Engineering 3-Hydroxypropionic Acid Production from Glucose in Yarrowia lipolytica through Malonyl-CoA Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain and Plasmid Construction
| Strain | Description | Reference |
|---|---|---|
| E. coli JM109 | For construction of recombinant plasmids | Vazyme |
| Y. lipolytica Po1f | MATa, Leu2−, Ura3− | [32] |
| Po1f-wMCR | Po1f cells harboring p1312-wMCR | this study |
| Po1f-tMCR | Po1f cells harboring p1312-tMCR | this study |
| Po1f-NC-00 | Po1f cells harboring p1312-tMCRN-tMCRC | this study |
| Po1f-NC-01 | Po1f cells harboring p1312-NC-GAPN | this study |
| Po1f-NC-02 | Po1f cells harboring p1312-NC-GAPN | this study |
| Po1f-NC-03 | Po1f cells harboring p1312-NC-GAPN and p1269-ACC1 | this study |
| Po1f-NC-04 | Po1f cells harboring p1312-NC-GAPN and p1269-ACC1-ACC1 | this study |
| Po1f-NC-05 | Po1f cells harboring p1312-NC-GAPN and p1269-ALD6 | this study |
| Po1f-NC-06 | Po1f cells harboring p1312-NC-GAPN and p1269-ACS | this study |
| Po1f-NC-07 | Po1f cells harboring p1312-NC-GAPN and p1269-ACS-ACS | this study |
| Po1f-NC-08 | Po1f cells harboring p1312-NC-GAPN and p1269-ACC1-ACS | this study |
| Po1f-NC-09 | Po1f-NC-08 transformed with JME4580-MLS1-sgRNA and fragment of donor-MLS1 | this study |
| Po1f-NC-10 | Po1f-NC-08 transformed with JME4580-CIT2-sgRNA and fragment of donor-CIT2 | this study |
| Po1f-NC-11 | Po1f-NC-09 transformed with JME4580-CIT2-sgRNA and fragment of donor-CIT2 | this study |
| Po1f-NC-12 | Po1f-NC-11 transformed with JME4580-HPDH-sgRNA and fragment of donor-HPDH | this study |
| Po1f-NC-13 | Po1f-NC-11 transformed with JME4580-MMSDH-sgRNA and fragment of donor-MMSDH | this study |
| Po1f-NC-14 | Po1f-NC-12 transformed with JME4580-MMSDH-sgRNA and fragment of donor-MMSDH | this study |
2.2. Materials and Yeast Cultivation
2.3. Enzyme Assay
2.4. Analytical Methods
2.5. Rna Isolation and Quantitative Real-Time PCR
2.6. Fed-Batch Fermentation
3. Results
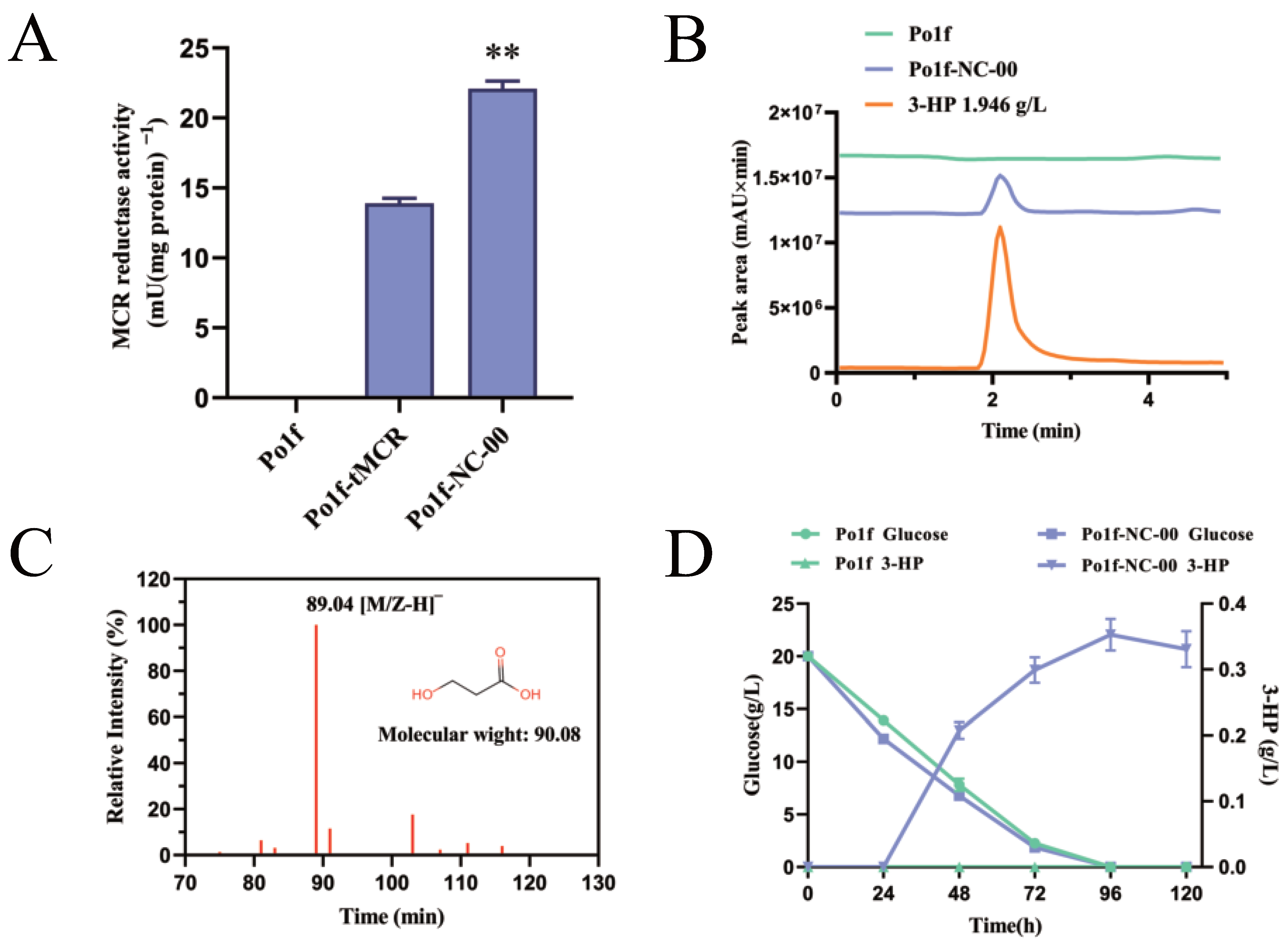
3.1. Construction of Malonyl-CoA Pathway for Synthesis of 3-HP in Y. lipolytica
3.2. Optimization of NADPH Supply to Increase 3-HP Production
