, J.F.; Lawrence, D.P.; Morales-Cruz, A.; Travadon, R.; Minio, A.; Hernandez-Martinez, R.; Rolshausen, P.E.; Baumgartner, K.; Cantu, D. Phylogenomics of plant-associated Botryosphaeriaceae species. Front. Microbiol. 2021, 12, 652802. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, A.J.; Chen, Y.Y.; Cheewangkoon, R.; Liu, J.K. Occurrence and morpho-molecular identification of Botryosphaeriales species from Guizhou Province, China. J. Fungi 2021, 7, 893. [Google Scholar] [CrossRef] [PubMed]
- Wijayawardene, N.N.; Hyde, K.D.; Dai, D.Q.; Sánchez-García, M.; Goto, B.T.; Saxena, R.K.; Erdoğdu, M.; Selçuk, F.; Rajeshkumar, K.C.; Aptroot, A.; et al. Outline of Fungi and fungus-like taxa—2021. Mycosphere 2022, 13, 53–453. [Google Scholar] [CrossRef]
- Batista, E.; Lopes, A.; Alves, A. What do we know about Botryosphaeriaceae? An overview of a worldwide cured dataset. Forests 2021, 12, 313. [Google Scholar] [CrossRef]
- Ekanayaka, A.H.; Dissanayake, A.J.; Jayasiri, S.C.; To-anun, C.; Jones, G.E.B.; Zhao, Q.; Hyde, K.D. Aplosporella thailandica; a novel species revealing the sexual-asexual connection in Aplosporellaceae (Botryosphaeriales). Mycosphere 2016, 7, 440–447. [Google Scholar] [CrossRef]
- Sharma, R.; Kulkarni, G.; Sonawane, M.S. Alanomyces, a new genus of Aplosporellaceae based on four loci phylogeny. Phytotaxa 2017, 297, 168–175. [Google Scholar] [CrossRef]
- Okane, I.; Lumyong, S.; Nakagiri, A.; Ito, T. Extensive host range of an endophytic fungus, Guignardia endophyllicola (anamorph, Phyllosticta capitalensis). Mycoscience 2003, 44, 353–363. [Google Scholar] [CrossRef]
- Glienke, C.; Pereira, O.L.; Stringari, D.; Fabris, J.; Kava-Cordeiro, V.; Galli-Terasawa, L.; Cunnington, J.; Shivas, R.G.; Groenewald, J.Z.; Crous, P.W. Endophytic and pathogenic Phyllosticta species, with reference to those associated with Citrus Black spot. Pers. Mol. Phylogeny Evol. Fungi 2011, 26, 47–56. [Google Scholar] [CrossRef]
- Wikee, S.; Udayanga, D.; Crous, P.W.; Chukeatirote, E.; McKenzie, E.H.C.; Bahkali, A.H.; Dai, D.; Hyde, K.D. Phyllosticta—An overview of current status of species recognition. Fungal Divers. 2011, 51, 43–61. [Google Scholar] [CrossRef]
- Wikee, S.; Lombard, L.; Crous, P.W.; Nakashima, C.; Motohashi, K.; Chukeatirote, E.; Alias, S.A.; McKenzie, E.H.C.; Hyde, K.D. Phyllosticta capitalensis, a widespread endophyte of plants. Fungal Divers. 2013, 60, 91–105. [Google Scholar] [CrossRef]
- Rashmi, M.; Kushveer, J.S.; Sarma, V.V. A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere 2019, 10, 798–1079. [Google Scholar] [CrossRef]
- Barr, M.E. Planistromellaceae, a new family in the Dothideales. Mycotaxon 1996, 60, 433–442. [Google Scholar]
- Halbwachs, H.; Brandl, R.; Bässler, C. Spore wall traits of ectomycorrhizal and saprotrophic agarics may mirror their distinct lifestyles. Fungal Ecol. 2015, 17, 197–204. [Google Scholar] [CrossRef]
- Wong, H.J.; Mohamad-Fauzi, N.; Rizman-Idid, M.; Convey, P.; Alias, S.A. Protective mechanisms and responses of micro-fungi towards ultraviolet-induced cellular damage. Polar Sci. 2019, 20, 19–34. [Google Scholar] [CrossRef]
- Ho, W.H.; Hyde, K.D. A new type of conidial septal pore in fungi. Fungal Divers. 2004, 15, 171–186. [Google Scholar]
- Belozerskaya, T.A.; Gessler, N.N.; Aver, A.A. Fungal Metabolites. In Fungal Metabolites Reference Series in Phytochemistry; Mérillon, J.-M., Ramawat, K.G., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 263–291. ISBN 978-3-31925-001-4. [Google Scholar]
- Nosanchuk, J.D.; Casadevall, A. The contribution of melanin to microbial pathogenesis. Cell. Microbiol. 2003, 5, 203–223. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.L.; Alves, A.; Pennycook, S.R.; Johnston, P.R.; Ramaley, A.; Akulov, A.; Crous, P.W. Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in the Botryosphaeriaceae. Persoonia 2008, 21, 29–55. [Google Scholar] [CrossRef]
- Phillips, A.J.L.; Alves, A. Taxonomy, phylogeny, and epitypification of Melanops tulasnei, the type species of Melanops. Fungal Divers. 2009, 38, 155–166. [Google Scholar]
- Ekman, S.; Andersen, H.L.; Wedin, M. The limitations of ancestral state reconstruction and the evolution of the ascus in the Lecanorales (Lichenized Ascomycota). Syst. Biol. 2008, 57, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Mardones, M.; Trampe-Jaschik, T.; Oster, S.; Elliott, M.; Urbina, H.; Schmitt, I.; Piepenbring, M. Phylogeny of the order Phyllachorales (Ascomycota, Sordariomycetes): Among and within order relationships based on five molecular loci. Pers. Mol. Phylogeny Evol. Fungi 2017, 39, 74–90. [Google Scholar] [CrossRef]
- Thiyagaraja, V.; Lücking, R.; Ertz, D.; Wanasinghe, D.N.; Karunarathna, S.C.; Camporesi, E.; Hyde, K.D. Evolution of non-lichenized, saprotrophic species of Arthonia (Ascomycota, Arthoniales) and resurrection of Naevia, with notes on Mycoporum. Fungal Divers. 2020, 102, 205–224. [Google Scholar] [CrossRef]
- Chethana, K.W.T.; Jayawardena, R.S.; Chen, Y.J.; Konta, S.; Tibpromma, S.; Phukhamsakda, C.; Abeywickrama, P.D.; Samarakoon, M.C.; Senwanna, C.; Mapook, A.; et al. Appressorial interactions with host and their evolution; Springer: Amsterdam, The Netherlands, 2021; Volume 110, ISBN 01-2-3456-789. [Google Scholar]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Dissanayake, A.J.; Bhunjun, C.S.; Maharachchikumbura, S.S.N.; Liu, J.K. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 2020, 11, 2652–2676. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest v2. Program distributed by the author. Evol. Biol. Cent. Uppsala Univ. 2004, 2, 1–2. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; ISBN 978-1-42449-752-2. [Google Scholar]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP: Phylogenetic analysis using parsimony, version 4.0 b10. Sinauer Assoc. Sunderl. 2002, 56, 1776–1788. [Google Scholar] [CrossRef]
- Rambaut, A. Fig. Tree. Tree Fig. Drawing Tool, v. 1.4.0. 2012. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 1 May 2021).
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef]
- Rambaut, A.; Suchard, M.A.; Xie, D.; Drummond, A.J.; Tracer Version 1.6. University of Edinburgh. 2013. Available online: http://tree.bio.ed.ac.uk/software/tracer (accessed on 10 April 2021).
