Emerging Data on the Diversity of Molecular Mechanisms Involving C/D snoRNAs
Abstract
:1. Introduction
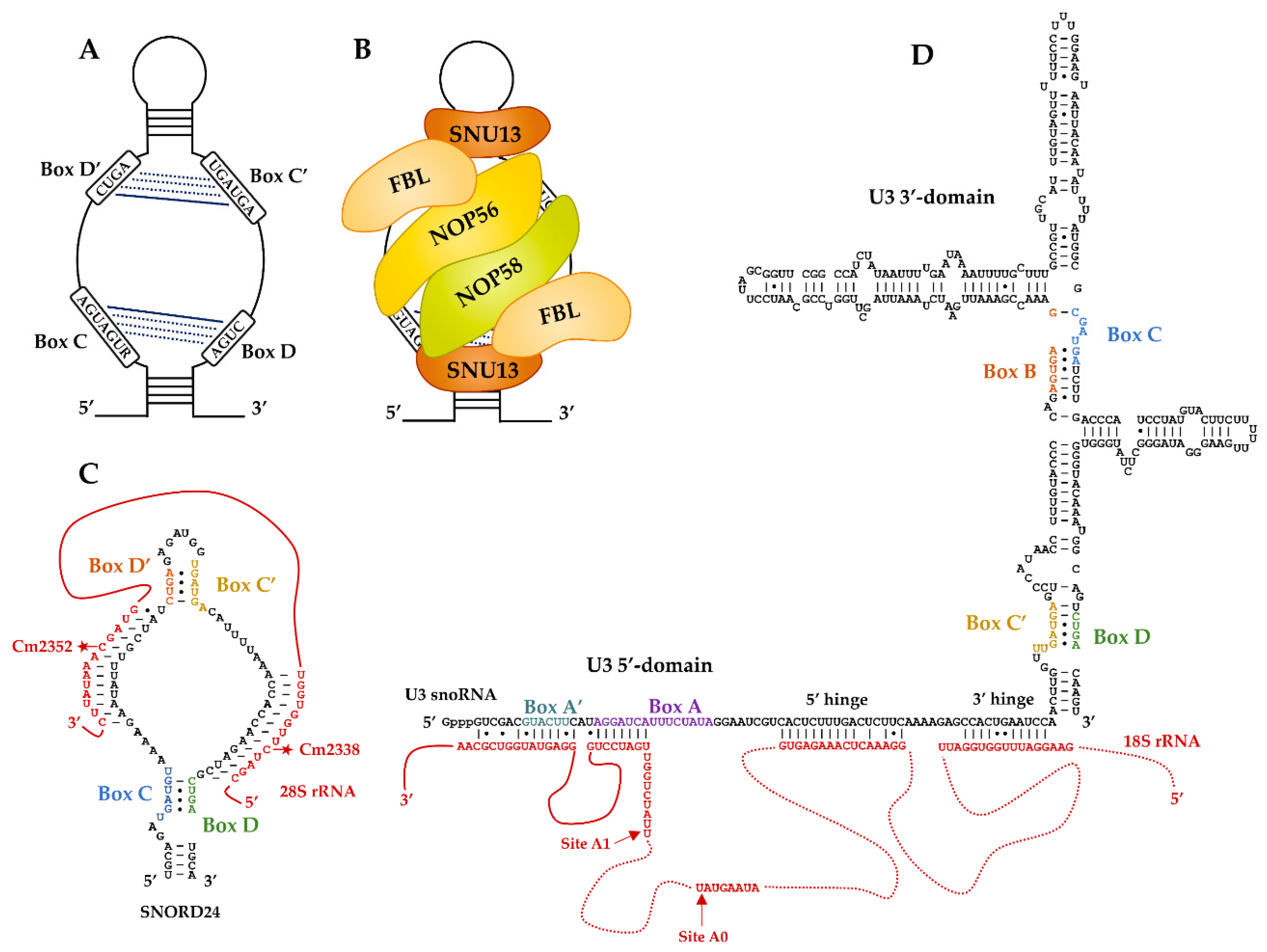
2. Molecular Features for the Canonical Paradigm of RNA-Guided Activity: The Box C/D RNP Methyltransferases
3. U3, the First Identified Atypical Box C/D RNP
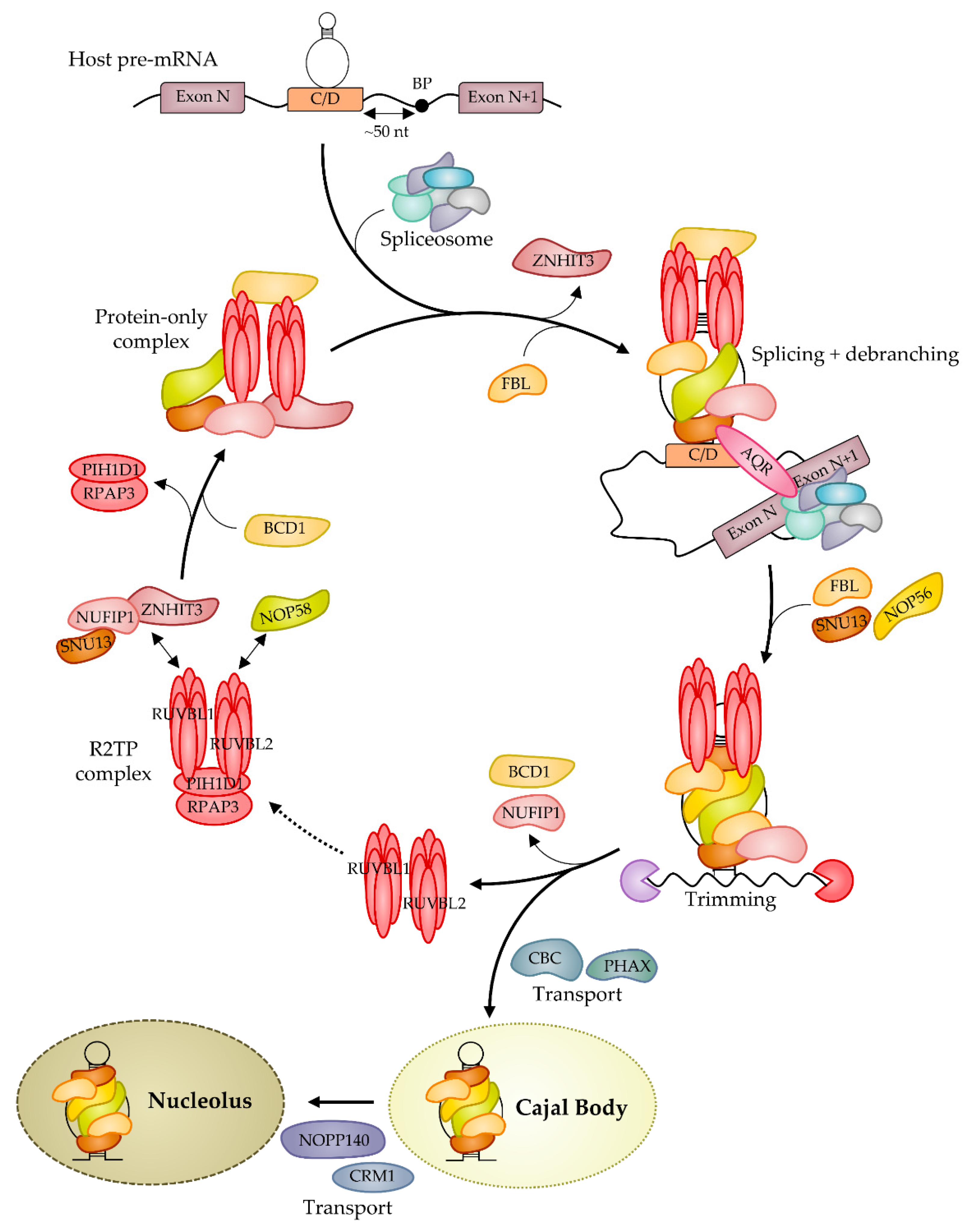
4. Biogenesis of Box C/D RNP: Assembly and Nuclear Journey
5. The Diversity of C/D snoRNA Maturation Forms
6. The Multiple Regulations of C/D snoRNP Biogenesis
7. The Strange Case of Alternative C/D snoRNPs
8. The Diversity of C/D snoRNA Expression
9. The Diversity of C/D snoRNA Molecular Partners and Targets
10. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Ambros, V. MicroRNAs: Tiny Regulators with Great Potential. Cell 2001, 107, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Weinberg, R.A.; Penman, S. Small Molecular Weight Monodisperse Nuclear RNA. J. Mol. Biol. 1968, 38, 289–304. [Google Scholar] [CrossRef]
- Nakamura, T.; Prestayko, A.W.; Busch, H. Studies on Nucleolar 4 to 6 S Ribonucleic Acid of Novikoff Hepatoma Cells. J. Biol. Chem. 1968, 243, 1368–1375. [Google Scholar] [CrossRef]
- Prestayko, A.W.; Tonato, M.; Busch, H. Low Molecular Weight RNA Associated with 28 s Nucleolar RNA. J. Mol. Biol. 1970, 47, 505–515. [Google Scholar] [CrossRef]
- Kass, S.; Tyc, K.; Steitz, J.A.; Sollner-Webb, B. The U3 Small Nucleolar Ribonucleoprotein Functions in the First Step of Preribosomal RNA Processing. Cell 1990, 60, 897–908. [Google Scholar] [CrossRef]
- Hughes, J.M. Functional Base-Pairing Interaction between Highly Conserved Elements of U3 Small Nucleolar RNA and the Small Ribosomal Subunit RNA. J. Mol. Biol. 1996, 259, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Tollervey, D. Base Pairing between U3 Small Nucleolar RNA and the 5′ End of 18S RRNA Is Required for Pre-RRNA Processing. Mol. Cell. Biol. 1999, 19, 6012–6019. [Google Scholar] [CrossRef] [Green Version]
- Dragon, F.; Gallagher, J.E.G.; Compagnone-Post, P.A.; Mitchell, B.M.; Porwancher, K.A.; Wehner, K.A.; Wormsley, S.; Settlage, R.E.; Shabanowitz, J.; Osheim, Y.; et al. A Large Nucleolar U3 Ribonucleoprotein Required for 18S Ribosomal RNA Biogenesis. Nature 2002, 417, 967–970. [Google Scholar] [CrossRef]
- Tyc, K.; Steitz, J.A. U3, U8 and U13 Comprise a New Class of Mammalian SnRNPs Localized in the Cell Nucleolus. EMBO J. 1989, 8, 3113–3119. [Google Scholar] [CrossRef]
- Kiss-László, Z.; Henry, Y.; Bachellerie, J.P.; Caizergues-Ferrer, M.; Kiss, T. Site-Specific Ribose Methylation of Preribosomal RNA: A Novel Function for Small Nucleolar RNAs. Cell 1996, 85, 1077–1088. [Google Scholar] [CrossRef] [Green Version]
- Balakin, A.G.; Smith, L.; Fournier, M.J. The RNA World of the Nucleolus: Two Major Families of Small RNAs Defined by Different Box Elements with Related Functions. Cell 1996, 86, 823–834. [Google Scholar] [CrossRef] [Green Version]
- Lafontaine, D.L.; Tollervey, D. Nop58p Is a Common Component of the Box C+D SnoRNPs That Is Required for SnoRNA Stability. RNA 1999, 5, 455–467. [Google Scholar] [CrossRef] [Green Version]
- Watkins, N.J.; Ségault, V.; Charpentier, B.; Nottrott, S.; Fabrizio, P.; Bachi, A.; Wilm, M.; Rosbash, M.; Branlant, C.; Lührmann, R. A Common Core RNP Structure Shared between the Small Nucleoar Box C/D RNPs and the Spliceosomal U4 SnRNP. Cell 2000, 103, 457–466. [Google Scholar] [CrossRef] [Green Version]
- Watkins, N.J.; Bohnsack, M.T. The Box C/D and H/ACA SnoRNPs: Key Players in the Modification, Processing and the Dynamic Folding of Ribosomal RNA. Wiley Interdiscip. Rev. RNA 2012, 3, 397–414. [Google Scholar] [CrossRef]
- Lafontaine, D.L.J. Noncoding RNAs in Eukaryotic Ribosome Biogenesis and Function. Nat. Struct. Mol. Biol. 2015, 22, 11–19. [Google Scholar] [CrossRef]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.-D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the Ribosome: The Influence of RRNA Modification on Eukaryotic Ribosome Biogenesis and Function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar] [CrossRef]
- Turowski, T.W.; Tollervey, D. Cotranscriptional Events in Eukaryotic Ribosome Synthesis. WIREs RNA 2015, 6, 129–139. [Google Scholar] [CrossRef]
- Henras, A.K.; Plisson-Chastang, C.; O’Donohue, M.-F.; Chakraborty, A.; Gleizes, P.-E. An Overview of Pre-Ribosomal RNA Processing in Eukaryotes. Wiley Interdiscip. Rev. RNA 2015, 6, 225–242. [Google Scholar] [CrossRef]
- Ganot, P.; Bortolin, M.L.; Kiss, T. Site-Specific Pseudouridine Formation in Preribosomal RNA Is Guided by Small Nucleolar RNAs. Cell 1997, 89, 799–809. [Google Scholar] [CrossRef] [Green Version]
- Ni, J.; Tien, A.L.; Fournier, M.J. Small Nucleolar RNAs Direct Site-Specific Synthesis of Pseudouridine in Ribosomal RNA. Cell 1997, 89, 565–573. [Google Scholar] [CrossRef] [Green Version]
- Kiss, A.M.; Jády, B.E.; Bertrand, E.; Kiss, T. Human Box H/ACA Pseudouridylation Guide RNA Machinery. Mol. Cell. Biol. 2004, 24, 5797–5807. [Google Scholar] [CrossRef] [Green Version]
- Decatur, W.A.; Fournier, M.J. RNA-Guided Nucleotide Modification of Ribosomal and Other RNAs. J. Biol. Chem. 2003, 278, 695–698. [Google Scholar] [CrossRef] [Green Version]
- Morrissey, J.P.; Tollervey, D. Yeast SnR30 Is a Small Nucleolar RNA Required for 18S RRNA Synthesis. Mol. Cell. Biol. 1993, 13, 2469–2477. [Google Scholar] [CrossRef] [Green Version]
- Atzorn, V.; Fragapane, P.; Kiss, T. U17/SnR30 Is a Ubiquitous SnoRNA with Two Conserved Sequence Motifs Essential for 18S RRNA Production. Mol. Cell. Biol. 2004, 24, 1769–1778. [Google Scholar] [CrossRef] [Green Version]
- Meier, U.T. RNA Modification in Cajal Bodies. RNA Biol. 2017, 14, 693–700. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Sloan, K.E. Modifications in Small Nuclear RNAs and Their Roles in Spliceosome Assembly and Function. Biol. Chem. 2018, 399, 1265–1276. [Google Scholar] [CrossRef]
- Bratkovič, T.; Božič, J.; Rogelj, B. Functional Diversity of Small Nucleolar RNAs. Nucleic Acids Res. 2020, 48, 1627–1651. [Google Scholar] [CrossRef] [Green Version]
- Bergeron, D.; Fafard-Couture, É.; Scott, M.S. Small Nucleolar RNAs: Continuing Identification of Novel Members and Increasing Diversity of Their Molecular Mechanisms of Action. Biochem. Soc. Trans. 2020, 48, 645–656. [Google Scholar] [CrossRef] [Green Version]
- Kufel, J.; Grzechnik, P. Small Nucleolar RNAs Tell a Different Tale. Trends Genet. 2019, 35, 104–117. [Google Scholar] [CrossRef] [Green Version]