3.3. Increasing Flux to 3-HP Production through Overexpression of MCR Pathway Genes
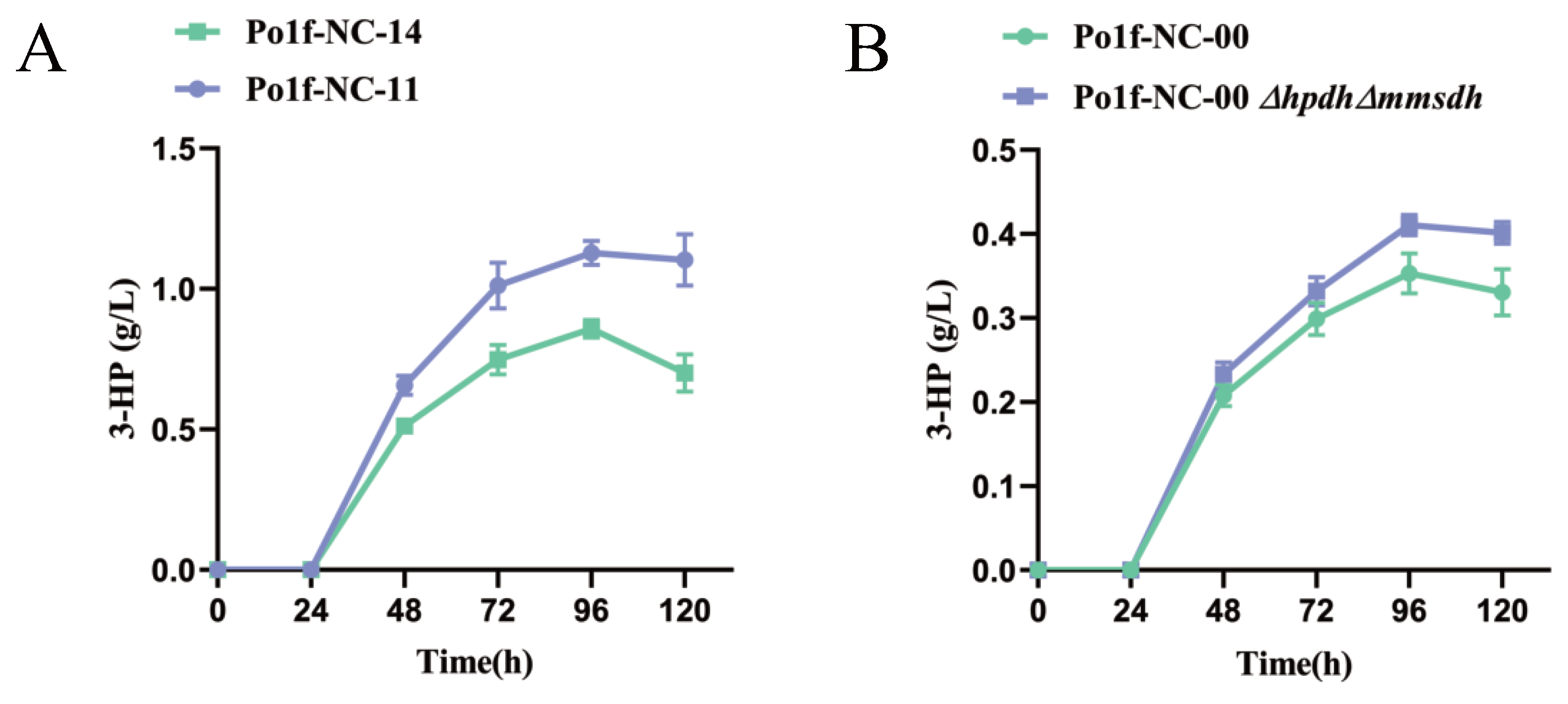
3.4. Knockout of Bypass Genes of Malonyl-CoA Pathway to Glyoxylate Cycle
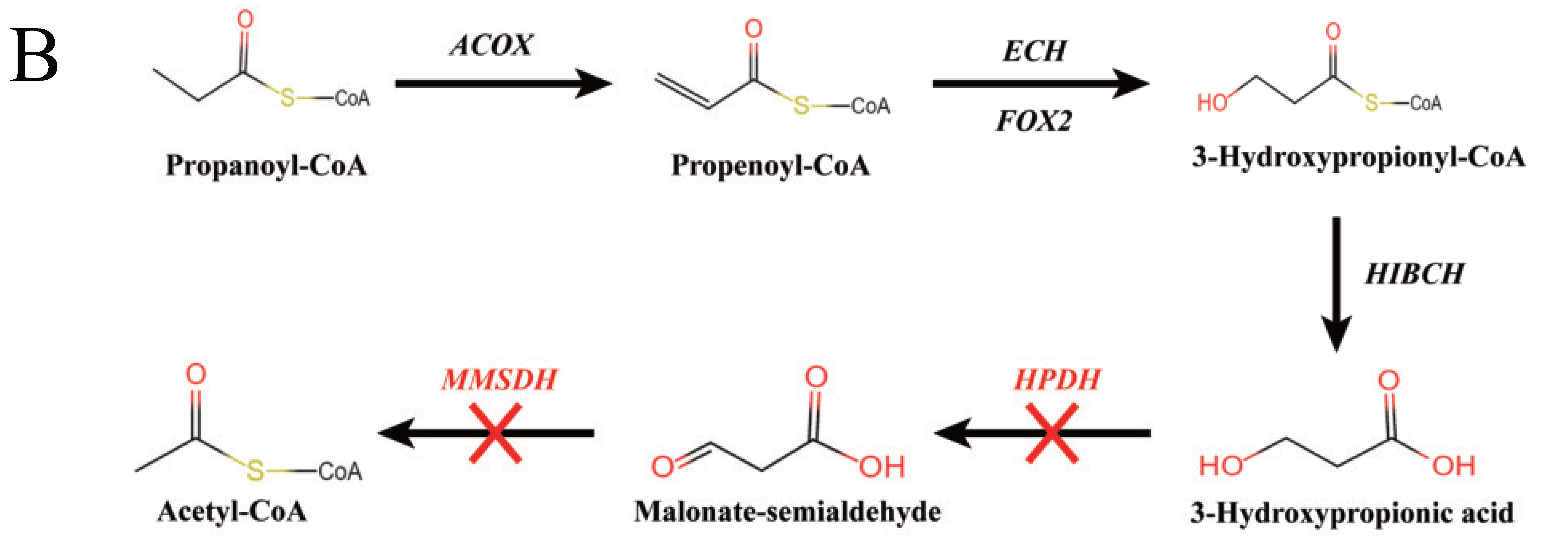
3.5. Knockout of 3-HP Degradation Related Genes in Malonate Semialdehyde Pathway
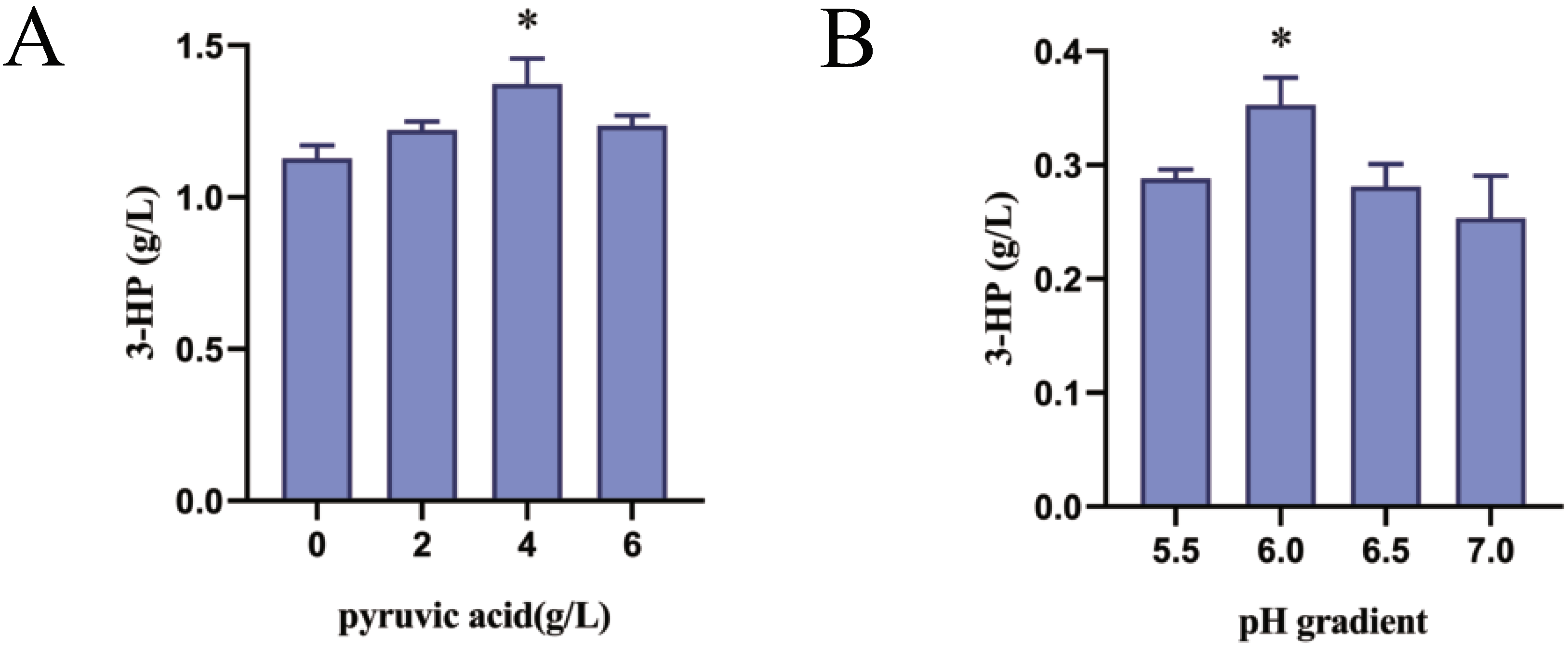
3.6. Effect of Pyruvic Acid Addition
3.7. Fermentation-Relevant Characteristics of 3-HP Synthesis by Y. lipolytica
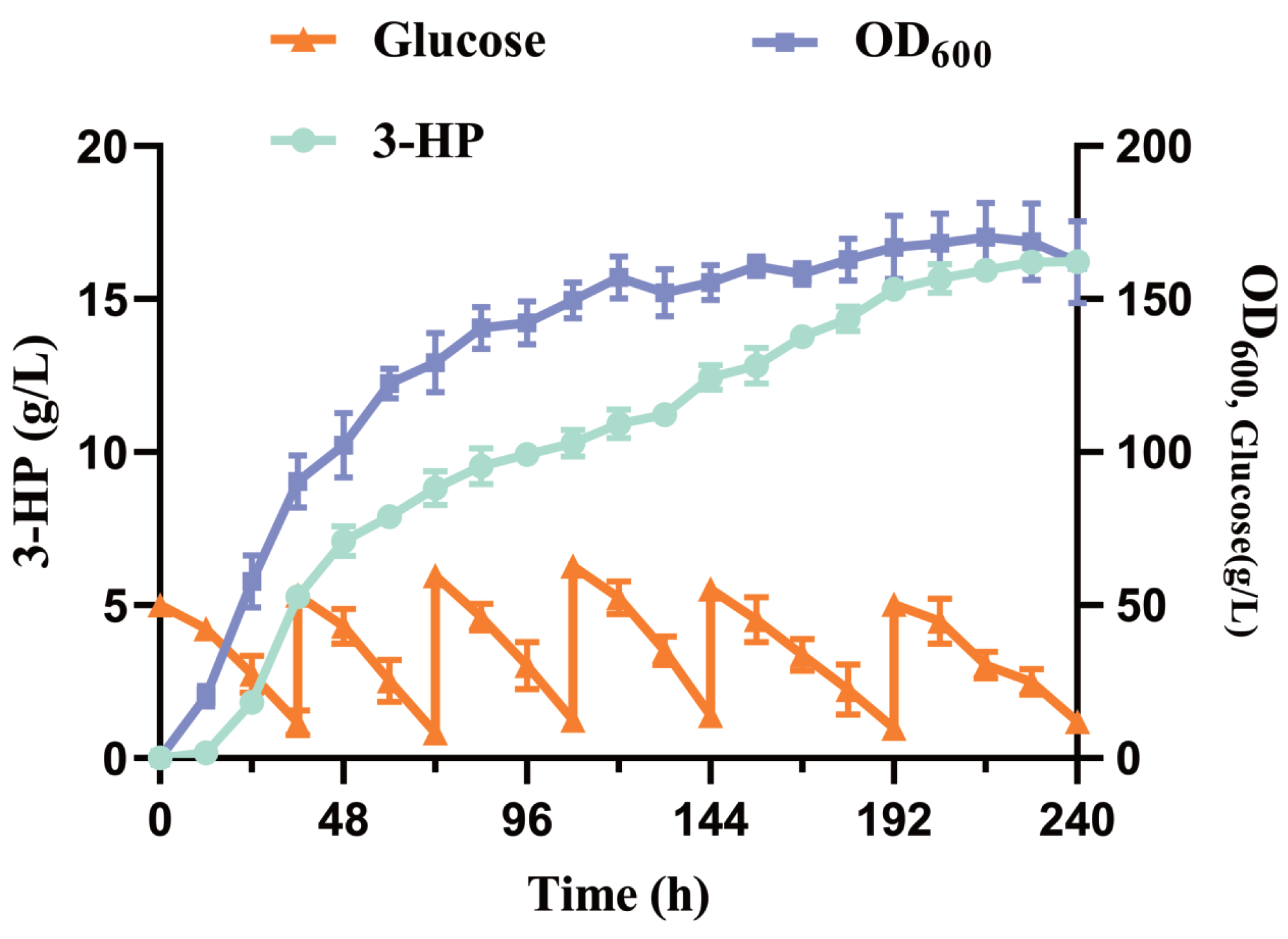
3.8. Fed-Batch Fermentation of 3-HP on Strain Po1f-NC-14
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zabed, H.M.; Akter, S.; Rupani, P.F.; Akor, J.; Zhang, Y.; Zhao, M.; Zhang, C.; Ragauskas, A.J.; Qi, X. Biocatalytic gateway to convert glycerol into 3-hydroxypropionic acid in waste-based biorefineries: Fundamentals, limitations, and potential research strategies. Biotechnol. Adv. 2022, 62, 108075. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.M.; Chen, W.J.; Yang, J.; Zhou, Y.M.; Hu, B.; Zhang, M.; Zhu, L.P.; Wang, G.Y.; Yang, S. Production of 3-hydroxypropionic acid in engineered Methylobacteriumextorquens AM1 Its Reassimilation through a Reductive Route. Microb. Cell Factories 2017, 16, 179. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cui, Z.; Sun, X.; Wang, Z.; Chen, T. Production of 3-Hydroxypropionic Acid from Renewable Substrates by Metabolically Engineered Microorganisms: A Review. Molecules 2023, 28, 1888. [Google Scholar] [CrossRef]
- McGregor, C.; Minton, N.P.; Kovács, K. Biosynthesis of poly (3HB-co-3HP) with variable monomer composition in recombinant Cupriavidus Necator H16. ACS Synth. Biol. 2021, 10, 3343–3352. [Google Scholar] [CrossRef] [PubMed]
- Bilal, M.; Xu, S.; Iqbal, H.M.; Cheng, H. Yarrowia lipolytica emerging biotechnological chassis forfunctional sugars biosynthesis. Crit. Rev. Food Sci. Nutr. 2021, 61, 535–552. [Google Scholar] [CrossRef]
- Jers, C.; Kalantari, A.; Garg, A.; Mijakovic, I. Production of 3-hydroxypropanoic acid from glycerol by metabolically engineered bacteria. Front. Bioeng. Biotechnol. 2019, 7, 124. [Google Scholar] [CrossRef]
