Quantitative Acetylome Analysis of Differentially Modified Proteins in Virulence-Differentiated Fusarium oxysporum f. sp. cucumerinum Isolates during Cucumber Colonization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials, Fungal Strains, and Interactions
2.2. Protein Extraction, Digestion, and Peptides Analysis
2.3. Bioinformatics Analyses
3. Results
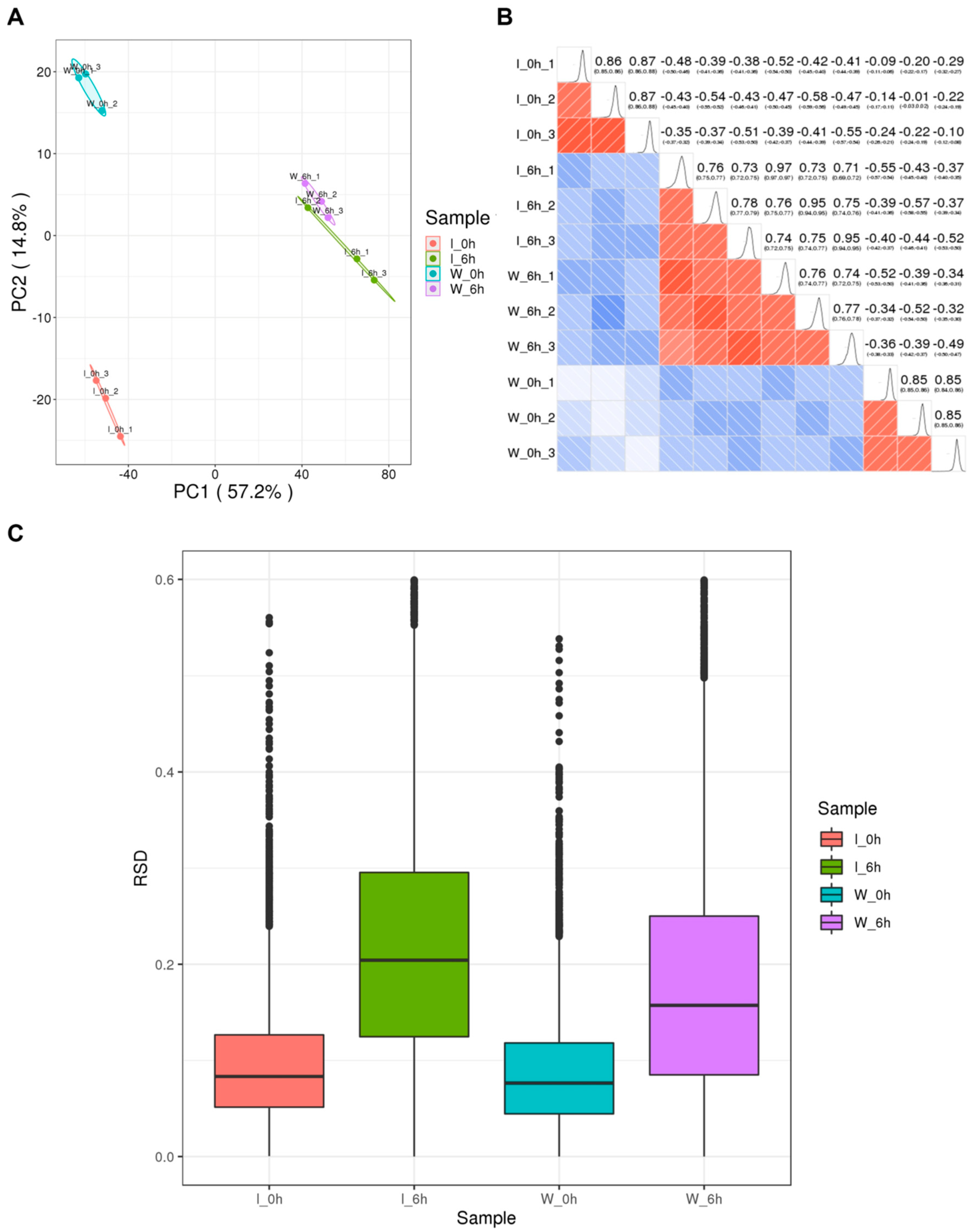
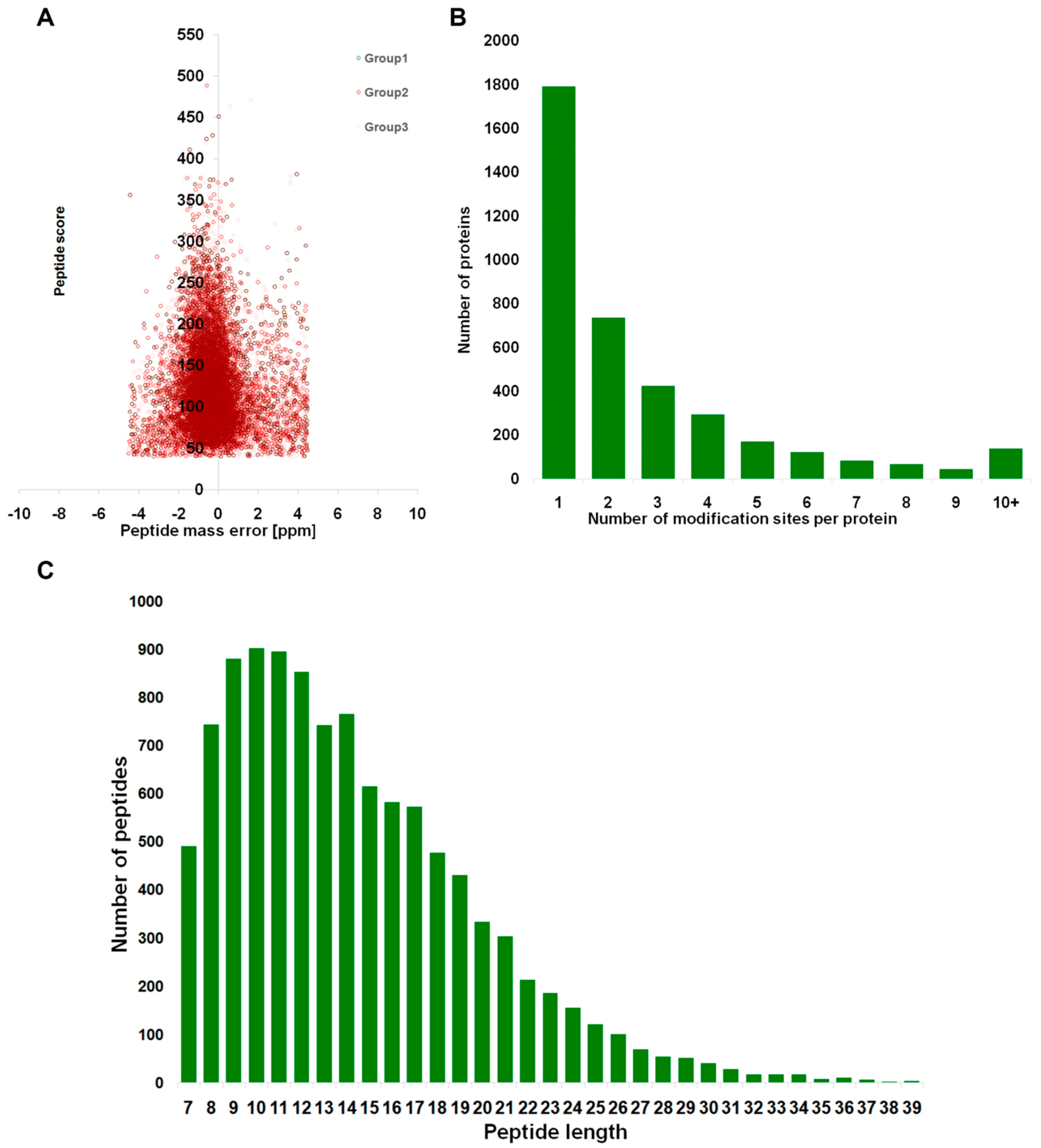
3.1. Comprehensive Analysis of Identified Lysine-Acetylated Proteins in Foc
3.2. Analysis of Lysine-Acetylated Motifs
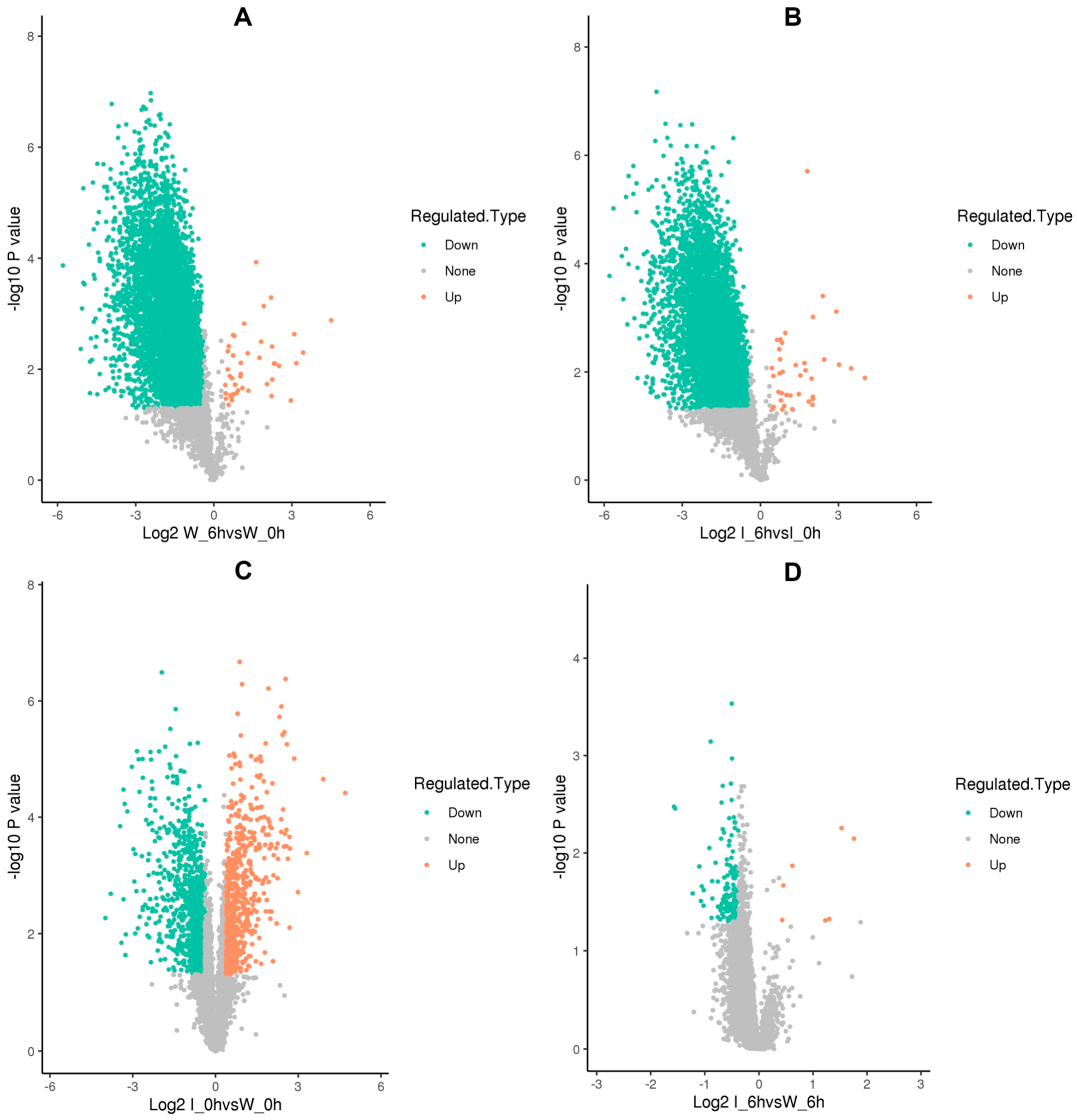
3.3. Characteristics of Differentially Lysine-Acetylated Sites and Proteins in Foc
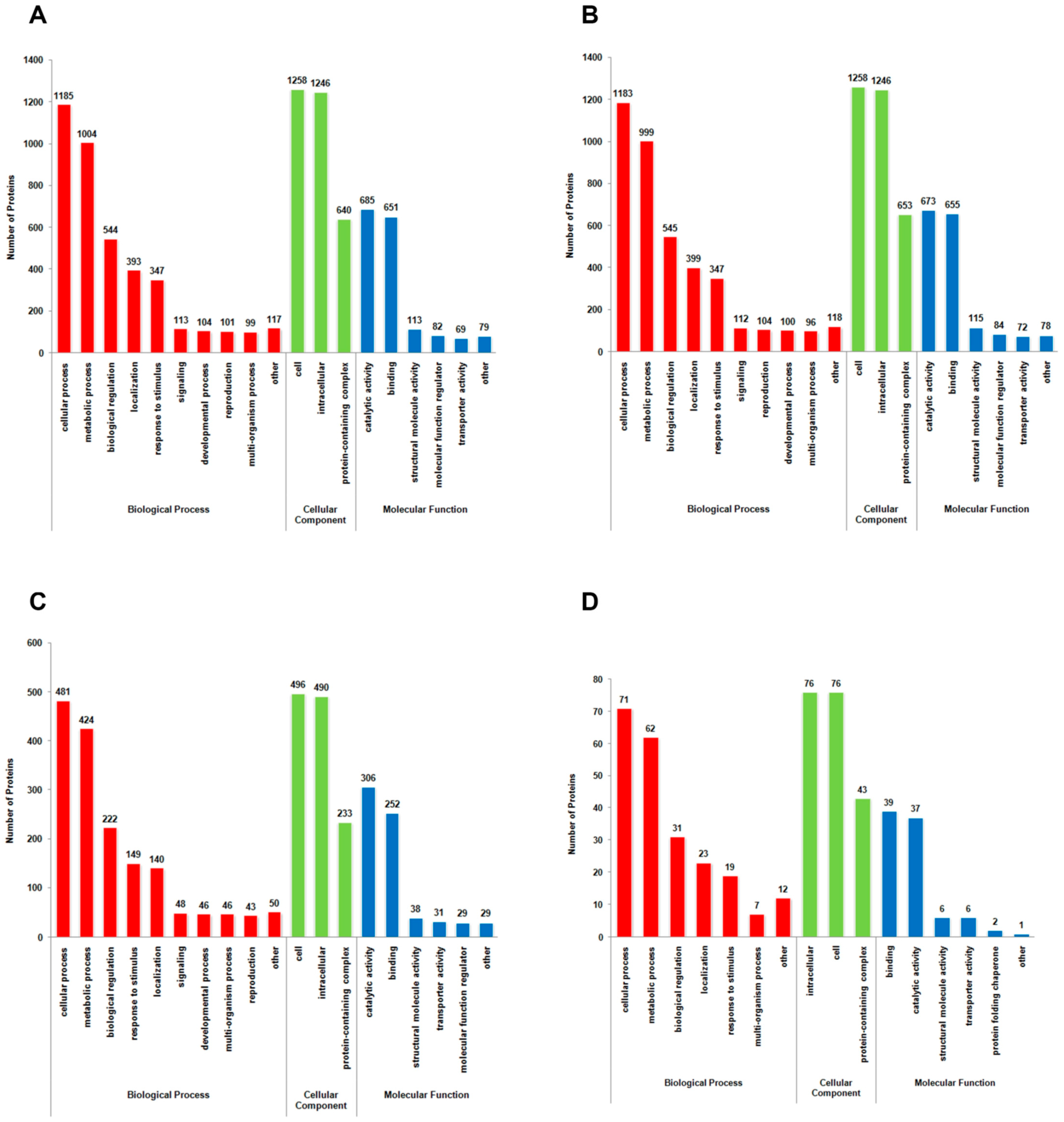
3.4. Functional Categorization and Subcellular Location of Differentially Lysine-Acetylated Proteins in Foc
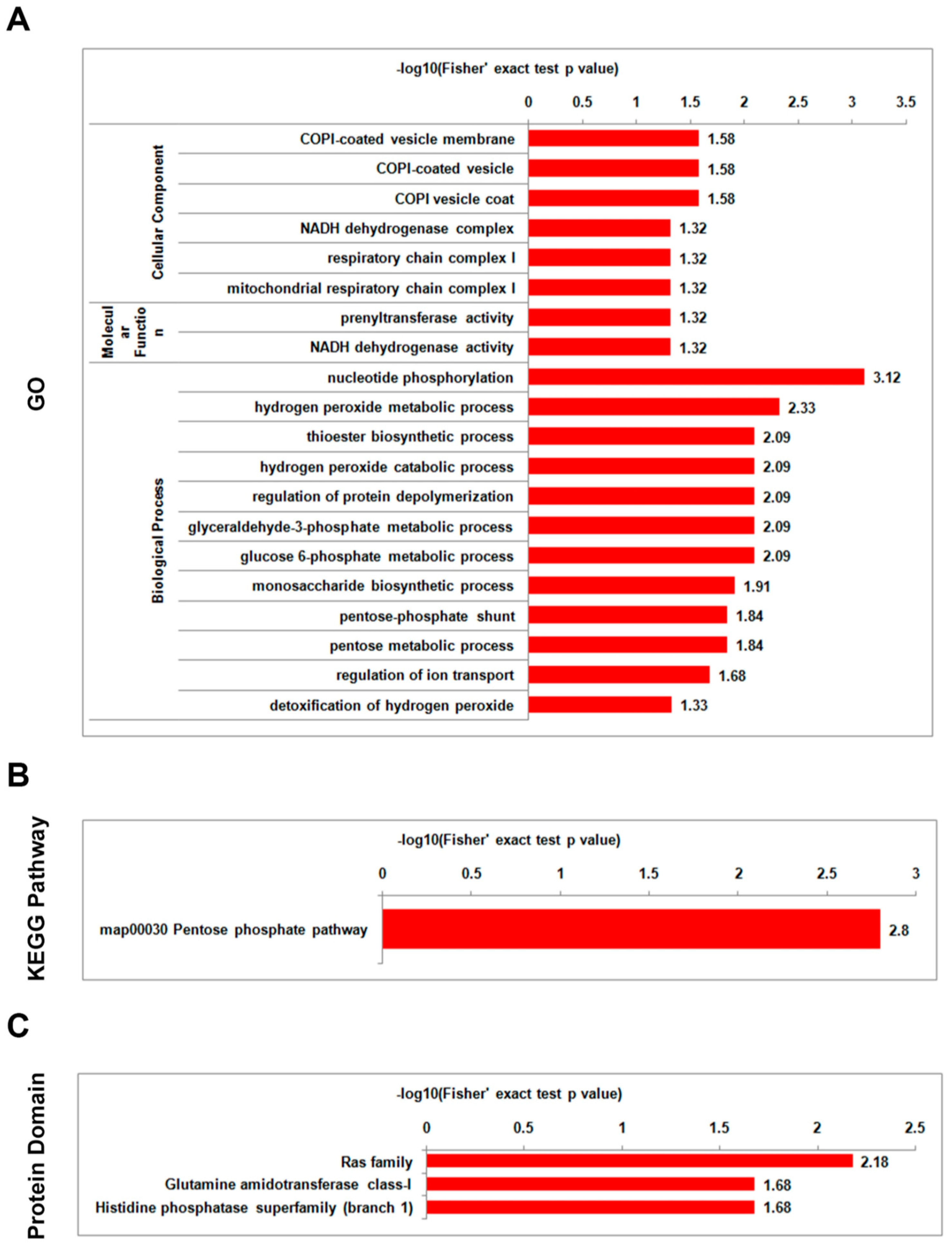
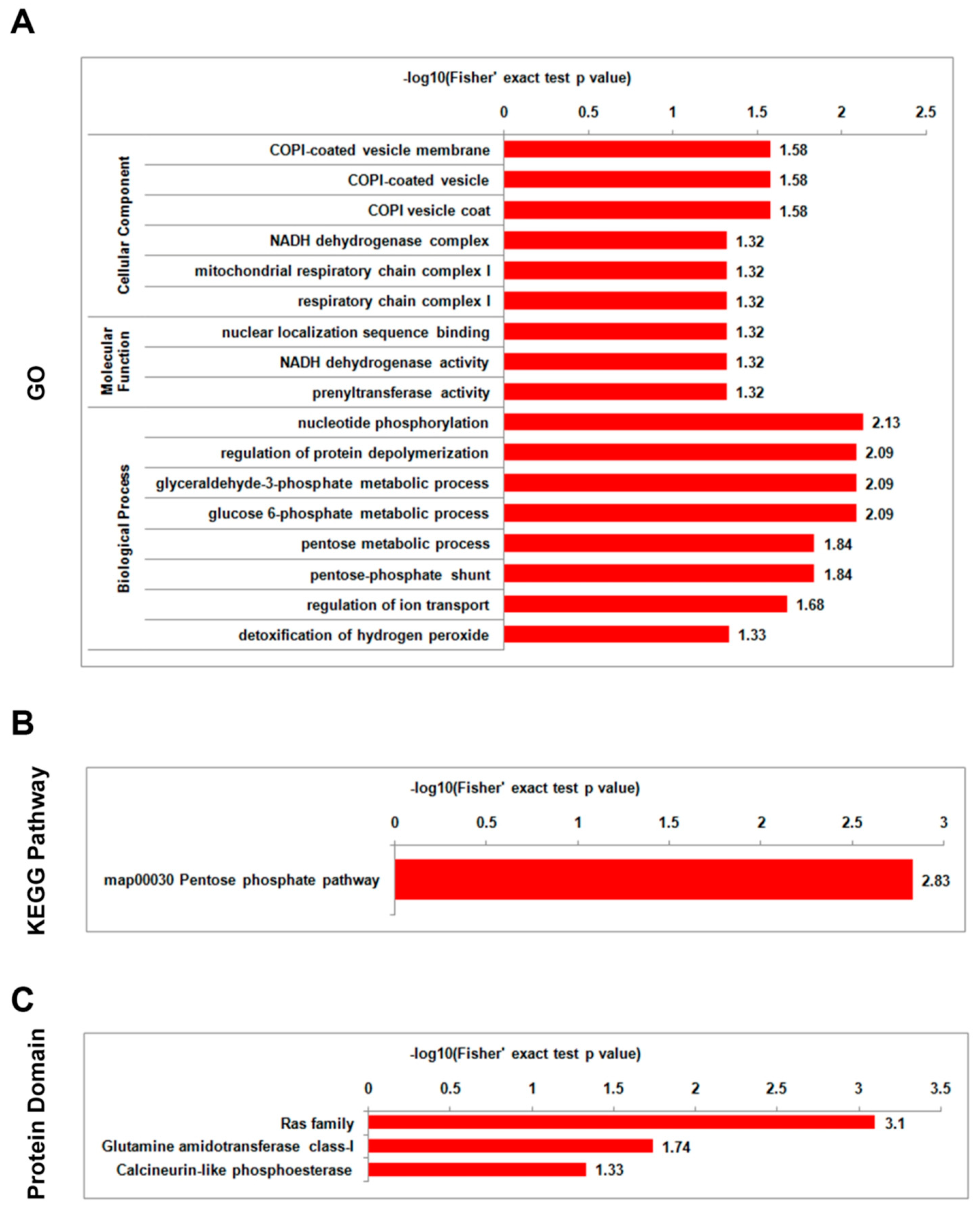
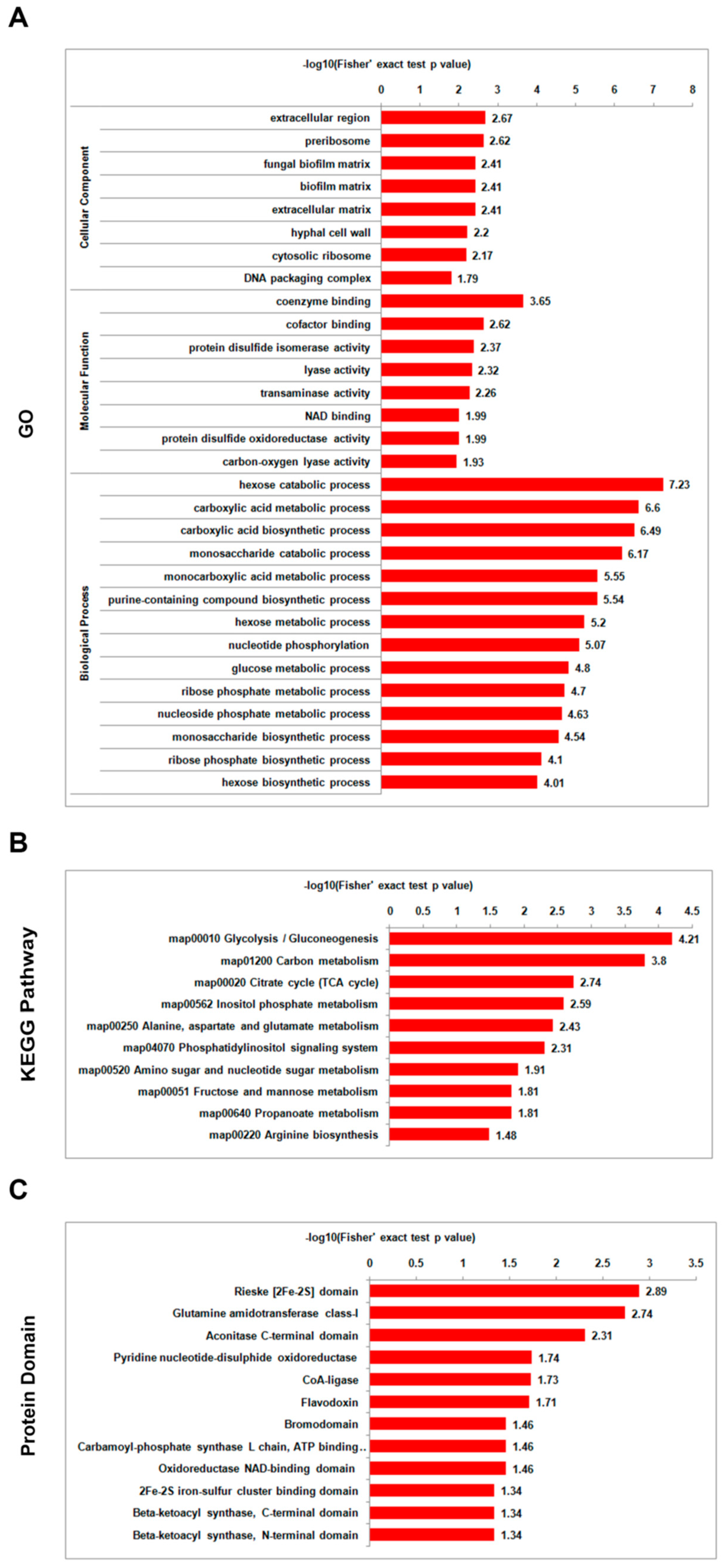
3.5. Analysis of Functional Enrichment of Differentially Lysine-Acetylated Proteins in Foc
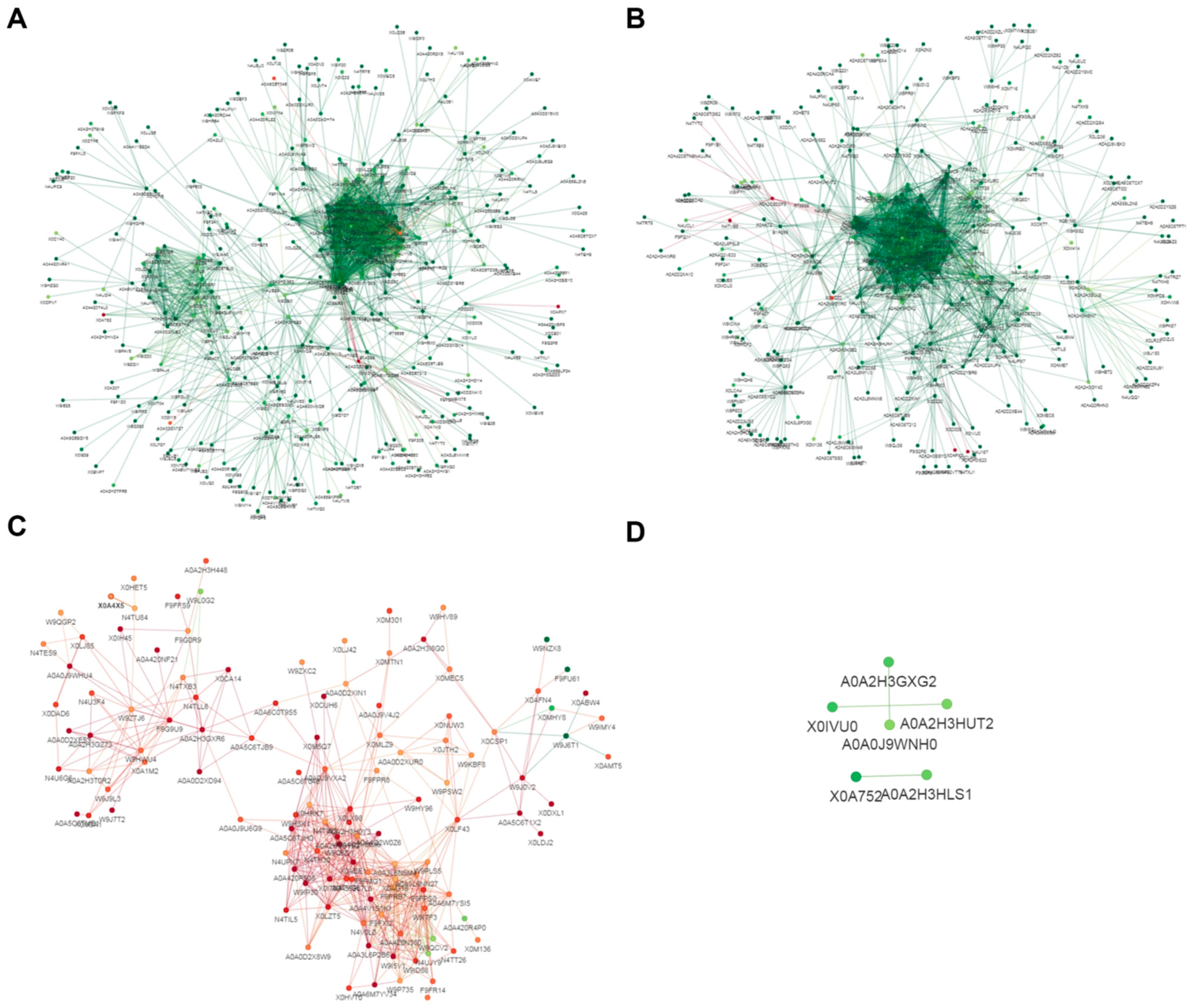
3.6. Analysis of Protein Interaction Network of Differentially Lysine-Acetylated Proteins in Foc
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Edel-Hermann, V.; Lecomte, C. Current status of Fusarium oxysporum formae speciales and races. Phytopathology 2019, 109, 512–530. [Google Scholar] [CrossRef]