- Boivin, V.; Faucher-Giguère, L.; Scott, M.; Abou-Elela, S. The Cellular Landscape of Mid-Size Noncoding RNA. Wiley Interdiscip. Rev. RNA 2019, 10, e1530. [Google Scholar] [CrossRef] [Green Version]
- Dupuis-Sandoval, F.; Poirier, M.; Scott, M.S. The Emerging Landscape of Small Nucleolar RNAs in Cell Biology. Wiley Interdiscip. Rev. RNA 2015, 6, 381–397. [Google Scholar] [CrossRef]
- Massenet, S.; Bertrand, E.; Verheggen, C. Assembly and Trafficking of Box C/D and H/ACA SnoRNPs. RNA Biol. 2017, 14, 680–692. [Google Scholar] [CrossRef] [Green Version]
- Bachellerie, J.P.; Cavaillé, J.; Hüttenhofer, A. The Expanding SnoRNA World. Biochimie 2002, 84, 775–790. [Google Scholar] [CrossRef]
- Omer, A.D.; Lowe, T.M.; Russell, A.G.; Ebhardt, H.; Eddy, S.R.; Dennis, P.P. Homologs of Small Nucleolar RNAs in Archaea. Science 2000, 288, 517–522. [Google Scholar] [CrossRef] [Green Version]
- Gaspin, C.; Cavaillé, J.; Erauso, G.; Bachellerie, J.P. Archaeal Homologs of Eukaryotic Methylation Guide Small Nucleolar RNAs: Lessons from the Pyrococcus Genomes. J. Mol. Biol. 2000, 297, 895–906. [Google Scholar] [CrossRef]
- Omer, A.D.; Ziesche, S.; Ebhardt, H.; Dennis, P.P. In Vitro Reconstitution and Activity of a C/D Box Methylation Guide Ribonucleoprotein Complex. Proc. Natl. Acad. Sci. USA 2002, 99, 5289–5294. [Google Scholar] [CrossRef] [Green Version]
- Rozhdestvensky, T.S.; Tang, T.H.; Tchirkova, I.V.; Brosius, J.; Bachellerie, J.-P.; Hüttenhofer, A. Binding of L7Ae Protein to the K-Turn of Archaeal SnoRNAs: A Shared RNA Binding Motif for C/D and H/ACA Box SnoRNAs in Archaea. Nucleic Acids Res. 2003, 31, 869–877. [Google Scholar] [CrossRef] [Green Version]
- Charpentier, B.; Muller, S.; Branlant, C. Reconstitution of Archaeal H/ACA Small Ribonucleoprotein Complexes Active in Pseudouridylation. Nucleic Acids Res. 2005, 33, 3133–3144. [Google Scholar] [CrossRef] [Green Version]
- Baker, D.L.; Youssef, O.A.; Chastkofsky, M.I.R.; Dy, D.A.; Terns, R.M.; Terns, M.P. RNA-Guided RNA Modification: Functional Organization of the Archaeal H/ACA RNP. Genes Dev. 2005, 19, 1238–1248. [Google Scholar] [CrossRef] [Green Version]
- Muller, S.; Leclerc, F.; Behm-Ansmant, I.; Fourmann, J.-B.; Charpentier, B.; Branlant, C. Combined in Silico and Experimental Identification of the Pyrococcus Abyssi H/ACA SRNAs and Their Target Sites in Ribosomal RNAs. Nucleic Acids Res. 2008, 36, 2459–2475. [Google Scholar] [CrossRef] [PubMed]
- Omer, A.D.; Ziesche, S.; Decatur, W.A.; Fournier, M.J.; Dennis, P.P. RNA-Modifying Machines in Archaea. Mol. Microbiol. 2003, 48, 617–629. [Google Scholar] [CrossRef]
- Dennis, P.P.; Omer, A. Small Non-Coding RNAs in Archaea. Curr. Opin. Microbiol. 2005, 8, 685–694. [Google Scholar] [CrossRef] [PubMed]
- van Nues, R.W.; Granneman, S.; Kudla, G.; Sloan, K.E.; Chicken, M.; Tollervey, D.; Watkins, N.J. Box C/D SnoRNP Catalysed Methylation Is Aided by Additional Pre-RRNA Base-Pairing. EMBO J. 2011, 30, 2420–2430. [Google Scholar] [CrossRef] [Green Version]
- Chagot, M.-E.; Quinternet, M.; Rothé, B.; Charpentier, B.; Coutant, J.; Manival, X.; Lebars, I. The Yeast C/D Box SnoRNA U14 Adopts a “Weak” K-Turn like Conformation Recognized by the Snu13 Core Protein in Solution. Biochimie 2019, 164, 70–82. [Google Scholar] [CrossRef]
- Klein, D.J.; Schmeing, T.M.; Moore, P.B.; Steitz, T.A. The Kink-Turn: A New RNA Secondary Structure Motif. EMBO J. 2001, 20, 4214–4221. [Google Scholar] [CrossRef]
- Moore, T.; Zhang, Y.; Fenley, M.O.; Li, H. Molecular Basis of Box C/D RNA-Protein Interactions; Cocrystal Structure of Archaeal L7Ae and a Box C/D RNA. Structure 2004, 12, 807–818. [Google Scholar] [CrossRef]
- Reichow, S.L.; Hamma, T.; Ferré-D’Amaré, A.R.; Varani, G. The Structure and Function of Small Nucleolar Ribonucleoproteins. Nucleic Acids Res. 2007, 35, 1452–1464. [Google Scholar] [CrossRef]
- Vidovic, I.; Nottrott, S.; Hartmuth, K.; Lührmann, R.; Ficner, R. Crystal Structure of the Spliceosomal 15.5 kD Protein Bound to a U4 SnRNA Fragment. Mol. Cell 2000, 6, 1331–1342. [Google Scholar] [CrossRef] [Green Version]
- Qu, G.; van Nues, R.W.; Watkins, N.J.; Maxwell, E.S. The Spatial-Functional Coupling of Box C/D and C’/D’ RNPs Is an Evolutionarily Conserved Feature of the Eukaryotic Box C/D SnoRNP Nucleotide Modification Complex. Mol. Cell. Biol. 2011, 31, 365–374. [Google Scholar] [CrossRef] [Green Version]
- Hamma, T.; Ferré-D’Amaré, A.R. Structure of Protein L7Ae Bound to a K-Turn Derived from an Archaeal Box H/ACA SRNA at 1.8 A Resolution. Structure 2004, 12, 893–903. [Google Scholar] [CrossRef]
- Charron, C.; Manival, X.; Cléry, A.; Senty-Ségault, V.; Charpentier, B.; Marmier-Gourrier, N.; Branlant, C.; Aubry, A. The Archaeal SRNA Binding Protein L7Ae Has a 3D Structure Very Similar to That of Its Eukaryal Counterpart While Having a Broader RNA-Binding Specificity. J. Mol. Biol. 2004, 342, 757–773. [Google Scholar] [CrossRef] [PubMed]
- Nolivos, S.; Carpousis, A.J.; Clouet-d’Orval, B. The K-Loop, a General Feature of the Pyrococcus C/D Guide RNAs, Is an RNA Structural Motif Related to the K-Turn. Nucleic Acids Res. 2005, 33, 6507–6514. [Google Scholar] [CrossRef] [Green Version]
- Watkins, N.J.; Dickmanns, A.; Lührmann, R. Conserved Stem II of the Box C/D Motif Is Essential for Nucleolar Localization and Is Required, Along with the 15.5K Protein, for the Hierarchical Assembly of the Box C/D SnoRNP. Mol. Cell. Biol. 2002, 22, 8342–8352. [Google Scholar] [CrossRef] [Green Version]
- Szewczak, L.B.W.; Gabrielsen, J.S.; Degregorio, S.J.; Strobel, S.A.; Steitz, J.A. Molecular Basis for RNA Kink-Turn Recognition by the H15.5K Small RNP Protein. RNA 2005, 11, 1407–1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cahill, N.M.; Friend, K.; Speckmann, W.; Li, Z.-H.; Terns, R.M.; Terns, M.P.; Steitz, J.A. Site-Specific Cross-Linking Analyses Reveal an Asymmetric Protein Distribution for a Box C/D SnoRNP. EMBO J. 2002, 21, 3816–3828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szewczak, L.B.W.; DeGregorio, S.J.; Strobel, S.A.; Steitz, J.A. Exclusive Interaction of the 15.5 KD Protein with the Terminal Box C/D Motif of a Methylation Guide SnoRNP. Chem. Biol. 2002, 9, 1095–1107. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Lai, S.; Jia, R.; Xu, A.; Zhang, L.; Lu, J.; Ye, K. Structural Basis for Site-Specific Ribose Methylation by Box C/D RNA Protein Complexes. Nature 2011, 469, 559–563. [Google Scholar] [CrossRef]