- Yu, Y.; Harris, A.J.; Blair, C.; He, X. RASP (Reconstruct Ancestral State in Phylogenies): A tool for historical biogeography. Mol. Phylogenet. Evol. 2015, 87, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Blair, C.; He, X. RASP 4: Ancestral state reconstruction tool for multiple genes and characters. Mol. Phylogenet. Evol. 2019, 37, 604–606. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ortega, S.; Garrido-Benavent, I.; Grube, M.; Olmo, R.; De los Ríos, A. Hidden diversity of marine borderline lichens and a new order of fungi: Collemopsidiales (Dothideomyceta). Fungal Divers. 2016, 80, 285–300. [Google Scholar] [CrossRef]
- Liu, J.K.; Hyde, K.D.; Jeewon, R.; Phillips, A.J.L.; Maharachchikumbura, S.S.N.; Ryberg, M.; Liu, Z.Y.; Zhao, Q. Ranking higher taxa using divergence times: A case study in Dothideomycetes. Fungal Divers. 2017, 84, 75–99. [Google Scholar] [CrossRef]
- Liu, N.G.; Ariyawansa, H.A.; Hyde, K.D.; Maharachchikumbura, S.S.N.; Zhao, R.L.; Phillips, A.J.L.; Jayawardena, R.S.; Thambugala, K.M.; Dissanayake, A.J.; Wijayawardene, N.N.; et al. Perspectives into the value of genera, families and orders in classification. Mycosphere 2016, 7, 1649–1668. [Google Scholar] [CrossRef]
- Lutzoni, F.; Nowak, M.D.; Alfaro, M.E.; Reeb, V.; Miadlikowska, J.; Krug, M.; Arnold, A.E.; Lewis, L.A.; Swofford, D.L.; Hibbett, D.; et al. Contemporaneous radiations of fungi and plants linked to symbiosis. Nat. Commun. 2018, 9, 5451. [Google Scholar] [CrossRef]
- Hyde, K.D.; Bao, D.F.; Hongsanan, S.; Chethana, K.W.T.; Yang, J.; Suwannarach, N. Evolution of freshwater Diaporthomycetidae (Sordariomycetes) provides evidence for five new orders and six new families. Fungal Divers. 2021, 107, 71–105. [Google Scholar] [CrossRef]
- Peace, A.L.; Phethean, J.J.J.; Franke, D.; Foulger, G.R.; Schiffer, C.; Welford, J.K.; McHone, G.; Rocchi, S.; Schnabel, M.; Doré, A.G. A review of Pangaea dispersal and Large Igneous Provinces—In search of a causative mechanism. Earth Sci. Rev. 2020, 206, 102902. [Google Scholar] [CrossRef]
- Heckman, D.S.; Geiser, D.M.; Eidell, B.R.; Stauffer, R.L.; Kardos, N.L.; Hedges, S.B. Molecular evidence for the early colonization of land by fungi and plants. Science 2001, 293, 1129–1133. [Google Scholar] [CrossRef]
- Hyde, K.D.; Soytong, K. The fungal endophyte dilemma. Fungal Divers. 2008, 33, 163–173. [Google Scholar]
- Promputtha, I.; Hyde, K.D.; McKenzie, E.H.C.; Peberdy, J.F.; Lumyong, S. Can leaf degrading enzymes provide evidence that endophytic fungi becoming saprobes? Fungal Divers. 2010, 41, 89–99. [Google Scholar] [CrossRef]
- Purahong, W.; Hyde, K.D. Effects of fungal endophytes on grass and non-grass litter decomposition rates. Fungal Divers. 2011, 47, 1–7. [Google Scholar] [CrossRef]
- Aime, M.C.; Toome, M.; McLaughlin, D.J. Pucciniomycotina. In Systematics and Evolution; Springer: Berlin/Heidelberg, Germany, 2014; pp. 271–294. ISBN 978-3-64255-318-9. [Google Scholar]
- Oberwinkler, F. Yeasts in Pucciniomycotina. Mycol. Prog. 2017, 16, 831–856. [Google Scholar] [CrossRef]
- Naranjo-Ortiz, M.A.; Gabaldón, T. Fungal evolution: Major ecological adaptations and evolutionary transitions. Biol. Rev. 2019, 94, 1443–1476. [Google Scholar] [CrossRef] [PubMed]
- Kalgutkar, R.M. Paleogene fungal palynomorphs from Bonnet Plume Formation, Yukon Territory. Contrib. Can. Paleontol. Geol. Surv. Can. Bull. 1993, 444, 51–105. [Google Scholar]
- Taylor, T.N.; Krings, M. Fossil microorganisms and land plants: Associations and interactions. Symbiosis 2005, 40, 119–135. [Google Scholar]
- Krings, M.; Dotzler, N.; Taylor, T.N.; Galtier, J. A Late Pennsylvanian fungal leaf endophyte from Grand-Croix, France. Rev. Palaeobot. Palynol. 2009, 156, 449–453. [Google Scholar] [CrossRef]
- Schoch, C.L.; Sung, G.H.; López-Giráldez, F.; Townsend, J.P.; Miadlikowska, J.; Hofstetter, V.; Robbertse, B.; Matheny, P.B.; Kauff, F.; Wang, Z.; et al. The Ascomycota tree of life: A phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst. Biol. 2009, 58, 224–239. [Google Scholar] [CrossRef]
- Savile, D.B.O. Possible interrelationships between fungal groups. In The Fungi: An Advanced Treatise Vol III: The Fungal Population; Ainsworth, G.C., Sussman, A.S., Eds.; Academic Press: New York, NY, USA, 1968. [Google Scholar]
- Lücking, R.; Huhndorf, S.; Pfister, D.H.; Plata, E.R.; Lumbsch, H.T. Fungi evolved right on track. Mycologia 2009, 101, 810–822. [Google Scholar] [CrossRef]
- Weissert, H. Mesozoic Pelagic Sediments: Archives for Ocean and Climate History During Green-House Conditions, 1st ed.; Hneke, H., Mulder, T., Eds.; Developments in Sedimentology; Elsevier’s Science & Technology Rights Department: Oxford, UK, 2011; Volume 63, pp. 765–792. [Google Scholar]
- Vandenberghe, N.; Hilgen, F.J.; Speijer, R.P.; Ogg, J.G.; Gradstein, F.M.; Hammer, O.; Hollis, C.J.; Hooker, J.J. The paleogene period. In The Geologic Time Scale; Elsevier: Amsterdam, The Netherlands, 2012; pp. 855–921. [Google Scholar] [CrossRef]
- Batista, E.; Lopes, A.; Alves, A. Botryosphaeriaceae species on forest trees in Portugal: Diversity, distribution and pathogenicity. Eur. J. Plant Pathol. 2020, 158, 693–720. [Google Scholar] [CrossRef]
- Rodriguez, R.J.; White, J.F.; Arnold, A.E.; Redman, R.S. Fungal endophytes: Diversity and functional roles: Tansley review. N. Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef]
- Raman, A.; Wheatley, W.; Popay, A. Endophytic fungus-vascular plant-insect interactions. Environ. Entomol. 2012, 41, 433–447. [Google Scholar] [CrossRef]
- Wheeler, D.L.; Dung, J.K.S.; Johnson, D.A. From pathogen to endophyte: An endophytic population of Verticillium dahliae evolved from a sympatric pathogenic population. New Phytol. 2019, 222, 497–510. [Google Scholar] [CrossRef] [PubMed]
- De Silva, N.I.; Phillips, A.J.L.; Liu, J.K.; Lumyong, S.; Hyde, K.D. Phylogeny and morphology of Lasiodiplodia species associated with Magnolia Forest plants. Sci. Rep. 2019, 9, 14355. [Google Scholar] [CrossRef] [PubMed]
- Samarakoon, M.C.; Hyde, K.D.; Sajeewa, S.N.; Stadler, M.; Jones, E.B.G.; Promputtha, I.; Camporesi, E.; Bulgakov, T.S.; Liu, J.K. Taxonomy, Phylogeny, Molecular Dating and Ancestral State Reconstruction of Xylariomycetidae (Sordariomycetes). Fungal Divers. 2021, 112, 1–88. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, N.; Cai, L. Typification and phylogenetic study of Phyllosticta ampelicida and P. vaccinii. Mycologia 2013, 105, 1030–1042. [Google Scholar] [CrossRef]
- Pavlic, D.; Wingfield, M.J.; Barber, P.; Slippers, B.; Hardy, G.E.S.J.; Burgess, T.I. Seven new species of the Botryosphaeriaceae from baobab and other native trees in Western Australia. Mycologia 2008, 100, 851–866. [Google Scholar] [CrossRef] [PubMed]
- Mahabale, T.S. On a fossil species of Diplodia from the Deccan Intertrappean series, MP, India. Palaeobot. 1968, 17, 295–297. [Google Scholar]
- Taylor, T.N.; Krings, M.; Taylor, E.L. Ascomycota. In Fossil Fungi; Academic Press: Cambridge, MA, USA, 2015; pp. 129–171. [Google Scholar]
- Saxena, R.K.; Wijayawardene, N.N.; Dai, D.Q.; Hyde, K.D.; Kirk, P.M. Diversity in fossil fungal spores. Mycosphere 2021, 12, 670–874. [Google Scholar] [CrossRef]
- Kalgutkar, R.M.; Nambudiri, E.M.V.; Tidwell, W.D. Diplodites sweetii sp. nov. from the Late Cretaceous (Maastrichtian) Deccan Intertrappean Beds of India. Rev. Palaeobot. Palynol. 1993, 77, 107–118. [Google Scholar] [CrossRef]
- Hagiwara, D.; Sakai, K.; Suzuki, S.; Umemura, M.; Nogawa, T.; Kato, N.; Osada, H.; Watanabe, A.; Kawamoto, S.; Gonoi, T.; et al. Temperature during conidiation affects stress tolerance, pigmentation, and trypacidin accumulation in the conidia of the airborne pathogen Aspergillus fumigatus. PLoS ONE 2017, 12, e0177050. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, K.A. Cenozoic Era: Facts About Climate, Animals & Plants. 2016. Available online: https://www.livescience.com/40352-cenozoic-era.html (accessed on 12 May 2022).
- Dou, Z.P.; Lu, M.; Wu, J.R.; He, W.; Zhang, Y. A new species and interesting records of Aplosporella from China. Sydowia 2017, 69, 1–7. [Google Scholar] [CrossRef]
- Fan, X.L.; Hyde, K.D.; Liu, J.K.; Liang, Y.M.; Tian, C.M. Multigene phylogeny and morphology reveal Phaeobotryon rhois sp. nov. (Botryosphaeriales, Ascomycota). Phytotaxa 2015, 205, 90–98. [Google Scholar] [CrossRef]
- Trakunyingcharoen, T.; Lombard, L.; Groenewald, J.Z.; Cheewangkoon, R.; To-Anun, C.; Crous, P.W. Caulicolous Botryosphaeriales from Thailand. Pers. Mol. Phylogeny Evol. Fungi 2015, 34, 87–99. [Google Scholar] [CrossRef]