- Michielse, C.B.; Rep, M. Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 2009, 10, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.L.; Lin, X.G.; Wang, J.H.; Shen, W.S.; Wu, S.; Peng, S.P.; Mao, T.T. Arbuscular mycorrhizal fungal inoculation enhances suppression of cucumber Fusarium wilt in greenhouse soils. Pedosphere 2010, 20, 586–593. [Google Scholar] [CrossRef]
- Martínez, R.; Aguilar, M.I.; Guirado, M.L.; Álvarez, A.; Gómez, J. First report of Fusarium wilt of cucumber caused by Fusarium oxysporum in Spain. Plant Pathol. 2003, 52, 410. [Google Scholar] [CrossRef]
- Zhou, X.; Wu, F. Dynamics of the diversity of fungal and Fusarium communities during continuous cropping of cucumber in the greenhouse. FEMS Microbiol. Ecol. 2012, 80, 469–478. [Google Scholar] [CrossRef]
- Barrett, L.G.; Thrall, P.H.; Dodds, P.N.; van der Merwe, M.; Linde, C.C.; Lawrence, G.J.; Burdon, J.J. Diversity and evolution of effector loci in natural populations of the plant pathogen Melampsora lini. Mol. Biol. Evol. 2009, 26, 2499–2513. [Google Scholar] [CrossRef]
- Ploetz, R.C. Fusarium wilt of banana. Phytopathology 2015, 105, 1512–1521. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Sun, M.; Lu, X.; Li, S. Serial passage through resistant and susceptible cucumber cultivars affects the virulence of Fusarium oxysporum f. sp. cucumerinum. Microbiol. Open 2019, 8, e00641. [Google Scholar] [CrossRef]
- Huang, X.Q.; Lu, X.H.; Sun, M.H.; Guo, R.J.; van Diepeningen, A.D.; Li, S.D. Transcriptome analysis of virulence-differentiated Fusarium oxysporum f. sp. cucumerinum isolates during cucumber colonisation reveals pathogenicity profiles. BMC Genom. 2019, 20, 570. [Google Scholar] [CrossRef]