- Aittaleb, M.; Rashid, R.; Chen, Q.; Palmer, J.R.; Daniels, C.J.; Li, H. Structure and Function of Archaeal Box C/D SRNP Core Proteins. Nat. Struct. Mol. Biol. 2003, 10, 256–263. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, J.; Huang, L.; Lilley, D.M.J.; Ye, K. Functional Organization of Box C/D RNA-Guided RNA Methyltransferase. Nucleic Acids Res. 2020, 48, 5094–5105. [Google Scholar] [CrossRef]
- Barandun, J.; Chaker-Margot, M.; Hunziker, M.; Molloy, K.R.; Chait, B.T.; Klinge, S. The Complete Structure of the Small-Subunit Processome. Nat. Struct. Mol. Biol. 2017, 24, 944–953. [Google Scholar] [CrossRef]
- Clerget, G.; Bourguignon-Igel, V.; Marmier-Gourrier, N.; Rolland, N.; Wacheul, L.; Manival, X.; Charron, C.; Kufel, J.; Méreau, A.; Senty-Ségault, V.; et al. Synergistic Defects in Pre-RRNA Processing from Mutations in the U3-Specific Protein Rrp9 and U3 SnoRNA. Nucleic Acids Res. 2020, 48, 3848–3868. [Google Scholar] [CrossRef]
- Kiss-László, Z.; Henry, Y.; Kiss, T. Sequence and Structural Elements of Methylation Guide SnoRNAs Essential for Site-Specific Ribose Methylation of Pre-RRNA. EMBO J. 1998, 17, 797–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapinaite, A.; Simon, B.; Skjaerven, L.; Rakwalska-Bange, M.; Gabel, F.; Carlomagno, T. The Structure of the Box C/D Enzyme Reveals Regulation of RNA Methylation. Nature 2013, 502, 519–523. [Google Scholar] [CrossRef]
- Tran, E.; Zhang, X.; Lackey, L.; Maxwell, E.S. Conserved Spacing between the Box C/D and C′/D′ RNPs of the Archaeal Box C/D SRNP Complex Is Required for Efficient 2′-O-Methylation of Target RNAs. RNA 2005, 11, 285–293. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-L.; Perasso, R.; Qu, L.-H.; Amar, L. Exploration of Pairing Constraints Identifies a 9 Base-Pair Core within Box C/D SnoRNA-RRNA Duplexes. J. Mol. Biol. 2007, 369, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Lin, J.; Ye, K. Box C/D Guide RNAs Recognize a Maximum of 10 Nt of Substrates. Proc. Natl. Acad. Sci. USA 2016, 113, 10878–10883. [Google Scholar] [CrossRef] [Green Version]
- Graziadei, A.; Gabel, F.; Kirkpatrick, J.; Carlomagno, T. The Guide SRNA Sequence Determines the Activity Level of Box C/D RNPs. eLife 2020, 9. [Google Scholar] [CrossRef]
- Wu, S.; Wang, Y.; Wang, J.; Li, X.; Li, J.; Ye, K. Profiling of RNA Ribose Methylation in Arabidopsis thaliana. Nucleic Acids Res. 2021, 49. [Google Scholar] [CrossRef] [PubMed]
- Marmier-Gourrier, N.; Cléry, A.; Senty-Ségault, V.; Charpentier, B.; Schlotter, F.; Leclerc, F.; Fournier, R.; Branlant, C. A Structural, Phylogenetic, and Functional Study of 15.5-KD/Snu13 Protein Binding on U3 Small Nucleolar RNA. RNA 2003, 9, 821–838. [Google Scholar] [CrossRef] [Green Version]
- Rothé, B.; Back, R.; Quinternet, M.; Bizarro, J.; Robert, M.-C.; Blaud, M.; Romier, C.; Manival, X.; Charpentier, B.; Bertrand, E.; et al. Characterization of the Interaction between Protein Snu13p/15.5K and the Rsa1p/NUFIP Factor and Demonstration of Its Functional Importance for SnoRNP Assembly. Nucleic Acids Res. 2014, 42, 2015–2036. [Google Scholar] [CrossRef] [Green Version]
- Rothé, B.; Manival, X.; Rolland, N.; Charron, C.; Senty-Ségault, V.; Branlant, C.; Charpentier, B. Implication of the Box C/D SnoRNP Assembly Factor Rsa1p in U3 SnoRNP Assembly. Nucleic Acids Res. 2017, 45, 7455–7473. [Google Scholar] [CrossRef] [Green Version]
- Lübben, B.; Marshallsay, C.; Rottmann, N.; Lührmann, R. Isolation of U3 SnoRNP from CHO Cells: A Novel 55 KDa Protein Binds to the Central Part of U3 SnoRNA. Nucleic Acids Res. 1993, 21, 5377–5385. [Google Scholar] [CrossRef] [Green Version]
- Venema, J.; Vos, H.R.; Faber, A.W.; van Venrooij, W.J.; Raué, H.A. Yeast Rrp9p Is an Evolutionarily Conserved U3 SnoRNP Protein Essential for Early Pre-RRNA Processing Cleavages and Requires Box C for Its Association. RNA 2000, 6, 1660–1671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pluk, H.; Soffner, J.; Lührmann, R.; van Venrooij, W.J. CDNA Cloning and Characterization of the Human U3 Small Nucleolar Ribonucleoprotein Complex-Associated 55-Kilodalton Protein. Mol. Cell. Biol. 1998, 18, 488–498. [Google Scholar] [CrossRef] [Green Version]
- Lukowiak, A.A.; Granneman, S.; Mattox, S.A.; Speckmann, W.A.; Jones, K.; Pluk, H.; Venrooij, W.J.; Terns, R.M.; Terns, M.P. Interaction of the U3-55k Protein with U3 SnoRNA Is Mediated by the Box B/C Motif of U3 and the WD Repeats of U3-55 k. Nucleic Acids Res. 2000, 28, 3462–3471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cléry, A.; Senty-Ségault, V.; Leclerc, F.; Raué, H.A.; Branlant, C. Analysis of Sequence and Structural Features That Identify the B/C Motif of U3 Small Nucleolar RNA as the Recognition Site for the Snu13p-Rrp9p Protein Pair. Mol. Cell. Biol. 2007, 27, 1191–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granneman, S.; Pruijn, G.J.M.; Horstman, W.; van Venrooij, W.J.; Luhrmann, R.; Watkins, N.J. The HU3-55K Protein Requires 15.5 K Binding to the Box B/C Motif as Well as Flanking RNA Elements for Its Association with the U3 Small Nucleolar RNA in Vitro. J. Biol. Chem. 2002, 277, 48490–48500. [Google Scholar] [CrossRef] [Green Version]
- Granneman, S.; Kudla, G.; Petfalski, E.; Tollervey, D. Identification of Protein Binding Sites on U3 SnoRNA and Pre-RRNA by UV Cross-Linking and High-Throughput Analysis of CDNAs. Proc. Natl. Acad. Sci. USA 2009, 106, 9613–9618. [Google Scholar] [CrossRef] [Green Version]
- Knox, A.A.; McKeegan, K.S.; Debieux, C.M.; Traynor, A.; Richardson, H.; Watkins, N.J. A Weak C’ Box Renders U3 SnoRNA Levels Dependent on HU3-55K Binding. Mol. Cell. Biol. 2011, 31, 2404–2412. [Google Scholar] [CrossRef] [Green Version]
- Watkins, N.J.; Lemm, I.; Ingelfinger, D.; Schneider, C.; Hossbach, M.; Urlaub, H.; Lührmann, R. Assembly and Maturation of the U3 SnoRNP in the Nucleoplasm in a Large Dynamic Multiprotein Complex. Mol. Cell 2004, 16, 789–798. [Google Scholar] [CrossRef]
- Chaker-Margot, M.; Barandun, J.; Hunziker, M.; Klinge, S. Architecture of the Yeast Small Subunit Processome. Science 2017, 355. [Google Scholar] [CrossRef] [PubMed]
- Kornprobst, M.; Turk, M.; Kellner, N.; Cheng, J.; Flemming, D.; Koš-Braun, I.; Koš, M.; Thoms, M.; Berninghausen, O.; Beckmann, R.; et al. Architecture of the 90S Pre-Ribosome: A Structural View on the Birth of the Eukaryotic Ribosome. Cell 2016, 166, 380–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Q.; Zhu, X.; Qi, J.; An, W.; Lan, P.; Tan, D.; Chen, R.; Wang, B.; Zheng, S.; Zhang, C.; et al. Molecular Architecture of the 90S Small Subunit Pre-Ribosome. eLife 2017, 6, e22086. [Google Scholar] [CrossRef] [PubMed]
