Conidia Fusion: A Mechanism for Fungal Adaptation to Nutrient-Poor Habitats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.1.1. Testing Nematode-Trapping Fungi (NTF)
2.1.2. Culture Medium
2.2. Experimental Methods
2.2.1. Strain Rejuvenation
2.2.2. Species Specificity of Conidia Fusion (CF)
2.2.3. Physiological Period of CF Formation
2.2.4. Effect of Nutrient Concentration on CFR
2.2.5. Effect of Conidia Number in Conidia Fusion Body (CFB) on the Germination of the CFB
2.2.6. Data Management and Analysis
3. Results
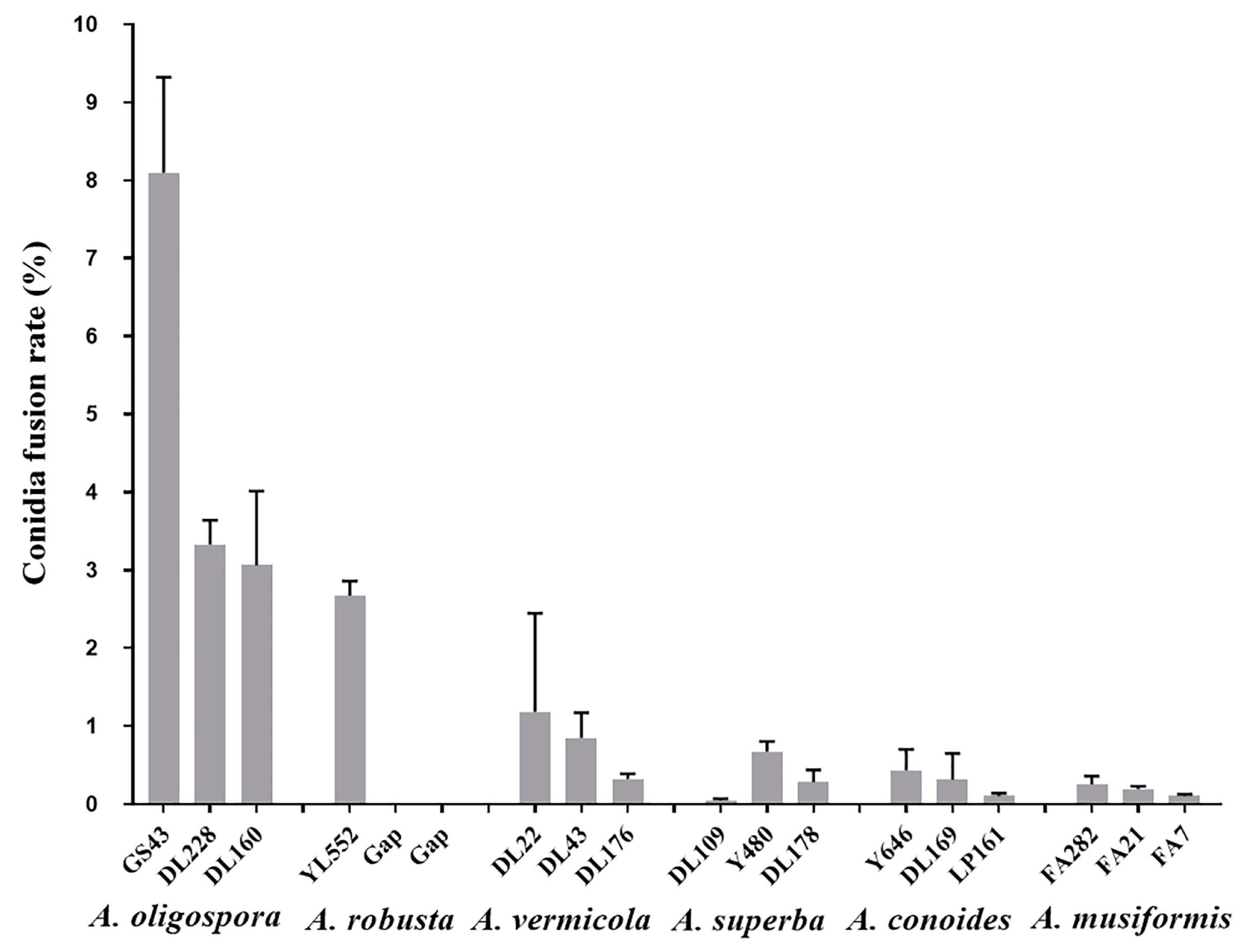
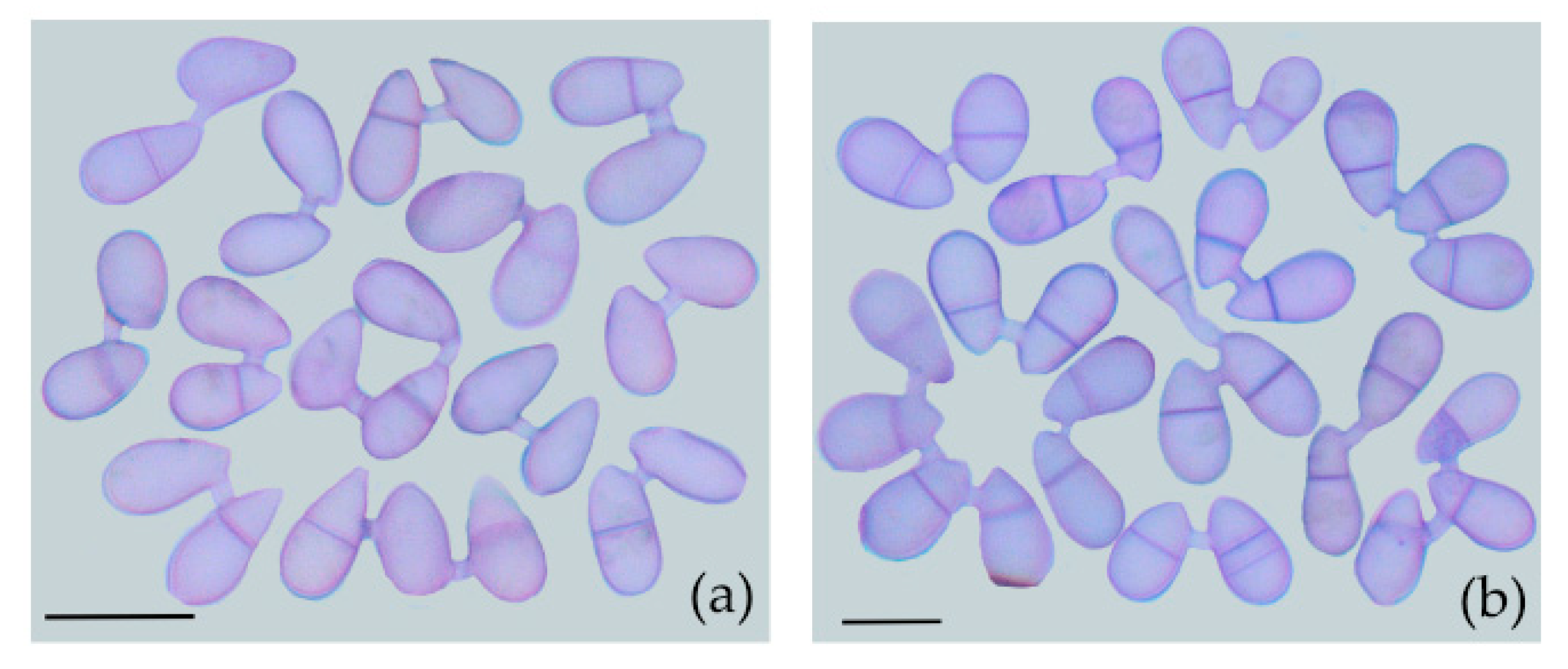
3.1. Species Specificity of Conidia Fusion (CF)
3.2. Physiological Period of CF
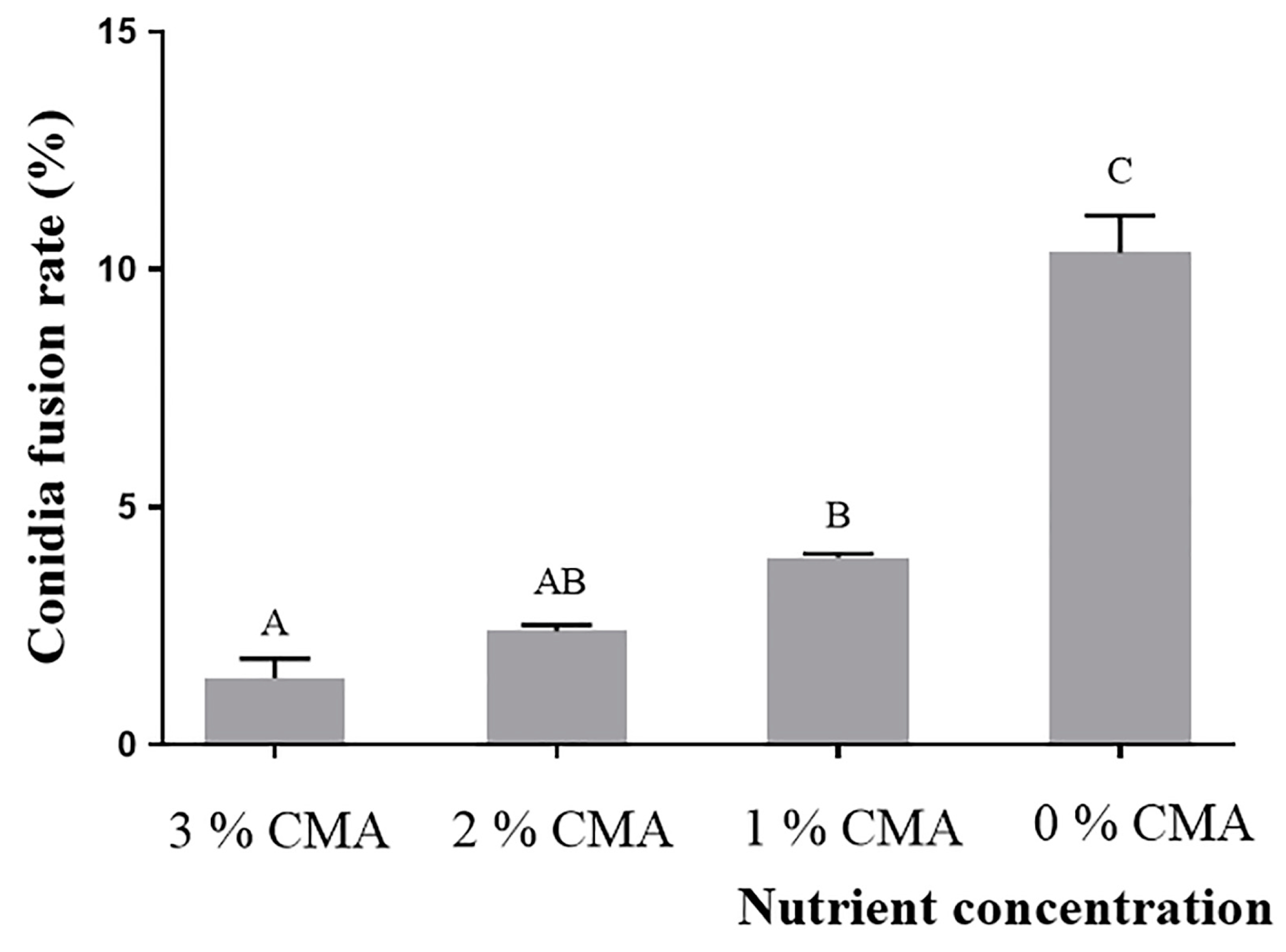
3.3. CFR of A. oligospora (DL228) Cultured at Different Nutrient Concentrations
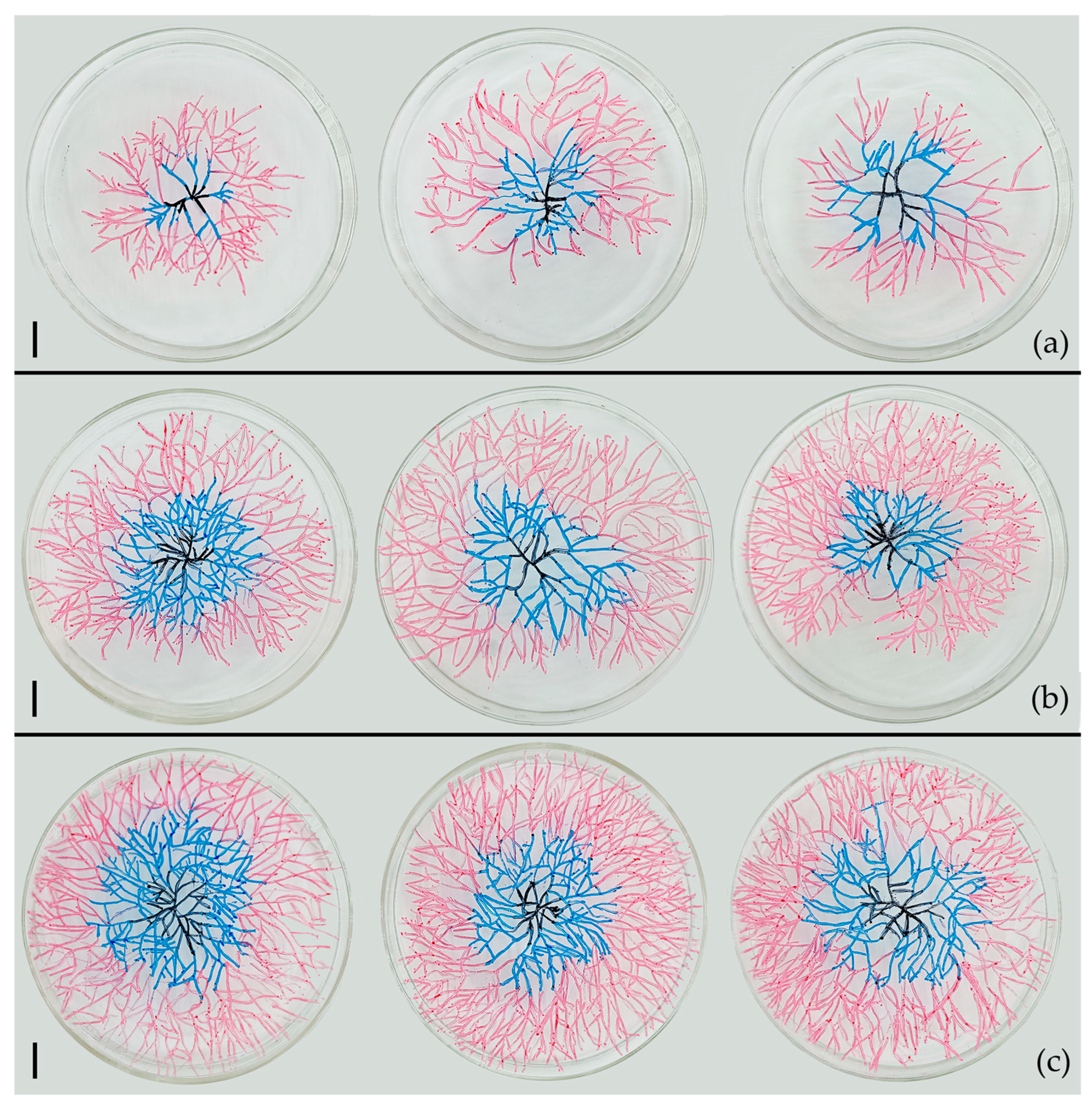
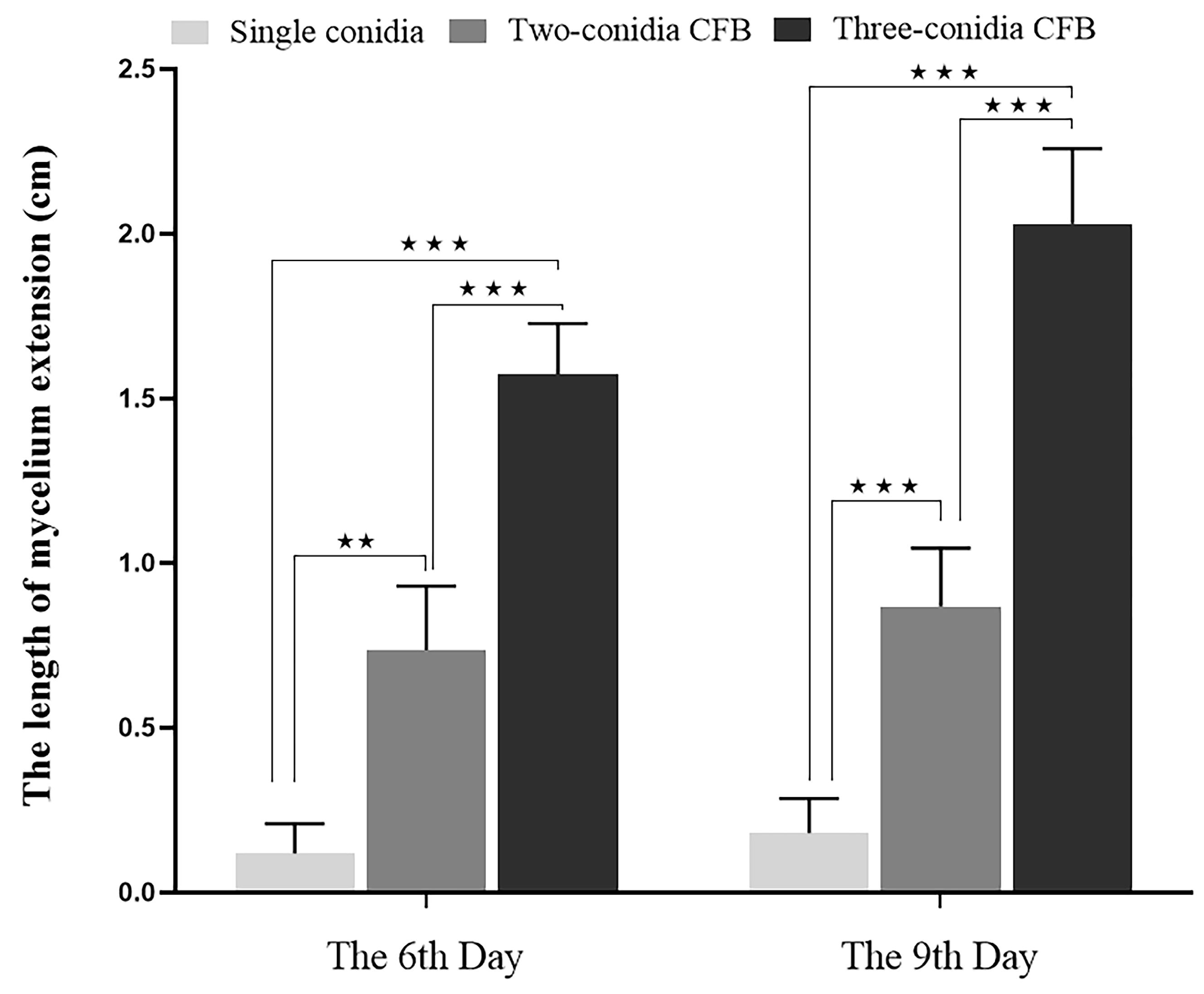
3.4. Influence of Conidia Number in Conidia Fusion Body (CFB) on the Germination of CFB
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Serna-Chavez, H.M.; Fierer, N.; van Bodegom, P.M. Global drivers and patterns of microbial abundance in soil. Glob. Ecol. Biogeogr. 2013, 22, 1162–1172. [Google Scholar] [CrossRef]
- Beule, L.; Guerra, V.; Lehtsaar, E.; Vaupel, A. Digging deeper: Microbial communities in subsoil are strongly promoted by trees in temperate agroforestry systems. Plant Soil 2022, 480, 423–437. [Google Scholar] [CrossRef]