- Choudhary, C.; Weinert, B.T.; Nishida, Y.; Verdin, E.; Mann, M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 536–550. [Google Scholar] [CrossRef]
- Kim, G.W.; Yang, X.J. Comprehensive lysine acetylomes emerging from bacteria to humans. Trends Biochem. Sci. 2011, 36, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.K.; Workman, J.L. Histone acetyltransferase complexes: One size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 2007, 8, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Kwon, S.; Lee, Y.H. Histone acetylation in fungal pathogens of plants. Plant Pathol. J. 2014, 30, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Narita, T.; Weinert, B.T.; Choudhary, C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 156–174. [Google Scholar] [CrossRef]
- Zhou, S.; Yang, Q.; Yin, C.; Liu, L.; Liang, W. Systematic analysis of the lysine acetylome in Fusarium graminearum. BMC Genom. 2016, 17, 1019. [Google Scholar] [CrossRef]
- Lv, B.; Yang, Q.; Li, D.; Liang, W.; Song, L. Proteome-wide analysis of lysine acetylation in the plant pathogen Botrytis cinerea. Sci. Rep. 2016, 6, 29313. [Google Scholar] [CrossRef]
- Lv, Y. Proteome-wide profiling of protein lysine acetylation in Aspergillus flavus. PLoS ONE 2017, 12, e0178603. [Google Scholar] [CrossRef]
- Li, D.; Lv, B.; Tan, L.; Yang, Q.; Liang, W. Acetylome analysis reveals the involvement of lysine acetylation in diverse biological processes in Phytophthora sojae. Sci. Rep. 2016, 6, 29897. [Google Scholar] [CrossRef]