- Raj, S.M.; Rathnasingh, C.; Jo, J.E.; Park, S. Production of 3-hydroxypropionic acid from glycerol by a novel recombinant Escherichia coli BL21 Strain. Process Biochem. 2008, 43, 1440–1446. [Google Scholar] [CrossRef]
- Mohan Raj, S.; Rathnasingh, C.; Jung, W.C.; Park, S. Effect of process parameters on 3-hydroxypropionic acid production from glycerol using a recombinant Escherichia coli. Appl. Microbiol. Biotechnol. 2009, 84, 649–657. [Google Scholar] [CrossRef]
- Rathnasingh, C.; Raj, S.M.; Jo, J.E.; Park, S. Development and evaluation of efficient recombinant Escherichia coli strains production 3-Hydroxypropionic Acid Glycerol. Biotechnol. Bioeng. 2009, 104, 729–739. [Google Scholar]
- Chu, H.S.; Kim, Y.S.; Lee, C.M.; Lee, J.H.; Jung, W.S.; Ahn, J.H.; Song, S.H.; Choi, I.S.; Cho, K.M. Metabolic engineering of 3-hydroxypropionic acid biosynthesis in Escherichia coli. Biotechnol. Bioeng. 2015, 112, 356–364. [Google Scholar] [CrossRef]
- Cheng, Z.; Jiang, J.; Wu, H.; Li, Z.; Ye, Q. Enhanced production of 3-hydroxypropionic acid from glucose via malonyl-CoA pathway by engineered Escherichia coli. Bioresour. Technol. 2016, 200, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ding, Y.; Zhang, R.; Liu, H.; Xian, M.; Zhao, G. Functional balance between enzymes in malonyl-CoA pathway for 3-hydroxypropionate biosynthesis. Metab. Eng. 2016, 34, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, X.; Ge, X.; Tian, P. High production of 3-hydroxypropionic acid in Kleb. Pneumoniae by systematic optimization of glycerol metabolism. Sci. Rep. 2016, 6, 26932. [Google Scholar] [CrossRef]
- Zhao, P.; Ma, C.; Xu, L.; Tian, P. Exploiting tandem repetitive promoters for high-level production of 3-hydroxypropionic acid. Appl. Microbiol. Biotechnol. 2019, 103, 4017–4031. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.Y.; Ding, Y.; Shi, T.Q.; Lin, L.; Huang, H.; Gao, Z.; Ji, X.J. Metabolic engineering of yeast for the production of 3-hydroxypropionic acid. Front. Microbiol. 2018, 9, 2185. [Google Scholar] [CrossRef]