- Yan, N.; Marschnerc, P.; Cao, W.H.; Zhou, C.Q.; Qin, W. Influence of salinity and water content on soil microorganisms. Int. Soil Water Conserv. Res. 2015, 3, 316–323. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.S.; Zhang, R.K.; Liu, Z.H.; Li, B.Z.; Yuan, Y.J. Microbial adaptation to enhance stress tolerance. Front. Microbiol. 2022, 13, 888746. [Google Scholar] [CrossRef] [PubMed]
- Mackelprang, R.; Vaishampayan, P.; Fisher, K. Adaptation to environmental extremes structures functional traits in biological soil crust and hypolithic microbial communities. Msystem 2022, 7, e0141921. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xiang, M.; Che, Y. The living strategy of nematophagous fungi. Mycoscience 2009, 50, 20–25. [Google Scholar] [CrossRef]
- Schiessl, K.T.; Janssen, E.M.L.; Kraemer, S.M.; Kraemer, M.N.; Ackermann, M. Magnitude and mechanism of siderophore-mediated competition at low iron solubility in the Pseudomonas aeruginosa pyochelin system. Front. Microbiol. 2017, 8, 1964. [Google Scholar] [CrossRef] [Green Version]
- Quispe-Cardenas, E.; Rogers, S. Microbial adaptation and response to high ammonia concentrations and precipitates during anaerobic digestion under psychrophilic and mesophilic conditions. Water Res. 2021, 204, 117596. [Google Scholar] [CrossRef]
- Blath, J.; Tóbiás, A. Invasion and fixation of microbial dormancy traits under competitive pressure. Stoch. Process. Appl. 2020, 130, 7363–7395. [Google Scholar] [CrossRef]