- Mapook, A.; Hyde, K.D.; McKenzie, E.H.; Jones, E.G.; Bhat, D.J.; Jeewon, R.; Stadler, M.; Samarakoon, M.C.; Malaithong, M.; Tanunchai, B.; et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed). Fungal Divers. 2020, 101, 1–175. [Google Scholar] [CrossRef]
- Du, Z.; Fan, X.L.; Yang, Q.; Hyde, K.D.; Tian, C.M. Aplosporella ginkgonis (Aplosporellaceae, Botryosphaeriales), a new species isolated from Ginkgo biloba in China. Mycosphere 2017, 8, 1246–1252. [Google Scholar] [CrossRef]
- Deepika, Y.S.; Mahadevakumar, S.; Amruthesh, K.N.; Lakshmidevi, N. A new collar rot disease of cowpea (Vigna unguiculata) caused by Aplosporella hesperidica in India. Lett. Appl. Microbiol. 2020, 71, 154–163. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Guarro, J.; Cheewangkoon, R.; Van Der Bank, M.; Swart, W.J.; Stchigel, A.M.; Roux, J.; Madrid, H.; Damm, U.; et al. Fungal Planet description sheets: 154–213. Persoonia 2013, 31, 188–296. [Google Scholar] [CrossRef] [PubMed]
- Slippers, B.; Roux, J.; Wingfield, M.J.; Van der Walt, F.J.J.; Jami, F.; Mehl, J.W.M.; Marais, G.J. Confronting the constraints of morphological taxonomy in the Botryosphaeriales. Pers. Mol. Phylogeny Evol. Fungi 2014, 33, 155–168. [Google Scholar] [CrossRef]
- Damm, U.; Fourie, P.H.; Crous, P.W. Aplosporella prunicola, a novel species of anamorphic Botryosphaeriaceae. Fungal Divers. 2007, 27, 35–43. [Google Scholar]
- Cosoveanu, A.; Cabrera, R. Endophytic fungi in species of Artemisia. J. Fungi 2018, 4, 53. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.; Barber, P.A.; Hardy, G.E.S.J.; Burgess, T.I. Botryosphaeriaceae from tuart (Eucalyptus gomphocephala) woodland, including descriptions of four new species. Mycol. Res. 2009, 113, 337–353. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Wingfield, M.J.; Le Roux, J.J.; Richardson, D.M.; Strasberg, D.; Shivas, R.G.; Alvarado, P.; Edwards, J.; Moreno, G.; Sharma, R.; et al. Fungal planet description sheets: 371–399. Pers. Mol. Phylogeny Evol. Fungi 2015, 35, 264–327. [Google Scholar] [CrossRef] [PubMed]
- Konta, S.; Hongsanan, S.; Phillips, A.J.L.; Jones, E.B.G.; Boonmee, S.; Hyde, K.D. Botryosphaeriaceae from palms in Thailand II—Two new species of Neodeightonia, N. rattanica and N. rattanicola from Calamus (rattan palm). Mycosphere 2016, 7, 950–961. [Google Scholar] [CrossRef]
- Abdollahzadeh, J.; Goltapeh, E.M.; Javadi, A.; Shams-Bakhsh, M.; Zare, R.; Phillips, A.J.L. Barriopsis iraniana and Phaeobotryon cupressi: Two new species of the Botryosphaeriaceae from trees in Iran. Pers. Mol. Phylogeny Evol. Fungi 2009, 23, 1–8. [Google Scholar] [CrossRef]
- Doilom, M.; Shuttleworth, L.A.; Roux, J.; Chukeatirote, E.; Hyde, K.D. Barriopsis tectonae sp. nov. a new species of Botryosphaeriaceae from Tectona grandis (teak) in Thailand. Phytotaxa 2014, 176, 81–91. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; Jeewon, R.; Maharachchikumbura, S.S.N.; Liu, J.K.; Bhat, D.J.; Jones, E.B.G.; McKenzie, E.H.C.; Camporesi, E.; Bulgakov, T.S.; et al. Fungal diversity notes 491–602: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2017, 83, 1–261. [Google Scholar] [CrossRef]
- Slippers, B.; Smit, W.A.; Crous, P.W.; Coutinho, T.A.; Wingfield, B.D.; Wingfield, M.J. Taxonomy, phylogeny and identification of Botryosphaeriaceae associated with pome and stone fruit trees in South Africa and other regions of the world. Plant Pathol. 2007, 56, 128–139. [Google Scholar] [CrossRef]
- Pan, M.; Zhu, H.; Bezerra, J.D.; Bonthond, G.; Tian, C.; Fan, X. Botryosphaerialean fungi causing canker and dieback of tree hosts from Mount Yudu in China. Mycol. Prog. 2019, 18, 1341–1361. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Camporesi, E.; Hyde, K.D.; Yan, J.Y.; Li, X.H. Saprobic Botryosphaeriaceae, including Dothiorella italica sp. nov., associated with urban and forest trees in Italy. Mycosphere 2017, 8, 1157–1176. [Google Scholar] [CrossRef]
- Goodarzian, K.; Ghanbary, M.A.T.; Babaeizad, V.; Mojerlou, S. Identification of root endophytic fungi from rangeland plants in Mazandaran province. Iran. J. For. Range Prot. Res. 2021, 18, 216–232. [Google Scholar]
- Ariyawansa, H.A.; Hyde, K.D.; Liu, J.K.; Wu, S.P.; Liu, Z.Y. Additions to Karst Fungi 1: Botryosphaeria minutispermatia sp. nov., from Guizhou Province, China. Phytotaxa 2016, 275, 35–44. [Google Scholar] [CrossRef]
- Zhou, Y.; Dou, Z.; He, W.; Zhang, X.; Zhang, Y. Botryosphaeria sinensia sp nov., a new species from China. Phytotaxa 2016, 245, 43–50. [Google Scholar] [CrossRef]
- Lee, S.Y.; Ten, L.N.; Back, C.G.; Jung, H.Y. First report of apple decline caused by Botryosphaeria sinensis in Korea. Korean J. Mycol. 2021, 49, 417–423. [Google Scholar] [CrossRef]
- Li, G.; Liu, F.; Li, J.; Liu, Q.; Chen, S. Characterization of Botryosphaeria dothidea and Lasiodiplodia pseudotheobromae from English Walnut in China. J. Phytopathol. 2016, 164, 348–353. [Google Scholar] [CrossRef]
- Mondragón-Flores, A.; Rodríguez-Alvarado, G.; Gómez-Dorantes, N.; Guerra-Santos, J.J.; Fernández-Pavía, S.P. Botryosphaeriaceae: A complex, diverse and cosmopolitan family of fungi. Rev. Mex. Cienc. Agrícolas 2021, 12, 643–654. [Google Scholar] [CrossRef]
- Zhou, J.; Diao, X.; Wang, T.; Chen, G.; Lin, Q.; Yang, X.; Xu, J. Phylogenetic diversity and antioxidant activities of culturable fungal endophytes associated with the mangrove species Rhizophora stylosa and R. mucronata in the South China Sea. PLoS ONE 2018, 13, e0197359. [Google Scholar] [CrossRef]
- Xu, C.; Wang, C.; Ju, L.; Zhang, R.; Biggs, A.R.; Tanaka, E.; Li, B.; Sun, G. Multiple locus genealogies and phenotypic characters reappraise the causal agents of apple ring rot in China. Fungal Divers. 2015, 71, 215–231. [Google Scholar] [CrossRef]
- Li, G.; Slippers, B.; Wingfield, M.J.; Chen, S. Variation in Botryosphaeriaceae from Eucalyptus plantations in YunNan Province in southwestern China across a climatic gradient. IMA Fungus 2020, 11, 22. [Google Scholar] [CrossRef]
- Li, G.Q.; Liu, F.F.; Li, J.Q.; Liu, Q.L.; Chen, S.F. Botryosphaeriaceae from Eucalyptus plantations and adjacent plants in China. Persoonia Mol. Phylogeny Evol. Fungi 2018, 40, 63–95. [Google Scholar] [CrossRef]
- Zhou, Y.P.; Zhang, M.; Dou, Z.; Zhang, Y. Botryosphaeria rosaceae sp. nov. and B. ramosa, new botryosphaeriaceous taxa from China. Mycosphere 2017, 8, 162–171. [Google Scholar] [CrossRef]
- Li, W.J.; McKenzie, E.H.C.; Liu, J.K.; Bhat, D.J.; Dai, D.Q.; Camporesi, E.; Tian, Q.; Maharachchikumbura, S.S.N.; Luo, Z.L.; Shang, Q.J.; et al. Taxonomy and phylogeny of hyaline-spored coelomycetes. Fungal Divers. 2020, 100, 279–801. [Google Scholar]
- Barakat, F.; Vansteelandt, M.; Triastuti, A.; Jargeat, P.; Jacquemin, D.; Graton, J.; Mejia, K.; Cabanillas, B.; Vendier, L.; Stigliani, J.L.; et al. Thiodiketopiperazines with two spirocyclic centers extracted from Botryosphaeria mamane, an endophytic fungus isolated from Bixa orellana L. Phytochemistry 2019, 158, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, F.; Diniz, I.; Pena, A.R.; Catarino, L.; Baldé, A.; Romeiras, M.; Batista, D. Diversity of the Botryosphaeriaceae family in Guinea-Bissau (West Africa): The beginning of a tale in cashew. In Proceedings of the 15th European Conference on Fungal Genetics, Rome, Italy, 17–20 February 2020; p. 125. [Google Scholar]
- Damm, U.; Crous, P.W.; Fourie, P.H. Botryosphaeriaceae as potential pathogens of Prunus species in South Africa, with descriptions of Diplodia africana and Lasiodiplodia plurivora sp. nov. Mycologia 2007, 99, 664–680. [Google Scholar] [CrossRef] [PubMed]
- Jami, F.; Slippers, B.; Wingfield, M.J.; Gryzenhout, M. Greater Botryosphaeriaceae diversity in healthy than associated diseased Acacia karroo tree tissues. Australas. Plant Pathol. 2013, 42, 421–430. [Google Scholar] [CrossRef]
- Hyde, K.D.; Chaiwan, N.; Norphanphoun, C.; Boonmee, S.; Camporesi, E.; Chethana, K.W.T.; Dayarathne, M.C.; De Silva, N.I.; Dissanayake, A.J.; Ekanayaka, A.H.; et al. Mycosphere notes 169–224. Mycosphere 2018, 9, 271–430. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Peduto Hand, F.; Gubler, W.D.; Trouillas, F.P. Botryosphaeriaceae species associated with dieback and canker disease of bay laurel in northern California with the description of Dothiorella californica sp. nov. Fungal Biol. 2017, 121, 347–360. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Hyde, K.D.; Jayasiri, S.C.; Buyck, B.; Chethana, K.W.T.; Dai, D.Q.; Dai, Y.C.; Daranagama, D.A.; Jayawardena, R.S.; Lücking, R.; et al. Fungal diversity notes 111–252—Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015, 75, 27–274. [Google Scholar] [CrossRef]