- Méreau, A.; Fournier, R.; Grégoire, A.; Mougin, A.; Fabrizio, P.; Lührmann, R.; Branlant, C. An in Vivo and in Vitro Structure-Function Analysis of the Saccharomyces Cerevisiae U3A SnoRNP: Protein-RNA Contacts and Base-Pair Interaction with the Pre-Ribosomal RNA. J. Mol. Biol. 1997, 273, 552–571. [Google Scholar] [CrossRef]
- Borovjagin, A.V.; Gerbi, S.A. An Evolutionary Intra-Molecular Shift in the Preferred U3 SnoRNA Binding Site on Pre-Ribosomal RNA. Nucleic Acids Res. 2005, 33, 4995–5005. [Google Scholar] [CrossRef] [Green Version]
- Marmier-Gourrier, N.; Cléry, A.; Schlotter, F.; Senty-Ségault, V.; Branlant, C. A Second Base Pair Interaction between U3 Small Nucleolar RNA and the 5’-ETS Region Is Required for Early Cleavage of the Yeast Pre-Ribosomal RNA. Nucleic Acids Res. 2011, 39, 9731–9745. [Google Scholar] [CrossRef]
- Dutca, L.M.; Gallagher, J.E.G.; Baserga, S.J. The Initial U3 SnoRNA:Pre-RRNA Base Pairing Interaction Required for Pre-18S RRNA Folding Revealed by in Vivo Chemical Probing. Nucleic Acids Res. 2011, 39, 5164–5180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beltrame, M.; Tollervey, D. Base Pairing between U3 and the Pre-Ribosomal RNA Is Required for 18S RRNA Synthesis. EMBO J. 1995, 14, 4350–4356. [Google Scholar] [CrossRef]
- Paul, A.; Tiotiu, D.; Bragantini, B.; Marty, H.; Charpentier, B.; Massenet, S.; Labialle, S. Bcd1p Controls RNA Loading of the Core Protein Nop58 during C/D Box SnoRNP Biogenesis. RNA 2019, 25, 496–506. [Google Scholar] [CrossRef]
- Hirose, T.; Steitz, J.A. Position within the Host Intron Is Critical for Efficient Processing of Box C/D SnoRNAs in Mammalian Cells. Proc. Natl. Acad. Sci. USA 2001, 98, 12914–12919. [Google Scholar] [CrossRef] [Green Version]
- Vincenti, S.; De Chiara, V.; Bozzoni, I.; Presutti, C. The Position of Yeast SnoRNA-Coding Regions within Host Introns Is Essential for Their Biosynthesis and for Efficient Splicing of the Host Pre-MRNA. RNA 2007, 13, 138–150. [Google Scholar] [CrossRef] [Green Version]
- Hirose, T.; Shu, M.-D.; Steitz, J.A. Splicing-Dependent and -Independent Modes of Assembly for Intron-Encoded Box C/D SnoRNPs in Mammalian Cells. Mol. Cell 2003, 12, 113–123. [Google Scholar] [CrossRef]
- Richard, P.; Kiss, T. Integrating SnoRNP Assembly with MRNA Biogenesis. EMBO Rep. 2006, 7, 590–592. [Google Scholar] [CrossRef] [Green Version]
- De, I.; Bessonov, S.; Hofele, R.; dos Santos, K.; Will, C.L.; Urlaub, H.; Lührmann, R.; Pena, V. The RNA Helicase Aquarius Exhibits Structural Adaptations Mediating Its Recruitment to Spliceosomes. Nat. Struct. Mol. Biol. 2015, 22, 138–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirose, T.; Ideue, T.; Nagai, M.; Hagiwara, M.; Shu, M.-D.; Steitz, J.A. A Spliceosomal Intron Binding Protein, IBP160, Links Position-Dependent Assembly of Intron-Encoded Box C/D SnoRNP to Pre-MRNA Splicing. Mol. Cell 2006, 23, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Morlando, M.; Ballarino, M.; Greco, P.; Caffarelli, E.; Dichtl, B.; Bozzoni, I. Coupling between SnoRNP Assembly and 3′ Processing Controls Box C/D SnoRNA Biosynthesis in Yeast. EMBO J. 2004, 23, 2392–2401. [Google Scholar] [CrossRef] [Green Version]
- Rothé, B.; Saliou, J.-M.; Quinternet, M.; Back, R.; Tiotiu, D.; Jacquemin, C.; Loegler, C.; Schlotter, F.; Peña, V.; Eckert, K.; et al. Protein Hit1, a Novel Box C/D SnoRNP Assembly Factor, Controls Cellular Concentration of the Scaffolding Protein Rsa1 by Direct Interaction. Nucleic Acids Res. 2014, 42, 10731–10747. [Google Scholar] [CrossRef] [PubMed]
- Bizarro, J.; Charron, C.; Boulon, S.; Westman, B.; Pradet-Balade, B.; Vandermoere, F.; Chagot, M.-E.; Hallais, M.; Ahmad, Y.; Leonhardt, H.; et al. Proteomic and 3D Structure Analyses Highlight the C/D Box SnoRNP Assembly Mechanism and Its Control. J. Cell Biol. 2014, 207, 463–480. [Google Scholar] [CrossRef] [PubMed]
- Boulon, S.; Marmier-Gourrier, N.; Pradet-Balade, B.; Wurth, L.; Verheggen, C.; Jády, B.E.; Rothé, B.; Pescia, C.; Robert, M.-C.; Kiss, T.; et al. The Hsp90 Chaperone Controls the Biogenesis of L7Ae RNPs through Conserved Machinery. J. Cell Biol. 2008, 180, 579–595. [Google Scholar] [CrossRef] [PubMed]
- McKeegan, K.S.; Debieux, C.M.; Boulon, S.; Bertrand, E.; Watkins, N.J. A Dynamic Scaffold of Pre-SnoRNP Factors Facilitates Human Box C/D SnoRNP Assembly. Mol. Cell. Biol. 2007, 27, 6782–6793. [Google Scholar] [CrossRef] [Green Version]
- McKeegan, K.S.; Debieux, C.M.; Watkins, N.J. Evidence That the AAA+ Proteins TIP48 and TIP49 Bridge Interactions between 15.5K and the Related NOP56 and NOP58 Proteins during Box C/D SnoRNP Biogenesis. Mol. Cell. Biol. 2009, 29, 4971–4981. [Google Scholar] [CrossRef] [Green Version]
- Bragantini, B.; Tiotiu, D.; Rothé, B.; Saliou, J.-M.; Marty, H.; Cianférani, S.; Charpentier, B.; Quinternet, M.; Manival, X. Functional and Structural Insights of the Zinc-Finger HIT Protein Family Members Involved in Box C/D SnoRNP Biogenesis. J. Mol. Biol. 2016, 428, 2488–2506. [Google Scholar] [CrossRef] [PubMed]
- Khoshnevis, S.; Dreggors, R.E.; Hoffmann, T.F.R.; Ghalei, H. A Conserved Bcd1 Interaction Essential for Box C/D SnoRNP Biogenesis. J. Biol. Chem. 2019, 294, 18360–18371. [Google Scholar] [CrossRef]
- Kiss, T.; Fayet, E.; Jády, B.E.; Richard, P.; Weber, M. Biogenesis and Intranuclear Trafficking of Human Box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 407–417. [Google Scholar] [CrossRef] [Green Version]
- Quinternet, M.; Rothé, B.; Barbier, M.; Bobo, C.; Saliou, J.-M.; Jacquemin, C.; Back, R.; Chagot, M.-E.; Cianférani, S.; Meyer, P.; et al. Structure/Function Analysis of Protein-Protein Interactions Developed by the Yeast Pih1 Platform Protein and Its Partners in Box C/D SnoRNP Assembly. J. Mol. Biol. 2015, 427, 2816–2839. [Google Scholar] [CrossRef] [PubMed]
- Quinternet, M.; Chagot, M.-E.; Rothé, B.; Tiotiu, D.; Charpentier, B.; Manival, X. Structural Features of the Box C/D SnoRNP Pre-Assembly Process Are Conserved through Species. Structure 2016, 24, 1693–1706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henri, J.; Chagot, M.-E.; Bourguet, M.; Abel, Y.; Terral, G.; Maurizy, C.; Aigueperse, C.; Georgescauld, F.; Vandermoere, F.; Saint-Fort, R.; et al. Deep Structural Analysis of RPAP3 and PIH1D1, Two Components of the HSP90 Co-Chaperone R2TP Complex. Structure 2018, 26, 1196–1209.e8. [Google Scholar] [CrossRef] [Green Version]