- Gabriela, R.M.; Read, N.D.; Wheals, A.E. Conidial anastomosis tubes in filamentous fungi. FEMS Microbiol. Lett. 2005, 249, 191–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzog, S.; Schumann, M.R.; Fleißner, A. Cell fusion in Neurospora crassa. Curr. Opin. Microbiol. 2015, 28, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Read, N.D.; Goryachev, A.B.; Lichius, A. The mechanistic basis of self-fusion between conidial anastomosis tubes during fungal colony initiation. Fungal Biol. Rev. 2012, 26, 1–11. [Google Scholar] [CrossRef]
- Goncalves, A.E.; Velho, A.C.; Stadnik, M.J. Formation of conidial anastomosis tubes and melanization of appressoria are antagonistic processes in Colletotrichum spp. from apple. Eur. J. Plant Pathol. 2016, 146, 497–506. [Google Scholar] [CrossRef]
- Fischer, M.S.; Glass, N.L. Communicate and fuse: How filamentous fungi establish and maintain an interconnected mycelial network. Front. Microbiol. 2019, 10, 619. [Google Scholar] [CrossRef] [Green Version]
- Tulasne, L.R.; Tulasne, C. Selecta Fungorum Carpologia; Imperial Press: Paris, France, 1863. [Google Scholar]
- Roca, M.G.; Davide, L.C.; Mendes-Costa, M.C.; Wheals, A. Conidial anastomosis tubes in Colletotrichum. Fungal Genet. Biol. 2003, 40, 138–145. [Google Scholar] [CrossRef]