- González-Domínguez, E.; Alves, A.; León, M.; Armengol, J. Characterization of Botryosphaeriaceae species associated with diseased loquat (Eriobotrya japonica) in Spain. Plant Pathol. 2017, 66, 77–89. [Google Scholar] [CrossRef]
- Osorio, J.A.; Crous, C.J.; de Beer, Z.W.; Wingfield, M.J.; Roux, J. Endophytic Botryosphaeriaceae, including five new species, associated with mangrove trees in South Africa. Fungal Biol. 2017, 121, 361–393. [Google Scholar] [CrossRef]
- Elena, G.; León, M.; Abad-Campos, P.; Armengol, J.; Mateu-Andrés, I.; Güemes-Heras, J. First report of Diplodia fraxini causing dieback of Fraxinus angustifolia in Spain. Plant Dis. 2018, 102, 2645–2646. [Google Scholar] [CrossRef]
- Hanifeh, S.; Ghoosta, Y.; Abbasi, S.; Phillips, A.J.L. First report of Diplodia malorum Fuckel the causal agent of canker disease of apple trees in Iran. Iran. J. Plant Pathol. 2013, 49, 83–84. [Google Scholar]
- Abdollahzadeh, J.; Hosseini, F.; Javadi, A. New records from Botryosphaeriaceae (Ascomycota) for mycobiota of Iran. Mycol. Iran. 2014, 1, 34–41. [Google Scholar]
- Linaldeddu, B.T.; Franceschini, A.; Alves, A.; Phillips, A.J.L. Diplodia quercivora sp. nov.: A new species of Diplodia found on declining Quercus canariensis trees in Tunisia. Mycologia 2013, 105, 1266–1274. [Google Scholar] [CrossRef]
- Van der Walt, F.J.J. Botryosphaeriaceae associated with native Acacia species in southern Africa with special reference to A. mellifera. Doctoral Dissertation, University of Pretoria, Pretoria, South Africa, 2008. [Google Scholar]
- Phillips, A.J.L.; Lopes, J.; Abdollahzadeh, J.; Bobev, S.; Alves, A. Resolving the Diplodia complex on apple and other Rosaceae hosts. Persoonia Mol. Phylogeny Evol. Fungi 2012, 29, 29–38. [Google Scholar] [CrossRef]
- Burgess, T.; Wingfield, B.D.; Wingfield, M.J. Comparison of genotypic diversity in native and introduced populations of Sphaeropsis sapinea isolated from Pinus radiata. Mycol. Res. 2001, 105, 1331–1339. [Google Scholar] [CrossRef]
- Chakusary, M.K.; Mohammadi, H.; Khodaparast, S.A. Diversity and pathogenicity of Botryosphaeriaceae species on forest trees in the north of Iran. Eur. J. For. Res. 2019, 138, 685–704. [Google Scholar] [CrossRef]
- Manzanos, T.; Aragones, A.; Iturritxa, E. Diplodia scrobiculata: A latent pathogen of Pinus radiata reported in northern Spain. Phytopathol. Mediterr. 2017, 56, 274–277. [Google Scholar]
- Úrbez-Torres, J.R.; Leavitt, G.M.; Guerrero, J.C.; Guevara, J.; Gubler, W.D. Identification and pathogenicity of Lasiodiplodia theobromae and Diplodia seriata, the causal agents of bot canker disease of grapevines in Mexico. Plant Dis. 2008, 92, 519–529. [Google Scholar] [CrossRef]
- De Errasti, A.; Carmarán, C.C.; Novas, M.V. Diversity and significance of fungal endophytes from living stems of naturalized trees from Argentina. Fungal Divers. 2010, 41, 29–40. [Google Scholar] [CrossRef]
- Alves, A.; Linaldeddu, B.T.; Deidda, A.; Scanu, B.; Phillips, A.J.L. The complex of Diplodia species associated with Fraxinus and some other woody hosts in Italy and Portugal. Fungal Divers. 2014, 67, 143–156. [Google Scholar] [CrossRef]
- Linaldeddu, B.T.; Bottecchia, F.; Bregant, C.; Maddau, L.; Montecchio, L. Diplodia fraxini and Diplodia subglobosa: The main species associated with cankers and dieback of Fraxinus excelsior in North-Eastern Italy. Forests 2020, 11, 883. [Google Scholar] [CrossRef]
- Yuan, H.S.; Lu, X.; Dai, Y.C.; Hyde, K.D.; Kan, Y.H.; Kušan, I.; He, S.H.; Liu, N.G.; Sarma, V.V.; Zhao, C.L.; et al. Fungal diversity notes 1277–1386: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2020, 104, 1–266. [Google Scholar] [CrossRef]
- Phookamsak, R.; Hyde, K.D.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Maharachchikumbura, S.S.N.; Raspé, O.; Karunarathna, S.C.; Wanasinghe, D.N.; Hongsanan, S.; et al. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019, 95, 1–273. [Google Scholar] [CrossRef]
- Zhang, M.; He, W.; Wu, J.R.; Zhang, Y. Two new species of Spencermartinsia (Botryosphaeriaceae, Botryosphaeriales) from China. Mycosphere 2016, 7, 942–949. [Google Scholar] [CrossRef]
- Xiao, X.E.; Wang, W.; Crous, P.W.; Wang, H.K.; Jiao, C.; Huang, F.; Pu, Z.X.; Zhu, Z.R.; Li, H.Y. Species of Botryosphaeriaceae associated with citrus branch diseases in China. Pers. Mol. Phylogeny Evol. Fungi 2021, 47, 106–135. [Google Scholar] [CrossRef]
- Hyde, K.D.; de Silva, N.I.; Jeewon, R.; Bhat, D.J.; Phookamsak, R.; Doilom, M.; Boonmee, S.; Jayawardena, R.S.; Maharachchikumbura, S.S.N.; Senanayake, I.C.; et al. AJOM new records and collections of fungi: 1–100. Asian J. Mycol. 2020, 3, 22–294. [Google Scholar] [CrossRef]
- De Wet, J.; Slippers, B.; Preisig, O.; Wingfield, B.D.; Tsopelas, P.; Wingfield, M.J. Molecular and morphological characterization of Dothiorella casuarini sp. nov. and other Botryosphaeriaceae with diplodia-like conidia. Mycologia 2009, 101, 503–511. [Google Scholar] [CrossRef]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.A.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Jeewon, R.; Phillips, A.J.L.; Bhat, D.J.; Wanasinghe, D.N.; Liu, J.K.; Lu, Y.Z.; et al. Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 2019, 10, 1–186. [Google Scholar] [CrossRef]
- You, C.J.; Liu, X.; Li, L.X.; Tsui, C.K.M.; Tian, C.M. Dothiorella magnoliae, a new species associated with dieback of Magnolia grandiflora from China. Mycosphere 2017, 8, 1031–1041. [Google Scholar] [CrossRef]
- Li, G.J.; Hyde, K.D.; Zhao, R.L.; Hongsanan, S.; Abdel-Aziz, F.A.; Abdel-Wahab, M.A.; Alvarado, P.; Alves-Silva, G.; Ammirati, J.F.; Ariyawansa, H.A.; et al. Fungal diversity notes 253–366: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 78, 1–237. [Google Scholar] [CrossRef]
- Li, W.; Liu, J.; Bhat, D.J.; Camporesi, E.; Xu, J.; Hyde, K.D. Introducing the novel species, Dothiorella symphoricarposicola, from Snowberry in Italy. Cryptogam. Mycol. 2014, 35, 257–270. [Google Scholar] [CrossRef]
- Linaldeddu, B.T.; Maddau, L.; Franceschini, A.; Alves, A.; Phillips, A.J.L. Botryosphaeriaceae species associated with lentisk dieback in Italy and description of Diplodia insularis sp. nov. Mycosphere 2016, 7, 962–977. [Google Scholar] [CrossRef]
- Tian, Q.; Li, W.J.; Hyde, K.D.; Camporesi, E.; Bhat, D.J.; Chomnunti, P.; Xu, J.C. Molecular taxonomy of five species of microfungi on Alnus spp. from Italy. Mycol. Prog. 2018, 17, 255–274. [Google Scholar] [CrossRef]
- Doll, D.A.; Rolshausen, P.E.; Pouzoulet, J.; Michailides, T.J. First report of Dothiorella iberica causing trunk and scaffold cankers of almond in California. Plant Dis. 2015, 99, 1185. [Google Scholar] [CrossRef]
- Váczy, K.Z.; Németh, M.Z.; Csikós, A.; Kovács, G.M.; Kiss, L. Dothiorella omnivora isolated from grapevine with trunk disease symptoms in Hungary. Eur. J. Plant Pathol. 2018, 150, 817–824. [Google Scholar] [CrossRef]
- Pavlic-Zupanc, D.; Piškur, B.; Slippers, B.; Wingfield, M.J.; Jurc, D. Molecular and morphological characterization of Dothiorella species associated with dieback of Ostrya carpinifolia in Slovenia and Italy. Phytopathol. Mediterr. 2015, 54, 241–252. [Google Scholar]
- Pitt, W.M.; Úrbez-Torres, J.R.; Trouillas, F.P. Dothiorella and Spencermartinsia, new species and records from grapevines in Australia. Australas. Plant Pathol. 2015, 44, 43–56. [Google Scholar] [CrossRef]
- Doilom, M.; Shuttleworth, L.A.; Roux, J.; Chukeatirote, E.; Hyde, K.D. Botryosphaeriaceae associated with Tectona grandis (teak) in Northern Thailand. Phytotaxa 2015, 233, 1–26. [Google Scholar] [CrossRef]
- Zhang, R.; Guo, X.; Sun, G.; Tang, M.; Gleason, M.L. Dothiorella viticola on Populus cathayana in China: A new record. Mycotaxon 2009, 109, 129–135. [Google Scholar] [CrossRef]
- Ramabulana, E.; Kunjeku, E.; Slippers, B.; Coetzee, M.P.A. Diversity of endophytes in the Botryosphaeriaceae differs on Anacardiaceae in disturbed and undisturbed ecosystems in South Africa. Forests 2022, 13, 341. [Google Scholar] [CrossRef]
- Douanla-Meli, C.; Scharnhorst, A. Palm foliage as pathways of pathogenic Botryosphaeriaceae fungi and host of new Lasiodiplodia species from Mexico. Pathogens 2021, 10, 1297. [Google Scholar] [CrossRef]
- Rojas, E.I.; Herre, E.A.; Mejia, L.C.; Arnold, A.E.; Chaverri, P.; Samuels, G.J. Endomelanconiopsis, a new anamorph genus in the Botryosphaeriaceae. Mycologia 2008, 100, 760–775. [Google Scholar] [CrossRef] [PubMed]