- Shi, S.; Chen, Y.; Siewers, V.; Nielsen, J. Improving production of malonyl coenzyme A-derived metabolites by abolishing Snf1-dependent regulation of Acc1. mBio 2014, 5, e01130-14. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Bao, J.; Kim, I.K.; Siewers, V.; Nielsen, J. Coupled incremental precursor and co-factor supply improves 3-hydroxypropionic acid production in Saccharomyces cerevisiae. Metab. Eng. 2014, 22, 104–109. [Google Scholar] [CrossRef] [PubMed]
- David, F.; Nielsen, J.; Siewers, V. Flux control at the malonyl-CoA node through hierarchical dynamic pathway regulation in Saccharomyces cerevisiae. ACS Synth. Biol. 2016, 5, 224–233. [Google Scholar] [CrossRef]
- Li, S.; Si, T.; Wang, M.; Zhao, H. Development of a synthetic malonyl-CoA sensor in Saccharomyces cerevisiae for intracellular metabolite monitoring and genetic screening. ACS Synth. Biol. 2015, 4, 1308–1315. [Google Scholar] [CrossRef]
- Fina, A.; Brêda, G.C.; Pérez-Trujillo, M.; Freire, D.M.G.; Almeida, R.V.; Albiol, J.; Ferrer, P. Benchmarking recombinant Pichia Pastor. 3-Hydroxypropionic Acidproduction Glycerol. Microb. Biotechnol. 2021, 14, 1671–1682. [Google Scholar] [CrossRef]
- Mamaev, D.; Zvyagilskaya, R. Yarrowia lipolytica: A Multitalented Yeast Species Ecol. FEMS Yeast Res. 2021, 21, foab008. [Google Scholar] [CrossRef] [PubMed]
- Madzak, C. Yarrowia lipolytica strains and their biotechnological applications: How natural biodiversity and metabolic engineering could contribute to cell factories improvement. J. Fungi 2021, 7, 548. [Google Scholar] [CrossRef] [PubMed]
- Madzak, C.; Gaillardin, C.; Beckerich, J.M. Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: A review. J. Biotechnol. 2004, 109, 63–81. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Hill, A.; Liu, L.; Knight, R.; Miller, J.; Pan, A.; Otoupal, P.; Alper, H.S. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 2014, 5, 3131. [Google Scholar] [CrossRef]
- Blazeck, J.; Liu, L.; Knight, R.; Alper, H.S. Heterologous production of pentane in the oleaginous yeast Yarrowia lipolytica. J. Biotechnol. 2013, 165, 184–194. [Google Scholar] [CrossRef]