- Ishikawa, F.H.; Souza, E.A.; Read, N.D.; Roca, M.G. Live-cell imaging of conidial fusion in the bean pathogen, Colletotrichum lindemuthianum. Fungal Biol. 2010, 114, 2–9. [Google Scholar] [CrossRef]
- Roca, M.G.; Arlt, J.; Jeffree, C.E.; Read, N.D. Cell biology of conidial anastomosis tubes in Neurospora crassa. Eukaryot. Cell. 2005, 4, 911–919. [Google Scholar] [CrossRef] [Green Version]
- Mehta, N.; Baghela, A. Quorum sensing-mediated inter-specific conidial anastomosis tube fusion between Colletotrichum gloeosporioides and C. siamense. IMA Fungus 2021, 12, 7. [Google Scholar] [CrossRef]
- Glass, N.L.; Fleissner, A. Re-Wiring the Network: Understanding the Mechanism and Function of Anastomosis in Filamentous Ascomycete Fungi; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar] [CrossRef]
- Kurian, S.M.; Di Pietro, A.; Read, N.D. Live-cell imaging of conidial anastomosis tube fusion during colony initiation in Fusarium oxysporum. PLoS ONE 2018, 13, e0195634. [Google Scholar] [CrossRef] [Green Version]
- Mehta, N.; Patil, R.; Baghela, A. Differential physiological prerequisites and gene expression profiles of conidial anastomosis tube and germ tube formation in Colletotrichum gloeosporioides. J. Fungi—Open Access Mycol. J. 2021, 7, 509. [Google Scholar] [CrossRef]