- Verkley, G.J.M.; van der Aa, H.A. Endomelanconium microsporum, a new coelomycete isolated from soil in Papua New Guinea. Mycologia 1997, 89, 967–970. [Google Scholar] [CrossRef]
- Jami, F.; Slippers, B.; Wingfield, M.J.; Gryzenhout, M. Botryosphaeriaceae species overlap on four unrelated, native South African hosts. Fungal Biol. 2014, 118, 168–179. [Google Scholar] [CrossRef]
- Thambugala, K.M.; Daranagama, D.A.; Camporesi, E.; Singtripop, C.; Liu, Z.Y.; Hyde, K.D. Multi-locus phylogeny reveals the sexual state of Tiarosporella in Botryosphaeriaceae. Cryptogam. Mycol. 2014, 35, 359–367. [Google Scholar] [CrossRef]
- Thynne, E.; McDonald, M.C.; Evans, M.; Wallwork, H.; Neate, S.; Solomon, P.S. Re-classification of the causal agent of white grain disorder on wheat as three separate species of Eutiarosporella. Australas. Plant Pathol. 2015, 44, 527–539. [Google Scholar] [CrossRef]
- Crous, P.W.; Muller, M.M.; Sanchez, R.M.; Giordano, L.; Bianchinotti, M.V.; Anderson, F.E.; Groenewald, J.Z. Resolving Tiarosporella spp. allied to Botryosphaeriaceae and Phacidiaceae. Phytotaxa 2015, 202, 73–93. [Google Scholar] [CrossRef]
- Jami, F.; Slippers, B.; Wingfield, M.; Gryzenhout, M. Five New Species of the Botryosphaeriaceae from Acacia karroo in South Africa. Cryptogam. Mycol. 2012, 33, 245–266. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, S.; Zhao, L.; Sun, X.; He, W.; Zhang, Y.; Dai, Y.C. Lasiodiplodia spp. associated with Aquilaria crassna in Laos. Mycol. Prog. 2019, 18, 683–701. [Google Scholar] [CrossRef]
- Aguiar, F.M.; Costa, R.V.; Silva, D.D.; Lana, U.G.P.; Gomes, E.A.; Cota, L.V. First report of Lasiodiplodia brasiliense causing maize stalk rot. Australas. Plant Dis. Notes 2018, 13, 41. [Google Scholar] [CrossRef]
- Jiang, N.; Phillips, A.J.L.; Zhang, Z.X.; Tian, C.M. Morphological and molecular identification of two novel species of Melanops in China. Mycosphere 2018, 9, 1187–1196. [Google Scholar] [CrossRef]
- Chen, S.F.; Fichtner, E.; Morgan, D.P.; Michailides, T.J. First report of Lasiodiplodia citricola and Neoscytalidium dimidiatum causing death of graft union of English walnut in California. Plant Dis. 2013, 97, 993. [Google Scholar] [CrossRef]
- Machado, A.R.; Pinho, D.B.; Pereira, O.L. Phylogeny, identification and pathogenicity of the Botryosphaeriaceae associated with collar and root rot of the biofuel plant Jatropha curcas in Brazil, with a description of new species of Lasiodiplodia. Fungal Divers. 2014, 67, 231–247. [Google Scholar] [CrossRef]
- Cruywagen, E.M.; Slippers, B.; Roux, J.; Wingfield, M.J. Phylogenetic species recognition and hybridisation in Lasiodiplodia: A case study on species from baobabs. Fungal Biol. 2016, 121, 420–436. [Google Scholar] [CrossRef]
- Abdollahzadeh, J.; Javadi, A.; Goltapeh, E.M.; Zare, R.; Phillips, A.J.L. Phylogeny and morphology of four new species of Lasiodiplodia from Iran. Pers. Mol. Phylogeny Evol. Fungi 2010, 25, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Correia, K.C.; Silva, M.A.; De Morais, M.A.; Armengol, J.; Phillips, A.J.L.; Câmara, M.P.S.; Michereff, S.J. Phylogeny, distribution and pathogenicity of Lasiodiplodia species associated with dieback of table grape in the main Brazilian exporting region. Plant Pathol. 2016, 65, 92–103. [Google Scholar] [CrossRef]
- Pavlic, D.; Slippers, B.; Coutinho, T.A.; Wingfield, M.J. Botryosphaeriaceae occurring on native Syzygium cordatum in South Africa and their potential threat to Eucalyptus. Plant Pathol. 2007, 56, 624–636. [Google Scholar] [CrossRef]
- Pavlic, D.; Slippers, B.; Coutinho, T.A.; Gryzenhout, M.; Wingfield, M.J. Lasiodiplodia gonubiensis sp. nov., a new Botryosphaeria anamorph from native Syzygium cordatum in South Africa. Stud. Mycol. 2004, 50, 313–322. [Google Scholar]
- Netto, M.S.; Lima, W.G.; Correia, K.C.; Da Silva, C.F.; Thon, M.; Martins, R.B.; Miller, R.N.; Michereff, S.J.; Câmara, M.P. Analysis of phylogeny, distribution, and pathogenicity of Botryosphaeriaceae species associated with gummosis of Anacardium in Brazil, with a new species of Lasiodiplodia. Fungal Biol. 2016, 121, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Custódio, F.A.; Machado, A.R.; Soares, D.J.; Pereira, O.L. Lasiodiplodia hormozganensis causing basal stem rot on Ricinus communis in Brazil. Australas. Plant Dis. Notes 2018, 13, 25. [Google Scholar] [CrossRef]
- Al-Sadi, A.M.; Al-Wehaibi, A.N.; Al-Shariqi, R.M.; Al-Hammadi, M.S.; Al-Hosni, I.A.; Al-Mahmooli, I.H.; Al-Ghaithi, A.G. Population genetic analysis reveals diversity in Lasiodiplodia species infecting date palm, citrus, and mango in Oman and the UAE. Plant Dis. 2013, 97, 1363–1369. [Google Scholar] [CrossRef]
- Dayarathne, M.C.; Jones, E.B.G.; Maharachchikumbura, S.S.N.; Devadatha, B.; Sarma, V.V.; Khongphinitbunjong, K.; Chomnunti, P.; Hyde, K.D. Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 2020, 11, 1–188. [Google Scholar] [CrossRef]
- Rodríguez-Gálvez, E.; Guerrero, P.; Barradas, C.; Crous, P.W.; Alves, A. Phylogeny and pathogenicity of Lasiodiplodia species associated with dieback of mango in Peru. Fungal Biol. 2017, 121, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Dou, Z.P.; He, W.; Zhang, Y. Does morphology matter in taxonomy of Lasiodiplodia? An answer from Lasiodiplodia hyalina sp. nov. Mycosphere 2017, 8, 1014–1027. [Google Scholar] [CrossRef]
- Coutinho, I.B.L.; Freire, F.C.O.; Lima, C.S.; Lima, J.S.; Gonçalves, F.J.T.; Machado, A.R.; Silva, A.M.S.; Cardoso, J.E. Diversity of genus Lasiodiplodia associated with perennial tropical fruit plants in northeastern Brazil. Plant Pathol. 2017, 66, 90–104. [Google Scholar] [CrossRef]
- Linaldeddu, B.T.; Deidda, A.; Scanu, B.; Franceschini, A.; Serra, S.; Berraf-Tebbal, A.; Zouaoui Boutiti, M.; Ben Jamâa, M.L.; Phillips, A.J.L. Diversity of Botryosphaeriaceae species associated with grapevine and other woody hosts in Italy, Algeria and Tunisia, with descriptions of Lasiodiplodia exigua and Lasiodiplodia mediterranea sp. nov. Fungal Divers. 2015, 71, 201–214. [Google Scholar] [CrossRef]
- Akgül, D.S.; Savaş, N.G.; Özarslandan, M. First Report of Wood Canker caused by Lasiodiplodia exigua and Neoscytalidium novaehollandiae on Grapevine in Turkey. Plant Dis. 2019, 103, 1036. [Google Scholar] [CrossRef]
- Zhang, Q.; Lu, Z.; Ding, Y.; Liu, H.; Feng, L. Canker on Machilus pauhoi caused by Lasiodiplodia margaritacea in the Fujian Province of China. Plant Dis. 2019, 103, 1417. [Google Scholar] [CrossRef]
- Meng, C.R.; Zhang, Q.; Yang, Z.F.; Geng, K.; Zeng, X.Y.; Chethana, K.W.T.; Wang, Y. Lasiodiplodia syzygii sp. nov. (Botryosphaeriaceae) causing post-harvest water-soaked brown lesions on Syzygium samarangense in Chiang Rai, Thailand. Biodivers. Data J. 2021, 9, e60604. [Google Scholar] [CrossRef] [PubMed]
- Tennakoon, D.S.; Kuo, C.H.; Maharachchikumbura, S.S.N.; Thambugala, K.M.; Gentekaki, E.; Phillips, A.J.L.; Bhat, D.J.; Wanasinghe, D.N.; de Silva, N.I.; Promputtha, I.; et al. Taxonomic and phylogenetic contributions to Celtis formosana, Ficus ampelas, F. septica, Macaranga tanarius and Morus australis leaf litter inhabiting microfungi. Fungal Divers. 2021, 108, 1–215. [Google Scholar] [CrossRef]
- Zaher, A.M.; Moharram, A.M.; Davis, R.; Panizzi, P.; Makboul, M.A.; Calderón, A.I. Characterisation of the metabolites of an antibacterial endophyte Botryodiplodia theobromae Pat. of Dracaena draco L. by LC–MS/MS. Nat. Prod. Res. 2015, 29, 2275–2281. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Tangjang, S.; Shukla, A.C. New Taxon of Fungal Endophytes from Phrynium capitatum Willd: A promising ethnomedicinal plant in Northeast India and its systematic and phylogenetic analysis. Sci. Technol. J. 2019, 7, 29–36. [Google Scholar] [CrossRef]
- Machado, A.R.; Pinho, D.B.; Soares, D.J.; Gomes, A.A.M.; Pereira, O.L. Bayesian analyses of five gene regions reveal a new phylogenetic species of Macrophomina associated with charcoal rot on oilseed crops in Brazil. Eur. J. Plant Pathol. 2019, 153, 89–100. [Google Scholar] [CrossRef]
- Sarr, M.P.; Ndiaye, M.B.; Groenewald, J.Z.; Crous, P.W. Genetic diversity in Macrophomina phaseolina, the causal agent of charcoal rot. Phytopathol. Mediterr. 2014, 53, 250–268. [Google Scholar]
- Zhao, L.; Cai, J.; He, W.; Zhang, Y. Macrophomina vaccinii sp. Nov. Causing blueberry stem blight in China. MycoKeys 2019, 55, 1–14. [Google Scholar] [CrossRef]
- Adamčík, S.; Cai, L.; Chakraborty, D.; Chen, X.H.; Cotter, H.V.T.; Dai, D.Q.; Dai, Y.C.; Das, K.; Deng, C.; Ghobad-Nejhad, M.; et al. Fungal biodiversity profiles 1-10. Cryptogam. Mycol. 2015, 36, 121–166. [Google Scholar] [CrossRef]