- Zhao, R.; Kakihara, Y.; Gribun, A.; Huen, J.; Yang, G.; Khanna, M.; Costanzo, M.; Brost, R.L.; Boone, C.; Hughes, T.R.; et al. Molecular Chaperone Hsp90 Stabilizes Pih1/Nop17 to Maintain R2TP Complex Activity That Regulates SnoRNA Accumulation. J. Cell Biol. 2008, 180, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Tycowski, K.T.; Shu, M.-D.; Kukoyi, A.; Steitz, J.A. A Conserved WD40 Protein Binds the Cajal Body Localization Signal of ScaRNP Particles. Mol. Cell 2009, 34, 47–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marnef, A.; Richard, P.; Pinzón, N.; Kiss, T. Targeting Vertebrate Intron-Encoded Box C/D 2′-O-Methylation Guide RNAs into the Cajal Body. Nucleic Acids Res. 2014, 42, 6616–6629. [Google Scholar] [CrossRef] [Green Version]
- Izumikawa, K.; Nobe, Y.; Ishikawa, H.; Yamauchi, Y.; Taoka, M.; Sato, K.; Nakayama, H.; Simpson, R.J.; Isobe, T.; Takahashi, N. TDP-43 Regulates Site-Specific 2′-O-Methylation of U1 and U2 SnRNAs via Controlling the Cajal Body Localization of a Subset of C/D ScaRNAs. Nucleic Acids Res. 2019, 47, 2487–2505. [Google Scholar] [CrossRef] [Green Version]
- Boulon, S.; Verheggen, C.; Jady, B.E.; Girard, C.; Pescia, C.; Paul, C.; Ospina, J.K.; Kiss, T.; Matera, A.G.; Bordonné, R.; et al. PHAX and CRM1 Are Required Sequentially to Transport U3 SnoRNA to Nucleoli. Mol. Cell 2004, 16, 777–787. [Google Scholar] [CrossRef]
- Machyna, M.; Kehr, S.; Straube, K.; Kappei, D.; Buchholz, F.; Butter, F.; Ule, J.; Hertel, J.; Stadler, P.F.; Neugebauer, K.M. The Coilin Interactome Identifies Hundreds of Small Noncoding RNAs That Traffic through Cajal Bodies. Mol. Cell 2014, 56, 389–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pradet-Balade, B.; Girard, C.; Boulon, S.; Paul, C.; Azzag, K.; Bordonné, R.; Bertrand, E.; Verheggen, C. CRM1 Controls the Composition of Nucleoplasmic Pre-SnoRNA Complexes to Licence Them for Nucleolar Transport. EMBO J. 2011, 30, 2205–2218. [Google Scholar] [CrossRef] [Green Version]
- Verheggen, C.; Mouaikel, J.; Thiry, M.; Blanchard, J.M.; Tollervey, D.; Bordonné, R.; Lafontaine, D.L.; Bertrand, E. Box C/D Small Nucleolar RNA Trafficking Involves Small Nucleolar RNP Proteins, Nucleolar Factors and a Novel Nuclear Domain. EMBO J. 2001, 20, 5480–5490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verheggen, C.; Bertrand, E. CRM1 Plays a Nuclear Role in Transporting SnoRNPs to Nucleoli in Higher Eukaryotes. Nucleus 2012, 3, 132–137. [Google Scholar] [CrossRef] [Green Version]
- Bortolin-Cavaillé, M.-L.; Cavaillé, J. The SNORD115 (H/MBII-52) and SNORD116 (H/MBII-85) Gene Clusters at the Imprinted Prader-Willi Locus Generate Canonical Box C/D SnoRNAs. Nucleic Acids Res. 2012, 40, 6800–6807. [Google Scholar] [CrossRef] [PubMed]
- Vitali, P.; Royo, H.; Marty, V.; Bortolin-Cavaillé, M.-L.; Cavaillé, J. Long Nuclear-Retained Non-Coding RNAs and Allele-Specific Higher-Order Chromatin Organization at Imprinted SnoRNA Gene Arrays. J. Cell Sci. 2010, 123 Pt 1, 70–83. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.-O.; Yin, Q.-F.; Wang, H.-B.; Zhang, Y.; Chen, T.; Zheng, P.; Lu, X.; Chen, L.-L.; Yang, L. Species-Specific Alternative Splicing Leads to Unique Expression of Sno-LncRNAs. BMC Genomics 2014, 15, 287. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Yin, Q.-F.; Luo, Z.; Yao, R.-W.; Zheng, C.-C.; Zhang, J.; Xiang, J.-F.; Yang, L.; Chen, L.-L. Unusual Processing Generates SPA LncRNAs That Sequester Multiple RNA Binding Proteins. Mol. Cell 2016, 64, 534–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Zhang, J.; Wang, M.; Li, X.; Gong, H.; Tang, H.; Chen, L.; Wan, L.; Liu, Q. Activity Dependent LoNA Regulates Translation by Coordinating RRNA Transcription and Methylation. Nat. Commun. 2018, 9, 1726. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, S.; Ardal, B.K.; Hollensen, A.K.; Damgaard, C.K.; Jensen, T.H. Box C/D SnoRNP Autoregulation by a Cis-Acting SnoRNA in the NOP56 Pre-MRNA. Mol. Cell 2018, 72, 99–111.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deschamps-Francoeur, G.; Garneau, D.; Dupuis-Sandoval, F.; Roy, A.; Frappier, M.; Catala, M.; Couture, S.; Barbe-Marcoux, M.; Abou-Elela, S.; Scott, M.S. Identification of Discrete Classes of Small Nucleolar RNA Featuring Different Ends and RNA Binding Protein Dependency. Nucleic Acids Res. 2014, 42, 10073–10085. [Google Scholar] [CrossRef]
- Zywicki, M.; Bakowska-Zywicka, K.; Polacek, N. Revealing Stable Processing Products from Ribosome-Associated Small RNAs by Deep-Sequencing Data Analysis. Nucleic Acids Res. 2012, 40, 4013–4024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saraiya, A.A.; Wang, C.C. SnoRNA, a Novel Precursor of MicroRNA in Giardia Lamblia. PLoS Pathog. 2008, 4, e10000224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, H.; Sun, Q.; Yao, Y. Characterization of Small RNAs Derived from TRNAs, RRNAs and SnoRNAs and Their Response to Heat Stress in Wheat Seedlings. PLoS ONE 2016, 11, e0150933. [Google Scholar] [CrossRef] [Green Version]
- Kishore, S.; Khanna, A.; Zhang, Z.; Hui, J.; Balwierz, P.J.; Stefan, M.; Beach, C.; Nicholls, R.D.; Zavolan, M.; Stamm, S. The SnoRNA MBII-52 (SNORD 115) Is Processed into Smaller RNAs and Regulates Alternative Splicing. Hum. Mol. Genet. 2010, 19, 1153–1164. [Google Scholar] [CrossRef] [Green Version]
- Ender, C.; Krek, A.; Friedländer, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A Human SnoRNA with MicroRNA-Like Functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Hutzinger, R.; Feederle, R.; Mrazek, J.; Schiefermeier, N.; Balwierz, P.J.; Zavolan, M.; Polacek, N.; Delecluse, H.-J.; Hüttenhofer, A. Expression and Processing of a Small Nucleolar RNA from the Epstein-Barr Virus Genome. PLoS Pathog. 2009, 5, e10000547. [Google Scholar] [CrossRef] [Green Version]
- Poole, A.R.; Vicino, I.; Adachi, H.; Yu, Y.-T.; Hebert, M.D. Regulatory RNPs: A Novel Class of Ribonucleoproteins That Potentially Contribute to Ribosome Heterogeneity. Biol. Open 2017, 6, 1342–1354. [Google Scholar] [CrossRef] [Green Version]
- Chow, R.D.; Chen, S. Sno-Derived RNAs Are Prevalent Molecular Markers of Cancer Immunity. Oncogene 2018, 37, 6442–6462. [Google Scholar] [CrossRef] [PubMed]