- Nicaud, J.M.; Madzak, C.; van den Broek, P.; Gysler, C.; Duboc, P.; Niederberger, P.; Gaillardin, C. Protein expression and secretion in the yeast Yarrowia lipolytica. FEMS Yeast Res. 2002, 2, 371–379. [Google Scholar]
- Juretzek, T.; Le Dall, M.T.; Mauersberger, S.; Gaillardin, C.; Barth, G.; Nicaud, J.M. Vectors for gene expression and amplification in the yeast Yarrowia lipolytica. Yeast 2001, 18, 97–113. [Google Scholar] [CrossRef]
- Blazeck, J.; Liu, L.; Redden, H.; Alper, H. Tuning gene expression in Yarrowia lipolytica A Hybrid Promot. Appl. Environ. Microbiol. 2011, 77, 7905–7914. [Google Scholar] [CrossRef]
- Pignède, G.; Wang, H.J.; Fudalej, F.; Seman, M.; Nicaud, J.M. Autocloning and amplification of LIP2 in Yarrowia lipolytica. Appl. Environ. Microbiol. 2000, 66, 3283–3289. [Google Scholar] [CrossRef]
- Gasmi, N.; Fudalej, F.; Kallel, H.; Nicaud, J.M. A molecular approach to optimize hIFN α2b expression and secretion in Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 2011, 89, 109–119. [Google Scholar] [CrossRef]
- Larroude, M.; Trabelsi, H.; Nicaud, J.M.; Rossignol, T. A set of Yarrowia lipolytica CRISPR/Cas9 vectors for exploiting wild-type strain diversity. Biotechnol. Lett. 2020, 42, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Barth, G.; Gaillardin, C. Yarrowia lipolytica. Nonconventional Yeasts in Biotechnology; Springer: Berlin/Heidelberg, Germany, 1996; pp. 313–388. [Google Scholar]
- Cao, X.; Wei, L.J.; Lin, J.Y.; Hua, Q. Enhancing linalool production by engineering oleaginous yeast Yarrowia lipolytica. Bioresour. Technol. 2017, 245, 1641–1644. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Shi, T.Q.; Wang, J.; Wei, P.; Ledesma-Amaro, R.; Ji, X.J. Engineering the lipid and fatty acid metabolism in Yarrowia lipolytica for sustainable production of high oleic oils. ACS Synth. Biol. 2022, 11, 1542–1554. [Google Scholar] [CrossRef] [PubMed]
- Fickers, P.; Cheng, H.; Sze Ki Lin, C. Sugar alcohols and organic acids synthesis in Yarrowia lipolytica: Where Are We? Microorganisms 2020, 8, 574. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, Q.; Xian, M.; Ding, Y.; Zhao, G. Dissection of malonyl-coenzyme A reductase of Chloroflexusaurantiacus Results in Enzyme Activity Improvement. PLoS ONE 2013, 8, e75554. [Google Scholar]