- Dai, D.Q.; Phookamsak, R.; Wijayawardene, N.N.; Li, W.J.; Bhat, D.J.; Xu, J.C.; Taylor, J.E.; Hyde, K.D.; Chukeatirote, E. Bambusicolous fungi. Fungal Divers. 2017, 82, 1–105. [Google Scholar] [CrossRef]
- Mukhtar, I.; Quan, X.; Khokhar, I.; Chou, T.; Huang, Q.; Jiang, S.; Yan, J.; Chen, B.; Huang, R.; Ashraf, H.J.; et al. First Report of Leaf Spot on Caryota mitis (Fishtail Palm) Caused by Neodeightonia palmicola in China. Plant Dis. 2019, 103, 2675. [Google Scholar] [CrossRef]
- Ligoxigakis, E.K.; Markakis, E.A.; Papaioannou, I.A.; Typas, M.A. First report of palm rot of Phoenix spp. caused by Neodeightonia phoenicum in Greece. Plant Dis. 2013, 97, 286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Song, X.L. Occurrence of leaf spot caused by Neodeightonia phoenicum on pygmy date plam (Phoenix roebelenii) in China. Plant Dis. 2021, 106, 2269. [Google Scholar] [CrossRef]
- Hipol, R.; Magtoto, L.; Tamang, S.M.; Amor, M. Antioxidant activities of fungal endophytes isolated from strawberry Fragaria x Ananassa fruit. Electron. J. Biol. 2014, 10, 107–112. [Google Scholar]
- Shetty, K.G.; Rivadeneira, D.V.; Jayachandran, K.; Walker, D.M. Isolation and molecular characterization of the fungal endophytic microbiome from conventionally and organically grown avocado trees in South Florida. Mycol. Prog. 2016, 15, 977–986. [Google Scholar] [CrossRef]
- Espinoza, J.G.; Briceño, E.X.; Chávez, E.R.; Úrbez-Torres, J.R.; Latorre, B.A. Neofusicoccum spp. associated with stem canker and dieback of blueberry in Chile. Plant Dis. 2009, 93, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, I.B.L.; Cardoso, J.E.; Lima, C.S.; Lima, J.S.; Gonçalves, F.J.T.; Silva, A.M.S.; Freire, F.C.O. An emended description of Neofusicoccum brasiliense and characterization of Neoscytalidium and Pseudofusicoccum species associated with tropical fruit plants in northeastern Brazil. Phytotaxa 2018, 358, 251–264. [Google Scholar] [CrossRef]
- Linaldeddu, B.T.; Alves, A.; Phillips, A.J.L. Sardiniella urbana gen. et sp. nov., a new member of the Botryosphaeriaceae isolated from declining Celtis australis trees in Sardinian streetscapes. Mycosphere 2016, 7, 893–905. [Google Scholar] [CrossRef]
- Demissie, A.G.; Darge, W.A.; Cafà, G. Neofusicoccum parvum causing Eucalyptus canker and die-back diseases in Ethiopia. Int. J. Plant Pathol. 2019, 11, 1–5. [Google Scholar] [CrossRef]
- Hokama, Y.M.; Savi, D.C.; Assad, B.; Aluizio, R.; Gomes-Figueiredo, J.A.; Adamoski, D.M.; Possiede, Y.M.; Glienke, C. Endophytic fungi isolated from Vochysia divergens in the pantanal, mato grosso do sul: Diversity, phylogeny and biocontrol of Phyllosticta citricarpa. In Endophytic Fungi: Diversity, Characterization and Biocontrol; Nova Publishers: Hauppauge, NY, USA, 2016; pp. 93–123. ISBN 978-1-53610-358-8. [Google Scholar]
- Crous, P.W.; Groenewald, J.Z.; Shivas, R.G.; Edwards, J.; Seifert, K.A.; Alfenas, A.C.; Alfenas, R.F.; Burgess, T.I.; Carnegie, A.J.; Hardy, G.E.S.J.; et al. Fungal planet description sheets: 69-91. Pers. Mol. Phylogeny Evol. Fungi 2011, 26, 108–156. [Google Scholar] [CrossRef]
- Chen, S.; Li, G.; Liu, F.; Michailides, T.J. Novel species of Botryosphaeriaceae associated with shoot blight of pistachio. Mycologia 2015, 107, 780–792. [Google Scholar] [CrossRef]
- Zhang, M.; Lin, S.; He, W.; Zhang, Y. Three species of Neofusicoccum (Botryosphaeriaceae, Botryosphaeriales) associated with woody plants from southern China. Mycosphere 2017, 8, 797–808. [Google Scholar] [CrossRef]
- Jami, F.; Slippers, B.; Wingfield, M.J.; Loots, M.T.; Gryzenhout, M. Temporal and spatial variation of Botryosphaeriaceae associated with Acacia karroo in South Africa. Fungal Ecol. 2015, 15, 51–62. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Zhang, W.; Li, X.; Zhou, Y.; Chethana, T.; Chukeatirote, E.; Hyde, K.D.; Yan, J.; Zhang, G.; Zhao, W. First report of Neofusicoccum mangiferae associated with grapevine dieback in China. Phytopathol. Mediterr. 2015, 54, 414–419. [Google Scholar]
- Krishnapillai, N.; Wilson Wijeratnam, R.S. First report of Neofusicoccum mediterraneum causing stem end rot on Karuthakolumban mangoes. Plant Dis. 2015, 99, 1858. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Groenewald, J.Z.; Cai, L.; Chen, Q.; Marincowitz, S.; Barnes, I.; Bensch, K.; Braun, U.; Camporesi, E.; Damm, U.; et al. Genera of phytopathogenic fungi: GOPHY 1. Stud. Mycol. 2017, 86, 99–216. [Google Scholar] [CrossRef] [PubMed]
- Pérez, F.S.; Meriño-Gergichevich, C.; Guerrero, C.J. Detection of Neofusicoccum nonquaesitum causing dieback and canker in highbush blueberry from Southern Chile. J. Soil Sci. Plant Nutr. 2014, 14, 581–588. [Google Scholar] [CrossRef]
- Berraf-Tebbal, A.; Guereiro, M.A.; Phillips, A.J.; Von Arx, J.A. Phylogeny of Neofusicoccum species associated with grapevine trunk diseases in Algeria, with description of Neofusicoccum algeriense sp. nov. Phytopathol. Mediterr. 2014, 53, 416–427. [Google Scholar]
- Ngobisa, A.I.C.N.; Abidin, M.A.Z.; Wong, M.Y.; Noordin, M.W.D.W. Neofusicoccum ribis associated with leaf blight on rubber (Hevea brasiliensis) in Peninsular Malaysia. Plant Pathol. J. 2013, 29, 10–16. [Google Scholar] [CrossRef]
- Boyogueno, A.D.B. Characterization of Botryosphaeriaceae and Cryphonectriaceae associated with Terminalia spp. In Africa; University of Pretoria: Pretoria, South Africa, 2010. [Google Scholar]
- Summerell, B.A.; Groenewald, J.Z.; Carnegie, A.; Summerbell, R.C.; Crous, P.W. Eucalyptus microfungi known from culture. 2. Alysidiella, Fusculina and Phlogicylindrium genera nova, with notes on some other poorly known taxa. Fungal Divers. 2006, 23, 323–350. [Google Scholar]
- Mohd, M.H.; Salleh, B.; Zakaria, L. Identification and molecular characterizations of Neoscytalidium dimidiatum causing stem canker of red-fleshed dragon fruit (Hylocereus polyrhizus) in Malaysia. J. Phytopathol. 2013, 161, 841–849. [Google Scholar] [CrossRef]
- Abdel-Motaal, F.F.; Nassar, M.S.M.; El-Zayat, S.A.; El-Sayed, M.A.; Shin-Ichi, I. Antifungal activity of endophytic fungi isolated from Egyptian henbane (Hyoscyamus muticus L.). Pakistan J. Bot. 2010, 42, 2883–2894. [Google Scholar]
- Huang, S.K.; Tangthirasunun, N.; Phillips, A.J.L.; Dai, D.Q.; Wanasinghe, D.N.; Wen, T.C.; Bahkali, A.H.; Hyde, K.D.; Kang, J.C. Morphology and phylogeny of Neoscytalidium orchidacearum sp. nov. (Botryosphaeriaceae). Mycobiology 2016, 44, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Suwannarach, N.; Kumla, J.; Lumyong, S. Leaf spot on cattleya orchid caused by Neoscytalidium orchidacearum in Thailand. Can. J. Plant Pathol. 2018, 40, 109–114. [Google Scholar] [CrossRef]
- Daranagama, D.A.; Thambugala, K.M.; Campino, B.; Alves, A.; Bulgakov, T.S.; Phillips, A.J.L.; Liu, X.; Hyde, K.D. Phaeobotryon negundinis sp. nov. (Botryosphaeriales) from Russia. Mycosphere 2016, 7, 933–941. [Google Scholar] [CrossRef]
- Úrbez-Torres, J.R. The status of Botryosphaeriaceae species infecting grapevines. Phytopathol. Mediterr. 2011, 50, 5–45. [Google Scholar]
- Zlatković, M.; Keča, N.; Wingfield, M.J.; Jami, F.; Slippers, B. Botryosphaeriaceae associated with the die-back of ornamental trees in the Western Balkans. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2016, 109, 543–564. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Carnegie, A.J.; Hardy, G.E.S.J.; Smith, D.; Summerell, B.A.; Cano-Lira, J.F.; Guarro, J.; Houbraken, J.; et al. Fungal planet description sheets: 558-624. Persoonia 2017, 38, 240–384. [Google Scholar] [CrossRef]
- Norphanphoun, C.; Hongsanan, S.; Gentekaki, E.; Chen, Y.J.; Kuo, C.H.; Hyde, K.D. Differentiation of species complexes in Phyllosticta enables better species resolution. Mycosphere 2020, 11, 2542–2628. [Google Scholar] [CrossRef]
- Sharma, R.; Kulkarni, G.; Shouche, Y.S. Pseudofusicoccum adansoniae isolated as an endophyte from Jatropha podagrica: New record for India. Mycotaxon 2013, 123, 39–45. [Google Scholar] [CrossRef]
- Senwanna, C.; Hongsanan, S.; Hyde, K.D.; Cheewangkoon, R.; Konta, S.; Wang, Y. First report of the sexual morph of Pseudofusicoccum adansoniae Pavlic, T.I.Burgess & M.J.Wingf. on Para rubber Chanokned. Cryptogam. Mycol. 2020, 41, 133–146. [Google Scholar]
- Li, L.; Mohd, M.H.; Mohamed Nor, N.M.I.; Subramaniam, S.; Latiffah, Z. Identification of Botryosphaeriaceae associated with stem-end rot of mango (Mangifera indica L.) in Malaysia. J. Appl. Microbiol. 2021, 130, 1273–1284. [Google Scholar] [CrossRef] [PubMed]