- Martens-Uzunova, E.S.; Hoogstrate, Y.; Kalsbeek, A.; Pigmans, B.; Vredenbregt-van den Berg, M.; Dits, N.; Nielsen, S.J.; Baker, A.; Visakorpi, T.; Bangma, C.; et al. C/D-Box SnoRNA-Derived RNA Production Is Associated with Malignant Transformation and Metastatic Progression in Prostate Cancer. Oncotarget 2015, 6, 17430–17444. [Google Scholar] [CrossRef] [Green Version]
- Mleczko, A.M.; Machtel, P.; Walkowiak, M.; Wasilewska, A.; Pietras, P.J.; Bąkowska-Żywicka, K. Levels of SdRNAs in Cytoplasm and Their Association with Ribosomes Are Dependent upon Stress Conditions but Independent from SnoRNA Expression. Sci. Rep. 2019, 9, 18397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, B.; Yegnasubramanian, S.; Wheelan, S.J.; Laiho, M. RNA-Seq of the Nucleolus Reveals Abundant SNORD44-Derived Small RNAs. PLoS ONE 2014, 9, e107519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taft, R.J.; Glazov, E.A.; Lassmann, T.; Hayashizaki, Y.; Carninci, P.; Mattick, J.S. Small RNAs Derived from SnoRNAs. RNA 2009, 15, 1233–1240. [Google Scholar] [CrossRef] [Green Version]
- Guillen-Chable, F.; Corona, U.R.; Pereira-Santana, A.; Bayona, A.; Rodríguez-Zapata, L.C.; Aquino, C.; Šebestová, L.; Vitale, N.; Hozak, P.; Castano, E. Fibrillarin Ribonuclease Activity Is Dependent on the GAR Domain and Modulated by Phospholipids. Cells 2020, 9, 1143. [Google Scholar] [CrossRef]
- Lemus-Diaz, N.; Ferreira, R.R.; Bohnsack, K.E.; Gruber, J.; Bohnsack, M.T. The Human Box C/D SnoRNA U3 Is a MiRNA Source and MiR-U3 Regulates Expression of Sortin Nexin 27. Nucleic Acids Res. 2020, 48, 8074–8089. [Google Scholar] [CrossRef]
- Zhong, F.; Zhou, N.; Wu, K.; Guo, Y.; Tan, W.; Zhang, H.; Zhang, X.; Geng, G.; Pan, T.; Luo, H.; et al. A SnoRNA-Derived PiRNA Interacts with Human Interleukin-4 Pre-MRNA and Induces Its Decay in Nuclear Exosomes. Nucleic Acids Res. 2015, 43, 10474–10491. [Google Scholar] [CrossRef] [Green Version]
- Langenberger, D.; Çakir, M.V.; Hoffmann, S.; Stadler, P.F. Dicer-Processed Small RNAs: Rules and Exceptions. J. Exp. Zool. B Mol. Dev. Evol. 2013, 320, 35–46. [Google Scholar] [CrossRef]
- Logan, M.K.; Burke, M.F.; Hebert, M.D. Altered Dynamics of ScaRNA2 and ScaRNA9 in Response to Stress Correlates with Disrupted Nuclear Organization. Biol. Open 2018, 7. [Google Scholar] [CrossRef] [Green Version]
- McLaurin, D.M.; Logan, M.K.; Lett, K.E.; Hebert, M.D. Molecular Determinants That Govern ScaRNA Processing by Drosha/DGCR8. Biol. Open 2020, 9. [Google Scholar] [CrossRef]
- Kishore, S.; Gruber, A.R.; Jedlinski, D.J.; Syed, A.P.; Jorjani, H.; Zavolan, M. Insights into SnoRNA Biogenesis and Processing from PAR-CLIP of SnoRNA Core Proteins and Small RNA Sequencing. Genome Biol. 2013, 14, R45. [Google Scholar] [CrossRef] [Green Version]
- Lykke-Andersen, S.; Chen, Y.; Ardal, B.R.; Lilje, B.; Waage, J.; Sandelin, A.; Jensen, T.H. Human Nonsense-Mediated RNA Decay Initiates Widely by Endonucleolysis and Targets SnoRNA Host Genes. Genes Dev. 2014, 28, 2498–2517. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Ashraf, S.; Wang, J.; Lilley, D.M. Control of Box C/D SnoRNP Assembly by N6-Methylation of Adenine. EMBO Rep. 2017, 18, 1631–1645. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, S.; Huang, L.; Lilley, D.M.J. Effect of Methylation of Adenine N6 on Kink Turn Structure Depends on Location. RNA Biol. 2019, 16, 1377–1385. [Google Scholar] [CrossRef] [Green Version]
- Westman, B.J.; Verheggen, C.; Hutten, S.; Lam, Y.W.; Bertrand, E.; Lamond, A.I. A Proteomic Screen for Nucleolar SUMO Targets Shows SUMOylation Modulates the Function of Nop5/Nop58. Mol. Cell 2010, 39, 618–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westman, B.J.; Lamond, A.I. A Role for SUMOylation in SnoRNP Biogenesis Revealed by Quantitative Proteomics. Nucleus 2011, 2, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Qin, W.; Lv, P.; Fan, X.; Quan, B.; Zhu, Y.; Qin, K.; Chen, Y.; Wang, C.; Chen, X. Quantitative Time-Resolved Chemoproteomics Reveals That Stable O-GlcNAc Regulates Box C/D SnoRNP Biogenesis. Proc. Natl. Acad. Sci. USA 2017, 114, E6749–E6758. [Google Scholar] [CrossRef] [Green Version]
- Kakihara, Y.; Makhnevych, T.; Zhao, L.; Tang, W.; Houry, W.A. Nutritional Status Modulates Box C/D SnoRNP Biogenesis by Regulated Subcellular Relocalization of the R2TP Complex. Genome Biol. 2014, 15, 404. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Wu, Z.J.; Groner, A.C.; He, H.H.; Cai, C.; Lis, R.T.; Wu, X.; Stack, E.C.; Loda, M.; Liu, T.; et al. EZH2 Oncogenic Activity in Castration-Resistant Prostate Cancer Cells Is Polycomb-Independent. Science 2012, 338, 1465–1469. [Google Scholar] [CrossRef] [Green Version]
- Yi, Y.; Li, Y.; Meng, Q.; Li, Q.; Li, F.; Lu, B.; Shen, J.; Fazli, L.; Zhao, D.; Li, C.; et al. A PRC2-Independent Function for EZH2 in Regulating RRNA 2’-O Methylation and IRES-Dependent Translation. Nat. Cell Biol. 2021, 23. [Google Scholar] [CrossRef]
- Nachmani, D.; Bothmer, A.H.; Grisendi, S.; Mele, A.; Bothmer, D.; Lee, J.D.; Monteleone, E.; Cheng, K.; Zhang, Y.; Bester, A.C.; et al. Germline NPM1 Mutations Lead to Altered RRNA 2′-O-Methylation and Cause Dyskeratosis Congenita. Nat. Genet. 2019, 51, 1518–1529. [Google Scholar] [CrossRef]
- Bragantini, B.; Charron, C.; Bourguet, M.; Paul, A.; Tiotiu, D.; Rothé, B.; Marty, H.; Terral, G.; Hessmann, S.; Decourty, L.; et al. The Box C/D SnoRNP Assembly Factor Bcd1 Interacts with the Histone Chaperone Rtt106 and Controls Its Transcription Dependent Activity. Nat. Commun. 2021, 12, 1859. [Google Scholar] [CrossRef]
- de los Santos-Velázquez, A.I.; de Oya, I.G.; Manzano-López, J.; Monje-Casas, F. Late RDNA Condensation Ensures Timely Cdc14 Release and Coordination of Mitotic Exit Signaling with Nucleolar Segregation. Curr. Biol. 2017, 27, 3248–3263.e5. [Google Scholar] [CrossRef] [PubMed]
- Doyon, Y.; Selleck, W.; Lane, W.S.; Tan, S.; Côté, J. Structural and Functional Conservation of the NuA4 Histone Acetyltransferase Complex from Yeast to Humans. Mol. Cell. Biol. 2004, 24, 1884–1896. [Google Scholar] [CrossRef] [Green Version]
- Kobor, M.S.; Venkatasubrahmanyam, S.; Meneghini, M.D.; Gin, J.W.; Jennings, J.L.; Link, A.J.; Madhani, H.D.; Rine, J. A Protein Complex Containing the Conserved Swi2/Snf2-Related ATPase Swr1p Deposits Histone Variant H2A.Z into Euchromatin. PLoS Biol. 2004, 2, E131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Cai, Y.; Jin, J.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Conaway, J.W.; Conaway, R.C. Subunit Organization of the Human INO80 Chromatin Remodeling Complex: An Evolutionarily Conserved Core Complex Catalyzes ATP-Dependent Nucleosome Remodeling. J. Biol. Chem. 2011, 286, 11283–11289. [Google Scholar] [CrossRef] [Green Version]