- Li, J.; Gao, M.; Gabriel, D.W.; Liang, W.; Song, L. Secretome-wide analysis of lysine acetylation in Fusarium oxysporum f. sp. lycopersici provides novel insights into infection-related proteins. Front. Microbiol. 2020, 11, 559440. [Google Scholar] [CrossRef]
- Lv, F.; Xu, Y.; Gabriel, D.W.; Wang, X.; Zhang, N.; Liang, W. Quantitative proteomic analysis reveals important roles of the acetylation of ER-resident molecular chaperones for conidiation in Fusarium oxysporum. Mol. Cell. Proteom. 2022, 21, 100231. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Jiang, N.; Lv, B.N.; Wu, H.X.; Li, S.D.; Sun, M.H. Tandem Mass Tag labelling quantitative acetylome analysis of differentially modified proteins during mycoparasitism of Clonostachys chloroleuca 67-1. Sci. Rep. 2021, 11, 22383. [Google Scholar] [CrossRef]
- Wang, Z.K.; Cai, Q.; Liu, J.; Ying, S.H.; Feng, M.G. Global insight into lysine acetylation events and their links to biological aspects in Beauveria bassiana, a fungal insect pathogen. Sci. Rep. 2017, 7, 44360. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Tong, M.; Bai, X.; Liu, X.; Cai, X.; Luo, X.; Zhang, P.; Cai, W.; Vallée, I.; Zhou, Y.; et al. Comprehensive proteomic analysis of lysine acetylation in the foodborne pathogen Trichinella spiralis. Front. Microbiol. 2017, 8, 2674. [Google Scholar] [CrossRef]
- Masi, A.; Mach, R.L.; Mach-Aigner, A.R. The pentose phosphate pathway in industrially relevant fungi: Crucial insights for bioprocessing. Appl. Microbiol. Biotechnol. 2021, 105, 4017–4031. [Google Scholar] [CrossRef]