- Boyd, D.A.; Cvitkovitch, D.G.; Hamilton, I.R. Sequence, expression, and function of the gene for the nonphosphorylating, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase of Streptococcus mutans. J. Bacteriol. 1995, 177, 2622–2627. [Google Scholar] [CrossRef]
- Zhang, L.; Tang, Y.; Guo, Z.p.; Ding, Z.Y.; Shi, G.Y. Improving the ethanol yield by reducing glycerol formation using cofactor regulation in Saccharomyces cerevisiae. Biotechnol. Lett. 2011, 33, 1375–1380. [Google Scholar] [CrossRef]
- Chen, Y.; Daviet, L.; Schalk, M.; Siewers, V.; Nielsen, J. Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism. Metab. Eng. 2013, 15, 48–54. [Google Scholar] [CrossRef]
- Xu, P.; Qiao, K.; Ahn, W.S.; Stephanopoulos, G. Engineering Yarrowia lipolyticaas a platform for synthesis of drop-in transportation fuels and oleochemicals. Proc. Natl. Acad. Sci. USA 2016, 113, 10848–10853. [Google Scholar] [CrossRef]
- Cao, X.; Lv, Y.B.; Chen, J.; Imanaka, T.; Wei, L.J.; Hua, Q. Metabolic engineering of oleaginous yeast Yarrowia lipolytica Forlimonene Overproduction. Biotechnol. Biofuels 2016, 9, 214. [Google Scholar] [CrossRef]
- Yu, W.; Cao, X.; Gao, J.; Zhou, Y.J. Overproduction of 3-hydroxypropionate in a super yeast chassis. Bioresour. Technol. 2022, 361, 127690. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Sun, G.; Zhang, X.; Nie, Q.; Zhao, Y.; Yang, J. Recent advances, challenges and metabolic engineering strategies in the biosynthesis of 3-hydroxypropionic acid. Biotechnol. Bioeng. 2022, 119, 2639–2668. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiang, X.; Cui, Z.; Wang, Z.; Qi, Q.; Hou, J. Engineering the oleaginous yeast Yarrowia lipolytica for production of α-farnesene. Biotechnol. Biofuels 2019, 12, 296. [Google Scholar] [CrossRef]