- Horst, R.K. Westcott’s Plant Disease Handbook; Springer: Berlin/Heidelberg, Germany, 2008; ISBN 978-1-40204-585-1. [Google Scholar]
- Monkai, J.; Liu, J.K.; Boonmee, S.; Chomnunti, P.; Chukeatirote, E.; Jones, E.B.G.; Wang, Y.; Hyde, K.D. Planistromellaceae (Botryosphaeriales). Cryptogam. Mycol. 2013, 34, 45–77. [Google Scholar] [CrossRef]
- Crous, P.W.; Giraldo, A.; Hawksworth, D.L.; Robert, V.; Kirk, P.M.; Guarro, J.; Robbertse, B.; Schoch, C.L.; Damm, U.; Trakunyingcharoen, T.; et al. The Genera of Fungi: Fixing the application of type species of generic names. IMA Fungus 2014, 5, 141–160. [Google Scholar] [CrossRef] [PubMed]
- Tanney, J.B.; Seifert, K.A. Pileospora piceae gen. et sp. nov. (Septorioideaceae, Botryosphaeriales) from Picea rubens. Mycol. Prog. 2019, 18, 163–174. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Carnegie, A.J.; Hardy, G.E.S.J.; Smith, D.; Summerell, B.A.; Guarro, J.; Houbraken, J.; Lombard, L.; et al. Fungal Planet description sheets: 625–715. Pers. -Mol. Phylogeny Evol. Fungi 2017, 32, 270–464. [Google Scholar] [CrossRef]
- Crous, S.; Taylor, J.E. Saccharata intermedia. Persoon 2009, 23, 198–199. [Google Scholar]
- Quaedvlieg, W.; Verkley, G.J.M.; Shin, H.D.; Barreto, R.W.; Alfenas, A.C.; Swart, W.J.; Groenewald, J.Z.; Crous, P.W. Sizing up septoria. Stud. Mycol. 2013, 75, 307–390. [Google Scholar] [CrossRef]
- Hyde, K.D.; Tennakoon, D.S.; Jeewon, R.; Bhat, D.J.; Maharachchikumbura, S.S.N.; Rossi, W.; Leonardi, M.; Lee, H.B.; Mun, H.Y.; Houbraken, J.; et al. Fungal diversity notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2019, 96, 1–242. [Google Scholar] [CrossRef]
- Zhang, Z.B.; Zeng, Q.G.; Yan, R.M.; Wang, Y.; Zou, Z.R.; Zhu, D. Endophytic fungus Cladosporium cladosporioides LF70 from Huperzia serrata produces Huperzine A. World J. Microbiol. Biotechnol. 2011, 27, 479–486. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; Bhat, J.D.; Mortimer, P.E.; Xu, J.; Promputtha, I.; Doilom, M.; Yang, J.B.; Tang, A.M.C.; Karunarathna, S.C. Identification of endophytic fungi from leaves of Pandanaceae based on their morphotypes and DNA sequence data from southern Thailand. MycoKeys 2018, 33, 25–67. [Google Scholar] [CrossRef]
- Bezerra, J.D.P.; Sandoval-Denis, M.; Paiva, L.M.; Silva, G.A.; Groenewald, J.Z.; Souza-Motta, C.M.; Crous, P.W. New endophytic Toxicocladosporium species from cacti in Brazil, and description of Neocladosporium gen. nov. IMA Fungus 2017, 8, 77–97. [Google Scholar] [CrossRef] [PubMed]
- Aghdam, S.A.; Fotouhifar, K. Identification of some endophytic fungi of cherry trees (Prunus avium) in Iran. Iran. J. Plant Prot. Sci. 2017, 48, 43–57. [Google Scholar] [CrossRef]
- Ali, A.; Bilal, S.; Khan, A.L.; Mabood, F.; Al-Harrasi, A.; Lee, I.J. Endophytic Aureobasidium pullulans BSS6 assisted developments in phytoremediation potentials of Cucumis sativus under Cd and Pb stress. J. Plant Interact. 2019, 14, 303–313. [Google Scholar] [CrossRef]
- Patil, M.; Patil, R.; Mohammad, S.; Maheshwari, V. Bioactivities of phenolics-rich fraction from Diaporthe arengae TATW2, an endophytic fungus from Terminalia arjuna (Roxb.). Biocatal. Agric. Biotechnol. 2017, 10, 396–402. [Google Scholar] [CrossRef]
- Bills, G.F.; Menéndez, V.G.; Platas, G. Kabatiella bupleuri sp. nov. (Dothideales), a pleomorphic epiphyte and endophyte of the Mediterranean plant Bupleurum gibraltarium (Apiaceae). Mycologia 2012, 104, 962–973. [Google Scholar] [CrossRef]
- Silva, A.C.; Henriques, J.; Diogo, E.; Ramos, A.P.; Bragança, H. First report of Sydowia polyspora causing disease on Pinus pinea shoots. For. Pathol. 2020, 50, 27–30. [Google Scholar] [CrossRef]
- Pang, K.L.; Hyde, K.D.; Alias, S.A.; Suetrong, S.; Guo, S.Y.; Idid, R.; Gareth Jones, E.B. Dyfrolomycetaceae, a new family in the Dothideomycetes, Ascomycota. Cryptogam. Mycol. 2013, 34, 223–232. [Google Scholar] [CrossRef]
- Li, W.L.; Maharachchikumbura, S.S.N.; Cheewangkoon, R.; Liu, J.K. Reassessment of Dyfrolomyces and four new species of Melomastia from Olive (Olea europaea) in Sichuan Province, China. J. Fungi 2022, 8, 76. [Google Scholar] [CrossRef]
- Fungi of Great Britain and Ireland. 2021. Available online: https://fungi.myspecies.info/all-fungi/hysterium-angustatum (accessed on 14 April 2022).
- Kohlmeyer, J.; Kohlmeyer, E. Marine Fungi from Tropical America and Africa. Mycologia 1971, 63, 831–861. [Google Scholar] [CrossRef]
- Heuchert, B.; Braun, U.; Diederich, P.; Ertz, D. Taxonomic monograph of the genus Taeniolella s. lat. (Ascomycota). Fungal Syst. Evol. 2018, 2, 69–261. [Google Scholar] [CrossRef]
- Hernández-Restrepo, M.; Bezerra, J.D.P.; Tan, Y.P.; Wiederhold, N.; Crous, P.W.; Guarro, J.; Gené, J. Re-evaluation of Mycoleptodiscus species and morphologically similar fungi. Persoonia Mol. Phylogeny Evol. Fungi 2019, 42, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Bao, D.F.; Hyde, K.D.; Luo, Z.L.; Su, H.Y.; Nalumpang, S. Minutisphaera aquaticum sp. nov. increases the known diversity of Minutisphaeraceae. Asian J. Mycol. 2019, 2, 306–314. [Google Scholar] [CrossRef]
- Ferrer, A.; Miller, A.N.; Shearer, C.A. Minutisphaera and Natipusilla: Two new genera of freshwater Dothideomycetes. Mycologia 2011, 103, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Mapook, A.; Hyde, K.D.; Dai, D.Q.; Li, J.; Jones, E.B.G.; Bahkali, A.H.; Boonmee, S. Muyocopronales, ord. Nov., (Dothideomycetes, Ascomycota) and a reappraisal of Muyocopron species from northern Thailand. Phytotaxa 2016, 265, 225–237. [Google Scholar] [CrossRef]
- Selbmann, L.; De Hoog, G.S.; Zucconi, L.; Isola, D.; Ruisi, S.; Gerrits van den Ende, A.H.G.; Ruibal, C.; De Leo, F.; Urzì, C.; Onofri, S. Drought meets acid: Three new genera in a dothidealean clade of extremotolerant fungi. Stud. Mycol. 2008, 61, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Oerke, E.C.; Leucker, M.; Steiner, U. Sensory assessment of Cercospora beticola sporulation for phenotyping the partial disease resistance of sugar beet genotypes. Plant Methods 2019, 15, 133. [Google Scholar] [CrossRef]
- Shivas, R.G.; McTaggart, A.R.; Young, A.J.; Crous, P.W. Zasmidium scaevolicola. Fungal Planet 47. Persoonia 2010, 24, 132–133. [Google Scholar]
- Farr, D.F.; Miller, M.E.; Bruton, B.D. Rhizopycnis vagum gen. et sp. nov., a New coelomycetous fungus from roots of melons and sugarcane. Mycologia 1998, 90, 290–296. [Google Scholar] [CrossRef]
- McKenzie, E. Alternaria alternata (Alternaria alternata). 2013. Available online: http://www.padil.gov.au. (accessed on 20 August 2022).
- Tymon, L.S.; Peever, T.L.; Johnson, D.A. Identification and enumeration of small-spored Alternaria species associated with potato in the U.S. Northwest. Plant Dis. 2016, 100, 465–472. [Google Scholar] [CrossRef]
- Dai, D.; Bhat, D.J.; Liu, J.; Chukeatirote, E.; Zhao, R.; Hyde, K.D. Bambusicola, a new genus from bamboo with asexual and sexual morphs. Cryptogam. Mycol. 2012, 33, 363–379. [Google Scholar] [CrossRef]
- Manamgoda, D.S.; Rossman, A.Y.; Castlebury, L.A.; Crous, P.W.; Madrid, H.; Chukeatirote, E.; Hyde, K.D. The genus Bipolaris. Stud. Mycol. 2014, 79, 221–288. [Google Scholar] [CrossRef] [PubMed]
- Khiralla, A.; Mohamed, I.E.; Tzanova, T.; Schohn, H.; Slezack-Deschaumes, S.; Hehn, A.; André, P.; Carre, G.; Spina, R.; Lobstein, A.; et al. Endophytic fungi associated with Sudanese medicinal plants show cytotoxic and antibiotic potential. FEMS Microbiol. Lett. 2016, 363, fnw089. [Google Scholar] [CrossRef] [PubMed]
- Ahmadpour, A.; Heidarian, Z.; Karami, S.; Tsukiboshi, T.; Zhang, M.; Javan-Nikkhah, M. New species of Bipolaris and Curvularia on grass species in Iran. Rostaniha 2012, 13, 69–82. [Google Scholar]
- Boonmee, S.; Ko, T.W.K.; Chukeatirote, E.; Hyde, K.D.; Chen, H.; Cai, L.; McKenzie, E.H.C.; Jones, E.B.G.; Kodsueb, R.; Hassan, B.A. Two new Kirschsteiniothelia species with Dendryphiopsis anamorphs cluster in Kirschsteiniotheliaceae fam. nov. Mycologia 2012, 104, 698–714. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Guarro, J.; Sutton, D.A.; Acharya, K.; Barber, P.A.; Boekhout, T.; Dimitrov, R.A.; Dueñas, M.; Dutta, A.K.; et al. Fungal Planet description sheets. Persoonia 2015, 34, 167–266. [Google Scholar] [CrossRef] [PubMed]
- Budziszewska, J.; Szypuła, W.; Wilk, M.; Wrzosek, M. Paraconiothyrium babiogorense sp. nov., a new endophyte from fir club moss Huperzia selago (Huperziaceae). Mycotaxon 2011, 115, 457–468. [Google Scholar] [CrossRef]
- CEMAS (Center for Asturian Mycological Studies). 2022. Available online: https://www.centrodeestudiosmicologicosasturianos.org/?p=47642 (accessed on 10 May 2022).