- Jiang, X.; Xiang, M.; Liu, X. Nematode-Trapping Fungi. Microbiol. Spectr. 2017, 5, 963–974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, H.; Zhao, Y.; Zhou, J.; Feng, H.; Jiang, D.; Zhang, K.Q.; Yang, J. Trapping devices of nematode-trapping fungi: Formation, evolution, and genomic perspectives. Biol. Rev. Camb. Philos. Soc. 2017, 92, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.C.; Li, X.M.; Zhao, N.; Yang, L.; Zhang, K.Q.; Yang, J.K. Regulatory mechanism of trap formation in the nematode-trapping fungi. J. Fungi 2022, 8, 406. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, E.; An, Z.; Liu, X.Z. Evolution of nematode-trapping cells of predatory fungi of the Orbiliaceae based on evidence from rRNA-encoding DNA and multiprotein sequences. Proc. Natl. Acad. Sci. USA 2007, 104, 8379–8384. [Google Scholar] [CrossRef]
- Yang, E.; Xu, L.; Yang, Y.; Zhang, X.Y.; Xiang, M.C.; Wang, C.S.; An, Z.Q.; Liu, X.Z. Origin and evolution of carnivorism in the Ascomycota (fungi). Proc. Natl. Acad. Sci. USA 2012, 109, 10960–10965. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Yu, Z.; Yang, J.; Li, J.; Liang, L.M.; Zhang, K.Q. Expansion of adhesion genes drives pathogenic adaptation of nematode-trapping fungi. iScience 2020, 23, 101057. [Google Scholar] [CrossRef]
- Soliman, M.S.; El-Deriny, M.M.; Ibrahim, D.S.S.; Zakaria, H.; Ahmed, Y. Suppression of root-knot nematode Meloidogyne incognita on tomato plants using the nematode trapping fungus Arthrobotrys oligospora Fresenius. J. Appl. Microbiol. 2021, 131, 2402–2415. [Google Scholar] [CrossRef]