- Houry, W.A.; Bertrand, E.; Coulombe, B. The PAQosome, an R2TP-Based Chaperone for Quaternary Structure Formation. Trends Biochem. Sci. 2018, 43, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Kressler, D.; Doère, M.; Rojo, M.; Linder, P. Synthetic Lethality with Conditional Dbp6 Alleles Identifies Rsa1p, a Nucleoplasmic Protein Involved in the Assembly of 60S Ribosomal Subunits. Mol. Cell. Biol. 1999, 19, 8633–8645. [Google Scholar] [CrossRef] [Green Version]
- Soeno, Y.; Taya, Y.; Stasyk, T.; Huber, L.A.; Aoba, T.; Hüttenhofer, A. Identification of Novel Ribonucleo-Protein Complexes from the Brain-Specific SnoRNA MBII-52. RNA 2010, 16, 1293–1300. [Google Scholar] [CrossRef] [Green Version]
- Falaleeva, M.; Pages, A.; Matuszek, Z.; Hidmi, S.; Agranat-Tamir, L.; Korotkov, K.; Nevo, Y.; Eyras, E.; Sperling, R.; Stamm, S. Dual Function of C/D Box Small Nucleolar RNAs in RRNA Modification and Alternative Pre-MRNA Splicing. Proc. Natl. Acad. Sci. USA 2016, 113, E1625–E1634. [Google Scholar] [CrossRef] [Green Version]
- Diao, L.-T.; Xiao, Z.-D.; Leng, X.-M.; Li, B.; Li, J.-H.; Luo, Y.-P.; Li, S.-G.; Yu, C.-H.; Zhou, H.; Qu, L.-H. Conservation and Divergence of Transcriptional Coregulations between Box C/D SnoRNA and Ribosomal Protein Genes in Ascomycota. RNA 2014, 20, 1376–1385. [Google Scholar] [CrossRef] [Green Version]
- Nabavi, S.; Nazar, R.N. U3 SnoRNA Promoter Reflects the RNA’s Function in Ribosome Biogenesis. Curr. Genet. 2008, 54, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Herter, E.K.; Stauch, M.; Gallant, M.; Wolf, E.; Raabe, T.; Gallant, P. SnoRNAs Are a Novel Class of Biologically Relevant Myc Targets. BMC Biol. 2015, 13, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walz, S.; Lorenzin, F.; Morton, J.; Wiese, K.E.; von Eyss, B.; Herold, S.; Rycak, L.; Dumay-Odelot, H.; Karim, S.; Bartkuhn, M.; et al. Activation and Repression by Oncogenic MYC Shape Tumour-Specific Gene Expression Profiles. Nature 2014, 511, 483–487. [Google Scholar] [CrossRef]
- Dieci, G.; Preti, M.; Montanini, B. Eukaryotic SnoRNAs: A Paradigm for Gene Expression Flexibility. Genomics 2009, 94, 83–88. [Google Scholar] [CrossRef] [Green Version]
- Pelczar, P.; Filipowicz, W. The Host Gene for Intronic U17 Small Nucleolar RNAs in Mammals Has No Protein-Coding Potential and Is a Member of the 5′-Terminal Oligopyrimidine Gene Family. Mol. Cell. Biol. 1998, 18, 4509–4518. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.M.; Steitz, J.A. Classification of Gas5 as a Multi-Small-Nucleolar-RNA (SnoRNA) Host Gene and a Member of the 5’-Terminal Oligopyrimidine Gene Family Reveals Common Features of SnoRNA Host Genes. Mol. Cell. Biol. 1998, 18, 6897–6909. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, R.; Suzuki, Y.; Takeuchi, N.; Wakaguri, H.; Ueda, T.; Sugano, S.; Nakai, K. Comprehensive Detection of Human Terminal Oligo-Pyrimidine (TOP) Genes and Analysis of Their Characteristics. Nucleic Acids Res. 2008, 36, 3707–3715. [Google Scholar] [CrossRef]
- Jalal, C.; Uhlmann-Schiffler, H.; Stahl, H. Redundant Role of DEAD Box Proteins P68 (Ddx5) and P72/P82 (Ddx17) in Ribosome Biogenesis and Cell Proliferation. Nucleic Acids Res. 2007, 35, 3590–3601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismael, H.; Altmeyer, S.; Stahl, H. Regulation of the U3-, U8-, and U13snoRNA Expression by the DEAD Box Proteins Ddx5/Ddx17 with Consequences for Cell Proliferation and Survival. Noncoding RNA 2016, 2, 11. [Google Scholar] [CrossRef]
- Gong, J.; Li, Y.; Liu, C.; Xiang, Y.; Li, C.; Ye, Y.; Zhang, Z.; Hawke, D.H.; Park, P.K.; Diao, L.; et al. A Pan-Cancer Analysis of the Expression and Clinical Relevance of Small Nucleolar RNAs in Human Cancer. Cell Rep. 2017, 21, 1968–1981. [Google Scholar] [CrossRef] [Green Version]
- Valleron, W.; Ysebaert, L.; Berquet, L.; Fataccioli, V.; Quelen, C.; Martin, A.; Parrens, M.; Lamant, L.; de Leval, L.; Gisselbrecht, C.; et al. Small Nucleolar RNA Expression Profiling Identifies Potential Prognostic Markers in Peripheral T-Cell Lymphoma. Blood 2012, 120, 3997–4005. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Xu, T.; Ganapathy, S.; Shadfan, M.; Long, M.; Huang, T.H.-M.; Thompson, I.; Yuan, Z.-M. Elevated SnoRNA Biogenesis Is Essential in Breast Cancer. Oncogene 2014, 33, 1348–1358. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.T.; Farzaneh, F. Are SnoRNAs and SnoRNA Host Genes New Players in Cancer? Nat. Rev. Cancer 2012, 12, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.-Y.; Guo, P.; Boyd, J.; Sun, X.; Li, Q.; Zhou, W.; Dong, J.-T. Implication of SnoRNA U50 in Human Breast Cancer. J. Genet. Genomics 2009, 36, 447–454. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.-Y.; Rodriguez, C.; Guo, P.; Sun, X.; Talbot, J.T.; Zhou, W.; Petros, J.; Li, Q.; Vessella, R.L.; Kibel, A.S.; et al. SnoRNA U50 Is a Candidate Tumor Suppressor Gene at 6q14.3 with a Mutation Associated with Clinically Significant Prostate Cancer. Hum. Mol. Genet. 2008, 17, 1031–1042. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Ye, M.-H.; Lv, S.-G.; Wang, Q.-X.; Wu, M.-J.; Xiao, B.; Kang, C.-S.; Zhu, X.-G. SNORD47, a Box C/D SnoRNA, Suppresses Tumorigenesis in Glioblastoma. Oncotarget 2017, 8, 43953–43966. [Google Scholar] [CrossRef]
- Zhu, W.; Niu, J.; He, M.; Zhang, L.; Lv, X.; Liu, F.; Jiang, L.; Zhang, J.; Yu, Z.; Zhao, L.; et al. SNORD89 Promotes Stemness Phenotype of Ovarian Cancer Cells by Regulating Notch1-c-Myc Pathway. J. Transl. Med. 2019, 17, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Zhao, L.; Wu, H.; Tian, G.; Dai, S.; Zhao, R.; Shan, B. C/D-Box Snord105b Promotes Tumorigenesis in Gastric Cancer via ALDOA/C-Myc Pathway. CPB 2018, 45, 2471–2482. [Google Scholar] [CrossRef] [Green Version]
- Cavaillé, J. Box C/D Small Nucleolar RNA Genes and the Prader-Willi Syndrome: A Complex Interplay. WIREs RNA 2017, 8, e1417. [Google Scholar] [CrossRef]