- Blixt, E.; Djurle, A.; Yuen, J.; Olson, Å. Fungicide sensitivity in Swedish isolates of Phaeosphaeria nodorum. Plant Pathol. 2009, 58, 655–664. [Google Scholar] [CrossRef]
- Jiang, S.H.; Wei, X.L.; Wei, J.C. Two new species of Strigula (lichenised Dothideomycetes, Ascomycota) from China, with a key to the Chinese foliicolous species. MycoKeys 2017, 19, 31–42. [Google Scholar] [CrossRef]
- Schubert, K.; Rischel, A.; Braun, D.U. A monograph of Fusicladium s.lat. (Hyphomycetes). Schlechtendalia 2013, 9, 1–132. [Google Scholar]
- Shen, M.; Zhang, J.Q.; Zhao, L.L.; Groenewald, J.Z.; Crous, P.W.; Zhang, Y. Venturiales. Stud. Mycol. 2020, 96, 185–308. [Google Scholar] [CrossRef]
- Ibrahim, M.; Schlegel, M.; Sieber, T.N. Venturia orni sp. nov., a species distinct from Venturia fraxini, living in the leaves of Fraxinus ornus. Mycol. Prog. 2016, 15, 29. [Google Scholar] [CrossRef]
- Zhao, G.; Liu, X.; Wu, W. Helicosporous hyphomycetes from China. Fungal Divers. 2007, 26, 313–524. [Google Scholar]
- Sati, S.C.; Pathak, R. New root endophytic water borne conidial fungi from Kumaun Himalaya. Curr. Bot. 2017, 8, 12. [Google Scholar] [CrossRef]

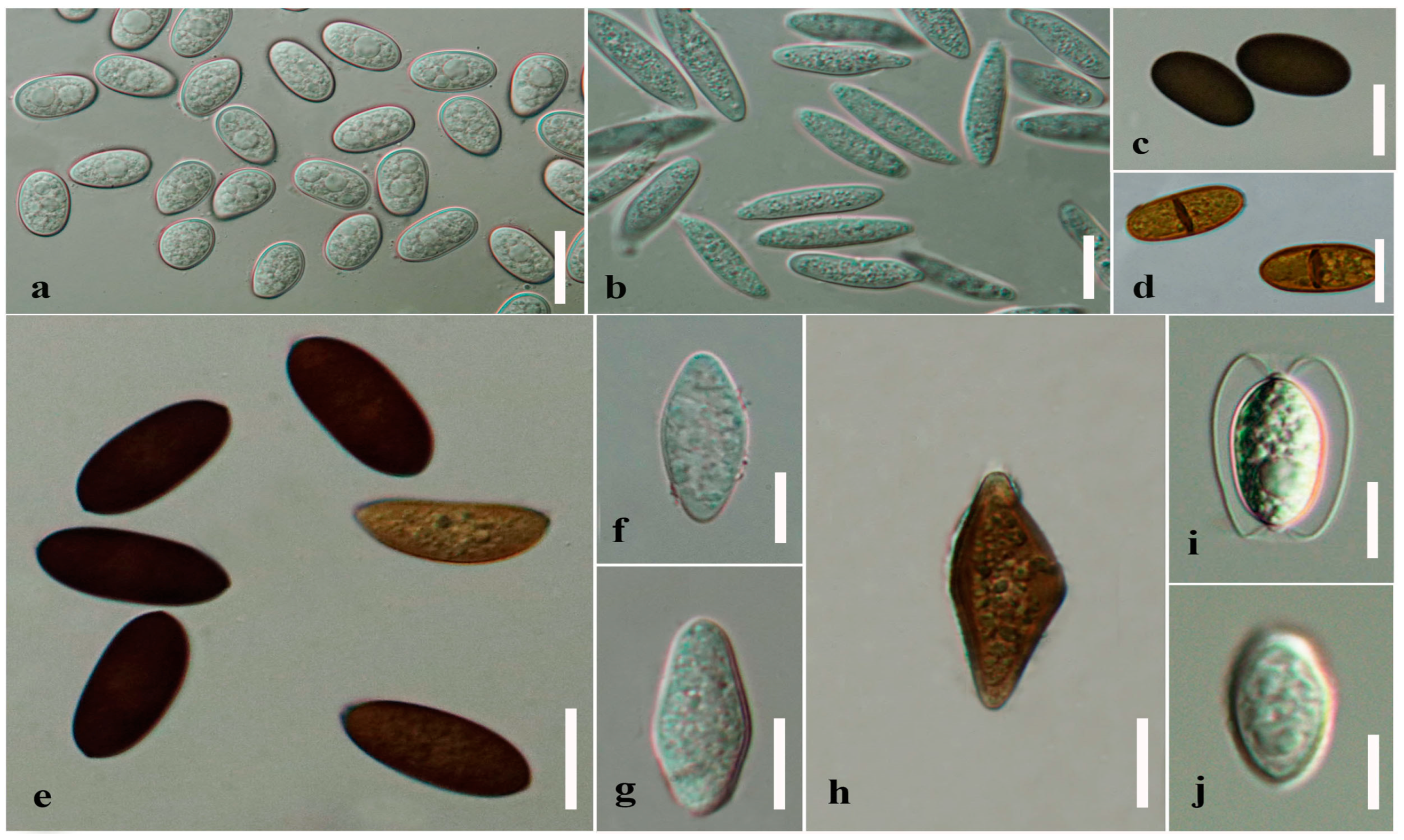







| Character | Aplosporellaceae | Botryosphaeriaceae | Melanopsaceae | Phyllostictaceae | Planistromellaceae | Saccharataceae | References | ||
|---|---|---|---|---|---|---|---|---|---|
| Colour | Ascospore | Pigmented | [3,4,7,36,49] | ||||||
| Hyaline | |||||||||
| Conidia | Pigmented | [3,4,50] | |||||||
| Hyaline | |||||||||
| Septation | Ascospore | Septate | [7] | ||||||
| Aseptate | |||||||||
| Conidia | Septate | [7,11,19,50] | |||||||
| Aseptate |
| Character | Parameter |
|---|---|
| Conidial colour | Hyaline (A), pigmented (B) and no asexual morph recorded (C) |
| Conidial septation | Aseptate (A), septate (B) and no asexual morph recorded (C) |
| Nutritional mode | Saprobes (A), pathogens (B) and endophytes (C) |
| Family | Divergence Times of Crown Age (Mya) | Divergence Times of Stem Age (Mya) |
|---|---|---|
| Aplosporellaceae | 42.8 (20.1–68.9) | 72.4 (46.9–101.2) |
| Botryosphaeriaceae | 69.9 (50.5–89.5) | 81.1 (60.9–102.1) |
| Melanopsaceae | 16.8 (5.1–36.8) | 72.9 (49.3–95.7) |
| Phyllostictaceae | 68. 6 (48.4–88.4) | 81.1 (60.9–102.1) |
| Planistromellaceae | 53.9 (34.5–72.7) | 72.9 (49.3–95.7) |
| Saccharataceae | 52.9 (31.3–79.5) | 72.4 (46.9–101.2) |
| Study | Slippers et al. [4] | Liu et al. [70] | Phillips et al. [7] | This Study | |
|---|---|---|---|---|---|
| No. of taxa | 140 | 364 | 100 | 306 | |
| Gene regions | SSU, LSU, ITS, tef1, β-tubulin and mtSSU (mitochondrial ribosomal small subunit) | LSU, SSU, tef1 and rpb2 | ITS and LSU | ITS and LSU | |
| Calibration/s | Mean = 0.000113 (SD = 0.000006) | Mean = 582.5 Mya (SD = 50.15 Mya) Fossil data 100 Mya (SD = 150 Mya) fossil Metacapnodiaceae | Mean = 110 Mya (SD = 5 Mya) | Mean = 110 Mya (SD = 5 Mya) | |
| Divergence time of crown age (Mya) | Aplosporellaceae | - | - | 40 | 43 |
| Botryosphaeriaceae | 44 | 44 | 61 | 70 | |
| Melanopsaceae | - | - | Not estimated | 17 | |
| Phyllostictaceae | 26 | 27 | 63 | 69 | |
| Planistromellaceae | 38 | 25 | 52 | 54 | |
| Saccharataceae | - | 28 | 50 | 53 | |
| Divergence time of stem age (Mya) | Aplosporellaceae | 57 | - | 94 | 72 |
| Botryosphaeriaceae | 87 | 52 | 94 | 81 | |
| Melanopsaceae | 75 | - | 74 | 73 | |
| Phyllostictaceae | 87 | 50 | 81 | 81 | |
| Planistromellaceae | 75 | 85 | 81 | 73 | |
| Saccharataceae | - | 114 | 74 | 72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rathnayaka, A.R.; Chethana, K.W.T.; Phillips, A.J.L.; Liu, J.-K.; Samarakoon, M.C.; Jones, E.B.G.; Karunarathna, S.C.; Zhao, C.-L. Re-Evaluating Botryosphaeriales: Ancestral State Reconstructions of Selected Characters and Evolution of Nutritional Modes. J. Fungi 2023, 9, 184. https://doi.org/10.3390/jof9020184
Rathnayaka AR, Chethana KWT, Phillips AJL, Liu J-K, Samarakoon MC, Jones EBG, Karunarathna SC, Zhao C-L. Re-Evaluating Botryosphaeriales: Ancestral State Reconstructions of Selected Characters and Evolution of Nutritional Modes. Journal of Fungi. 2023; 9(2):184. https://doi.org/10.3390/jof9020184
Chicago/Turabian StyleRathnayaka, Achala R., K. W. Thilini Chethana, Alan J. L. Phillips, Jian-Kui Liu, Milan C. Samarakoon, E. B. Gareth Jones, Samantha C. Karunarathna, and Chang-Lin Zhao. 2023. "Re-Evaluating Botryosphaeriales: Ancestral State Reconstructions of Selected Characters and Evolution of Nutritional Modes" Journal of Fungi 9, no. 2: 184. https://doi.org/10.3390/jof9020184
APA StyleRathnayaka, A. R., Chethana, K. W. T., Phillips, A. J. L., Liu, J.-K., Samarakoon, M. C., Jones, E. B. G., Karunarathna, S. C., & Zhao, C.-L. (2023). Re-Evaluating Botryosphaeriales: Ancestral State Reconstructions of Selected Characters and Evolution of Nutritional Modes. Journal of Fungi, 9(2), 184. https://doi.org/10.3390/jof9020184