- Kumar, D.; Gouda, S.A. Evaluation of mycoparasitic efficacy of nematode-trapping fungi against Rhizoctonia solani inciting sheath blight disease in rice (Oryza sativa L.). Biol. Control 2018, 122, 31–40. [Google Scholar] [CrossRef]
- Lin, H.C.; Hsueh, Y.P. Laboratory maintenance and culturing of the nematode-trapping fungus Arthrobotrys oligospora. Curr. Protoc. 2021, 1, e41. [Google Scholar] [CrossRef]
- Wang, S.; Liu, X. Tools and basic procedures of gene manipulation in nematode-trapping fungi. Mycology 2023, 14, 75–90. [Google Scholar] [CrossRef]
- Zhang, K.Q.; Mo, M.H. Flora Fungorum Sinicorum (Vol.33): Arthrobotrys et Gengra Cetera Cognata; Science Press: Beijing, China, 2006; pp. 1–157. [Google Scholar]
- Hay, F.S. Unusual germination of spores of Arthrobotrys conoides and A. cladodes. Mycol. Res. 1995, 99, 981–982. [Google Scholar] [CrossRef]
- Zhang, K.Q.; Hyde, K.D. Nematode-Trapping Fungi; Springer Science & Business: Berlin, Germany, 2014. [Google Scholar]
- Yu, Z.F.; Mo, M.H.; Zhang, Y.; Zhang, K.Q. Taxonomy of nematode-trapping fungi from Orbiliaceae, Ascomycota. In Nematode-Trapping Fungi, 2nd ed.; Zhang, K.Q., Hyde, K.D., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 41–210. [Google Scholar]
- Venkatesh, N.; Greco, C.; Drott, M.T.; Ludwikoski, I.; Keller, N.M.; Keller, N.P. Bacterial hitchhikers derive benefits from fungal housing. Curr. Biol. 2022, 32, 1523–1533. [Google Scholar] [CrossRef]