- Chen, X.; Yang, X.; Shen, Y.; Hou, J.; Bao, X. Increasing malonyl-CoA derived product through controlling the transcription regulators of phospholipid synthesis in Saccharomyces cerevisiae. ACS Synth. Biol. 2017, 6, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.L.; Shi, T.Q.; Lin, L.; Ledesma-Amaro, R.; Ji, X.J. Advancing Yarrowia lipolytica as a superior biomanufacturing platform by tuning gene expression using promoter engineering. Bioresour. Technol. 2022, 347, 126717. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Carothers, J.M.; Keasling, J.D. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat. Biotechnol. 2012, 30, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Peng, Q.Q.; Chen, Y.Y.; Song, P.; Ji, X.J.; Huang, H.; Shi, T.Q. High-yield α-humulene production in Yarrowia lipolytica from waste cooking oil based on transcriptome analysis and metabolic engineering. Microb. Cell Factories 2022, 21, 271. [Google Scholar] [CrossRef]



| Plasmid | Description | Reference |
|---|---|---|
| pINA1312 | Y. lipolytica-integrative plasmid, hp4d promoter, XPR2 terminator, ura3d1 selection marker, Km | [26] |
| pINA1269 | Y. lipolytica-integrative plasmid, hp4d promoter, XPR2 terminator, LEU2 selection marker, Amp | [26] |
| pTrc99a-wMCR | pTrc99a plasmid contains wild MCR gene from C. aurantiacus | [12] |
| p1312-wMCR | pINA1312 plasmid contains wild MCR gene from C. aurantiacus | this study |
| p1312-tMCR | pINA1312 plasmid contains codon-optimized MCR gene from C. aurantiacus | this study |
| p1312-tMCRN-tMCRC | pINA1312 plasmid contains codon-optimized MCR-N gene cassette and MCR-C gene cassette | this study |
| p1312-NC-GAPN | pINA1312 plasmid contains codon-optimized MCR-N/C gene cassettes and codon optimized GAPN gene cassette from Bacillus cereus | this study |
| p1312-NC-GAPN | pINA1312 plasmid contains codon-optimized MCR-N/C gene cassettes and codon optimized GAPN gene cassette from Streptococcus mutans | this study |
| p1269-ACC1 | pINA1269 plasmid contains ACC1 gene from Y. lipolytica | this study |
| p1269-ACC1-ACC1 | pINA1269 plasmid contains two ACC1 gene cassettes | this study |
| p1269-ACS | pINA1269 plasmid contains codon-optimized mutant ACS gene from Salmonella enterica | this study |
| p1269-ACS-ACS | pINA1269 plasmid contains two ACS gene cassettes | this study |
| p1269-ALD6 | pINA1269 plasmid contains ALD6 gene from Y. lipolytica | this study |