- Fromm, S.; Senkler, J.; Eubel, H.; Peterhänsel, C.; Braun, H.P. Life without complex I: Proteome analyses of an Arabidopsis mutant lacking the mitochondrial NADH dehydrogenase complex. J. Exp. Bot. 2016, 67, 3079–3093. [Google Scholar] [CrossRef] [PubMed]
- Formosa, L.E.; Dibley, M.G.; Stroud, D.A.; Ryan, M.T. Building a complex complex: Assembly of mitochondrial respiratory chain complex I. Semin. Cell Dev. Biol. 2018, 76, 154–162. [Google Scholar] [CrossRef]
- Garner, A.L. Nucleosides, nucleotides and nucleic acids as therapeutics: A virtual special issue. ACS Pharmacol. Transl. Sci. 2021, 4, 1714–1715. [Google Scholar] [CrossRef]
- Arakel, E.C.; Schwappach, B. Formation of COPI-coated vesicles at a glance. J. Cell Sci. 2018, 131, jcs209890. [Google Scholar] [CrossRef]
- Xie, C.; Shang, Q.; Mo, C.; Xiao, Y.; Wang, G.; Xie, J.; Jiang, D.; Xiao, X. Early secretory pathway-associated proteins SsEmp24 and SsErv25 are involved in morphogenesis and pathogenicity in a filamentous phytopathogenic fungus. mbio 2021, 12, e03173-21. [Google Scholar] [CrossRef]
- Phasha, M.M.; Wingfield, M.J.; Wingfield, B.D.; Coetzee, M.P.A.; Hallen-Adams, H.; Fru, F.; Swalarsk-Parry, B.S.; Yilmaz, N.; Duong, T.A.; Steenkamp, E.T. Ras2 is important for growth and pathogenicity in Fusarium circinatum. Fungal Genet. Biol. 2021, 150, 103541. [Google Scholar] [CrossRef] [PubMed]