- Whittenbury, R.; McLee, A.G. Rhodopseudomonas palustris and Rh. viridis-photosynthetic budding bacteria. Arch. Mikrobiol. 1967, 59, 324–334. [Google Scholar] [CrossRef]
- Zhang, H.; Wei, Z.; Zhang, J.; Liu, X.F. Classification of dendrocola nematode-trapping fungi. For. Res. Engl. 2021, 32, 1295–1304. [Google Scholar] [CrossRef]
- Zhang, F.; Boonmee, S.; Yang, Y.Q.; Zhou, F.P.; Xiao, W.; Yang, X.Y. Arthrobotrys blastospora sp. nov. (Orbiliomycetes): A living fossil displaying morphological traits of mesozoic carnivorous fungi. J. Fungi 2023, 9, 451. [Google Scholar] [CrossRef]
- Yang, Y.Q.; Zhang, F.; Li, Z.Q.; Zhou, F.P.; Yang, X.Y.; Xiao, W. Morphological and multigene phylogenetic analyses reveal two new nematode trapping fungi (Arthrobotrys, Orbiliaceae) from Yunnan, China. Phytotaxa 2023, 591, 263–272. [Google Scholar] [CrossRef]
- Zhang, F.; Boonmee, S.; Monkai, J.; Yang, X.Y.; Xiao, W. Drechslerelladaliensis and D. xiaguanensis (Orbiliales, Orbiliaceae), two new nematode-trapping fungi from Yunnan, China. Biodivers. Data J. 2022, 10, e96642. [Google Scholar] [CrossRef]
- Li, Y.; Hyde, K.D.; Jeewon, R.; Cai, L.; Vijaykrishna, D.; Zhang, K. Phylogenetics and evolution of nematode-trapping fungi (Orbiliales) estimated from nuclear and protein coding genes. Mycologia 2005, 97, 1034–1046. [Google Scholar] [CrossRef]