| p1269-ACC1-ACS | pINA1269 plasmid contains ACC1 gene cassette and mutant ACS gene cassette | this study |
| JME4580 | Y. lipolytica CRISPR/Cas9 plasmid, HPHex marker for sgRNA cloning, RFP, Amp | Addgene |
| JME4580-MLS1-sgRNA | JME4580 plasmid contains sgRNA of MLS1 gene instead of RFP expression gene | this study |
| JME4580-CIT2-sgRNA | JME4580 plasmid contains sgRNA of CIT2 gene instead of RFP expression gene | this study |
| JME4580-HPDH-sgRNA | ME4580 plasmid contains sgRNA of HPDH gene instead of RFP expression gene | this study |
| JME4580-MMSDH-sgRNA | JME4580 plasmid contains sgRNA of MMSDH gene instead of RFP expression gene | this study |
| pCE-Zero | For constructing CRISPR/Cas9 donor fragment, Amp, Km, linearized | Vazyme |
| donor-MLS1 | pCE-Zero plasmid contains donor fragment of 1kb upstream and 1kb downstream of MLS1 gene (YALI0D19140g) | this study |
| donor-CIT2 | pCE-Zero plasmid contains donor fragment of 1kb upstream and 1kb downstream of CIT2 gene (YALI0E00638g) | this study |
| donor-HPDH | pCE-Zero plasmid contains donor fragment of 1kb upstream and 1kb downstream of HPDH gene (YALI0F02607g) | this study |
| donor-MMSDH | pCE-Zero plasmid contains donor fragment of 1kb upstream and 1kb downstream of MMSDH gene (YALI0C01859g) | this study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Sun, Y.; Wei, T.; Gong, D.; Wang, Q.; Zhan, Z.; Song, J. Engineering 3-Hydroxypropionic Acid Production from Glucose in Yarrowia lipolytica through Malonyl-CoA Pathway. J. Fungi 2023, 9, 573. https://doi.org/10.3390/jof9050573
Liu S, Sun Y, Wei T, Gong D, Wang Q, Zhan Z, Song J. Engineering 3-Hydroxypropionic Acid Production from Glucose in Yarrowia lipolytica through Malonyl-CoA Pathway. Journal of Fungi. 2023; 9(5):573. https://doi.org/10.3390/jof9050573
Chicago/Turabian StyleLiu, Shiyu, Yao Sun, Tianhui Wei, Dianliang Gong, Qi Wang, Zhe Zhan, and Jinzhu Song. 2023. "Engineering 3-Hydroxypropionic Acid Production from Glucose in Yarrowia lipolytica through Malonyl-CoA Pathway" Journal of Fungi 9, no. 5: 573. https://doi.org/10.3390/jof9050573