- Shay, R.; Wiegand, A.A.; Trail, F. Biofilm formation and structure in the filamentous fungus Fusarium graminearum, a plant pathogen. Microbiol. Spectr. 2022, 10, e00171-22. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.F.; Zarnowski, R.; Andes, D.R. The extracellular matrix of fungal biofilms. Adv. Exp. Med. Biol. 2016, 931, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, C.H.; Morelli, K.A.; Schultz, D.; Nadell, C.D.; Cramer, R.A. Fungal biofilm architecture produces hypoxic microenvironments that drive antifungal resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 22473–22483. [Google Scholar] [CrossRef]
- García-Olmedo, F.; Rodríguez-Palenzuela, P.; Molina, A.; Alamillo, J.M.; López-Solanilla, E.; Berrocal-Lobo, M.; Poza-Carrión, C. Antibiotic activities of peptides, hydrogen peroxide and peroxynitrite in plant defence. FEBS Lett. 2001, 498, 219–222. [Google Scholar] [CrossRef]
- Di Pietro, A.; García-MacEira, F.I.; Méglecz, E.; Roncero, M.I. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 2001, 39, 1140–1152. [Google Scholar] [CrossRef]
- Leal, G.A.; Gomes, L.H.; Albuquerque, P.S.; Tavares, F.C.; Figueira, A. Searching for Moniliophthora perniciosa pathogenicity genes. Fungal Biol. 2010, 114, 842–854. [Google Scholar] [CrossRef]
- López-Díaz, C.; Rahjoo, V.; Sulyok, M.; Ghionna, V.; Martín-Vicente, A.; Capilla, J.; Di Pietro, A.; López-Berges, M.S. Fusaric acid contributes to virulence of Fusarium oxysporum on plant and mammalian hosts. Mol. Plant Pathol. 2018, 19, 440–453. [Google Scholar] [CrossRef]
- He, X.; Li, W.; Zhang, W.; Jin, X.; Shenkute, A.G.; Aynalem, T.; Xu, S.; Wang, W. Transcriptome sequencing analysis provides insights into the response to Fusarium oxysporum in Lilium pumilum. Evol. Bioinform. 2019, 15, 1176934319838818. [Google Scholar] [CrossRef]