- Fisher, R.M.; Cornwallis, C.K.; West, S.A. Group formation, relatedness, and the evolution of multicellularity. Curr. Biol. CB 2013, 23, 1120–1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernardes, J.P.; John, U.; Woltermann, N.; Valiadi, M.; Hermann, R.J.; Becks, L. The evolution of convex trade-offs enables the transition towards multicellularity. Nat. Commun. 2021, 12, 4222. [Google Scholar] [CrossRef] [PubMed]
- Mian, I.S.; Rose, C. Communication theory and multicellular biology. Integr. Biol. 2011, 3, 350–367. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Zhang, F.; Yang, Y.; Zhou, F.; Boonmee, S.; Xiao, W.; Yang, X. Conidia Fusion: A Mechanism for Fungal Adaptation to Nutrient-Poor Habitats. J. Fungi 2023, 9, 755. https://doi.org/10.3390/jof9070755
Yang X, Zhang F, Yang Y, Zhou F, Boonmee S, Xiao W, Yang X. Conidia Fusion: A Mechanism for Fungal Adaptation to Nutrient-Poor Habitats. Journal of Fungi. 2023; 9(7):755. https://doi.org/10.3390/jof9070755
Chicago/Turabian StyleYang, Xinju, Fa Zhang, Yaoquan Yang, Faping Zhou, Saranyaphat Boonmee, Wen Xiao, and Xiaoyan Yang. 2023. "Conidia Fusion: A Mechanism for Fungal Adaptation to Nutrient-Poor Habitats" Journal of Fungi 9, no. 7: 755. https://doi.org/10.3390/jof9070755
APA StyleYang, X., Zhang, F., Yang, Y., Zhou, F., Boonmee, S., Xiao, W., & Yang, X. (2023). Conidia Fusion: A Mechanism for Fungal Adaptation to Nutrient-Poor Habitats. Journal of Fungi, 9(7), 755. https://doi.org/10.3390/jof9070755