| Eukaryotic Orthologous Group (KOG) Functional Description | Comparisons | |||
|---|---|---|---|---|
| W_6h vs. W_0h | I_6h vs. I_0h | I_0h vs. W_0h | I_6h vs. W_6h | |
| [E] Amino acid transport and metabolism | 143 | 139 | 75 | 6 |
| [C] Energy production and conversion | 132 | 128 | 72 | 10 |
| [O] Posttranslational modification, protein turnover, chaperones | 181 | 174 | 71 | 18 |
| [J] Translation, ribosomal structure, and biogenesis | 172 | 172 | 67 | 13 |
| [G] Carbohydrate transport and metabolism | 95 | 93 | 62 | 11 |
| [S] Function unknown | 135 | 133 | 57 | 6 |
| [I] Lipid transport and metabolism | 89 | 81 | 42 | 6 |
| [Q] Secondary metabolites biosynthesis, transport, and catabolism | 85 | 80 | 39 | 5 |
| [T] Signal transduction mechanisms | 106 | 98 | 36 | 1 |
| [U] Intracellular trafficking, secretion, and vesicular transport | 127 | 130 | 33 | 9 |
| [K] Transcription | 66 | 67 | 26 | 4 |
| [B] Chromatin structure and dynamics | 40 | 40 | 26 | 5 |
| [A] RNA processing and modification | 78 | 81 | 26 | 2 |
| [H] Coenzyme transport and metabolism | 52 | 50 | 23 | 4 |
| [F] Nucleotide transport and metabolism | 47 | 46 | 21 | 3 |
| [P] Inorganic ion transport and metabolism | 33 | 31 | 19 | 5 |
| [Z] Cytoskeleton | 50 | 49 | 16 | 1 |
| [D] Cell cycle control, cell division, chromosome partitioning | 38 | 40 | 13 | 2 |
| [M] Cell wall/membrane/envelope biogenesis | 17 | 18 | 9 | 2 |
| [L] Replication, recombination, and repair | 34 | 34 | 7 | 1 |
| [Y] Nuclear structure | 16 | 15 | 5 | 2 |
| [V] Defense mechanisms | 7 | 8 | 5 | 2 |
| [W] Extracellular structures | 2 | 2 | - | - |
| Subcellular Localization | Comparisons | |||
|---|---|---|---|---|
| W_6h vs. W_0h | I_6h vs. I_0h | I_0h vs. W_0h | I_6h vs. W_6h | |
| Cytoplasm | 642 | 627 | 328 | 64 |
| Nucleus | 568 | 558 | 217 | 31 |
| Mitochondria | 420 | 409 | 170 | 17 |
| Extracellular | 144 | 140 | 87 | 8 |
| Plasma membrane | 138 | 139 | 65 | 6 |
| Cytoplasm, nucleus | 113 | 111 | 51 | 5 |
| Cytoskeleton | 55 | 51 | - | - |
| Other | 17 | 15 | 21 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Lu, X.; Hao, J.; Li, S. Quantitative Acetylome Analysis of Differentially Modified Proteins in Virulence-Differentiated Fusarium oxysporum f. sp. cucumerinum Isolates during Cucumber Colonization. J. Fungi 2023, 9, 920. https://doi.org/10.3390/jof9090920
Zhou Y, Lu X, Hao J, Li S. Quantitative Acetylome Analysis of Differentially Modified Proteins in Virulence-Differentiated Fusarium oxysporum f. sp. cucumerinum Isolates during Cucumber Colonization. Journal of Fungi. 2023; 9(9):920. https://doi.org/10.3390/jof9090920
Chicago/Turabian StyleZhou, Ying, Xiaohong Lu, Jianjun Hao, and Shidong Li. 2023. "Quantitative Acetylome Analysis of Differentially Modified Proteins in Virulence-Differentiated Fusarium oxysporum f. sp. cucumerinum Isolates during Cucumber Colonization" Journal of Fungi 9, no. 9: 920. https://doi.org/10.3390/jof9090920