- Badrock, A.P.; Uggenti, C.; Wacheul, L.; Crilly, S.; Jenkinson, E.M.; Rice, G.I.; Kasher, P.R.; Lafontaine, D.L.J.; Crow, Y.J.; O’Keefe, R.T. Analysis of U8 SnoRNA Variants in Zebrafish Reveals How Bi-Allelic Variants Cause Leukoencephalopathy with Calcifications and Cysts. Am. J. Hum. Genet. 2020, 106, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Saxena, T.; Tandon, B.; Sharma, S.; Chameettachal, S.; Ray, P.; Ray, A.R.; Kulshreshtha, R. Combined MiRNA and MRNA Signature Identifies Key Molecular Players and Pathways Involved in Chikungunya Virus Infection in Human Cells. PLoS ONE 2013, 8, e79886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juranic Lisnic, V.; Babic Cac, M.; Lisnic, B.; Trsan, T.; Mefferd, A.; Das Mukhopadhyay, C.; Cook, C.H.; Jonjic, S.; Trgovcich, J. Dual Analysis of the Murine Cytomegalovirus and Host Cell Transcriptomes Reveal New Aspects of the Virus-Host Cell Interface. PLoS Pathog. 2013, 9, e1006311. [Google Scholar] [CrossRef] [PubMed]
- Fleming, D.S.; Miller, L.C. Identification of Small Non-Coding RNA Classes Expressed in Swine Whole Blood during HP-PRRSV Infection. Virology 2018, 517, 56–61. [Google Scholar] [CrossRef]
- Murray, J.L.; Sheng, J.; Rubin, D.H. A Role for H/ACA and C/D Small Nucleolar RNAs in Viral Replication. Mol. Biotechnol. 2014, 56, 429–437. [Google Scholar] [CrossRef]
- Stamm, S.; Lodmell, J.S. C/D Box SnoRNAs in Viral Infections: RNA Viruses Use Old Dogs for New Tricks. Noncoding RNA Res. 2019, 4, 46–53. [Google Scholar] [CrossRef]
- Deng, W.; Zhu, X.; Skogerbø, G.; Zhao, Y.; Fu, Z.; Wang, Y.; He, H.; Cai, L.; Sun, H.; Liu, C.; et al. Organization of the Caenorhabditis Elegans Small Non-Coding Transcriptome: Genomic Features, Biogenesis, and Expression. Genome Res. 2006, 16, 20–29. [Google Scholar] [CrossRef] [Green Version]
- Hazen, S.P.; Naef, F.; Quisel, T.; Gendron, J.M.; Chen, H.; Ecker, J.R.; Borevitz, J.O.; Kay, S.A. Exploring the Transcriptional Landscape of Plant Circadian Rhythms Using Genome Tiling Arrays. Genome Biol. 2009, 10, R17. [Google Scholar] [CrossRef] [Green Version]
- Cervantes, M.; Forné, I.; Ranjit, S.; Gratton, E.; Imhof, A.; Sassone-Corsi, P. BMAL1 Associates with NOP58 in the Nucleolus and Contributes to Pre-RRNA Processing. iScience 2020, 23, 101151. [Google Scholar] [CrossRef]
- Cavaillé, J.; Buiting, K.; Kiefmann, M.; Lalande, M.; Brannan, C.I.; Horsthemke, B.; Bachellerie, J.P.; Brosius, J.; Hüttenhofer, A. Identification of Brain-Specific and Imprinted Small Nucleolar RNA Genes Exhibiting an Unusual Genomic Organization. Proc. Natl. Acad. Sci. USA 2000, 97, 14311–14316. [Google Scholar] [CrossRef] [Green Version]
- Cavaillé, J.; Vitali, P.; Basyuk, E.; Hüttenhofer, A.; Bachellerie, J.P. A Novel Brain-Specific Box C/D Small Nucleolar RNA Processed from Tandemly Repeated Introns of a Noncoding RNA Gene in Rats. J. Biol. Chem. 2001, 276, 26374–26383. [Google Scholar] [CrossRef] [Green Version]
- Cavaillé, J.; Seitz, H.; Paulsen, M.; Ferguson-Smith, A.C.; Bachellerie, J.-P. Identification of Tandemly-Repeated C/D SnoRNA Genes at the Imprinted Human 14q32 Domain Reminiscent of Those at the Prader-Willi/Angelman Syndrome Region. Hum. Mol. Genet. 2002, 11, 1527–1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erales, J.; Marchand, V.; Panthu, B.; Gillot, S.; Belin, S.; Ghayad, S.E.; Garcia, M.; Laforêts, F.; Marcel, V.; Baudin-Baillieu, A.; et al. Evidence for RRNA 2′-O-Methylation Plasticity: Control of Intrinsic Translational Capabilities of Human Ribosomes. Proc. Natl. Acad. Sci. USA 2017, 114, 12934–12939. [Google Scholar] [CrossRef] [Green Version]
- Hebras, J.; Krogh, N.; Marty, V.; Nielsen, H.; Cavaillé, J. Developmental Changes of RRNA Ribose Methylations in the Mouse. RNA Biol. 2020, 17, 150–164. [Google Scholar] [CrossRef]
- Belin, S.; Beghin, A.; Solano-Gonzàlez, E.; Bezin, L.; Brunet-Manquat, S.; Textoris, J.; Prats, A.-C.; Mertani, H.C.; Dumontet, C.; Diaz, J.-J. Dysregulation of Ribosome Biogenesis and Translational Capacity Is Associated with Tumor Progression of Human Breast Cancer Cells. PLoS ONE 2009, 4, e7147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcel, V.; Ghayad, S.E.; Belin, S.; Therizols, G.; Morel, A.-P.; Solano-Gonzàlez, E.; Vendrell, J.A.; Hacot, S.; Mertani, H.C.; Albaret, M.A.; et al. P53 Acts as a Safeguard of Translational Control by Regulating Fibrillarin and RRNA Methylation in Cancer. Cancer Cell 2013, 24, 318–330. [Google Scholar] [CrossRef] [Green Version]
- Krogh, N.; Jansson, M.D.; Häfner, S.J.; Tehler, D.; Birkedal, U.; Christensen-Dalsgaard, M.; Lund, A.H.; Nielsen, H. Profiling of 2′-O-Me in Human RRNA Reveals a Subset of Fractionally Modified Positions and Provides Evidence for Ribosome Heterogeneity. Nucleic Acids Res. 2016, 44, 7884–7895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janin, M.; Coll-SanMartin, L.; Esteller, M. Disruption of the RNA Modifications That Target the Ribosome Translation Machinery in Human Cancer. Mol. Cancer 2020, 19, 70. [Google Scholar] [CrossRef] [Green Version]
- Patil, P.; Kibiryeva, N.; Uechi, T.; Marshall, J.; O’Brien, J.E.; Artman, M.; Kenmochi, N.; Bittel, D.C. ScaRNAs Regulate Splicing and Vertebrate Heart Development. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2015, 1852, 1619–1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitali, P.; Kiss, T. Cooperative 2′-O-Methylation of the Wobble Cytidine of Human Elongator TRNAMet(CAT) by a Nucleolar and a Cajal Body-Specific Box C/D RNP. Genes Dev. 2019, 33, 741–746. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.K.; Gurha, P.; Tran, E.J.; Maxwell, E.S.; Gupta, R. Sequential 2′-O-Methylation of Archaeal Pre-TRNATrp Nucleotides Is Guided by the Intron-Encoded but Trans-Acting Box C/D Ribonucleoprotein of Pre-TRNA. J. Biol. Chem. 2004, 279, 47661–47671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clouet d’Orval, B.; Bortolin, M.L.; Gaspin, C.; Bachellerie, J.P. Box C/D RNA Guides for the Ribose Methylation of Archaeal TRNAs. The TRNATrp Intron Guides the Formation of Two Ribose-Methylated Nucleosides in the Mature TRNATrp. Nucleic Acids Res. 2001, 29, 4518–4529. [Google Scholar] [CrossRef] [Green Version]
- Dennis, P.P.; Omer, A.; Lowe, T. A Guided Tour: Small RNA Function in Archaea. Mol. Microbiol. 2001, 40, 509–519. [Google Scholar] [CrossRef] [Green Version]
- Elliott, B.A.; Ho, H.-T.; Ranganathan, S.V.; Vangaveti, S.; Ilkayeva, O.; Abou Assi, H.; Choi, A.K.; Agris, P.F.; Holley, C.L. Modification of Messenger RNA by 2′-O-Methylation Regulates Gene Expression in Vivo. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Deryusheva, S.; Gall, J.G. Small Cajal Body-Specific RNAs of Drosophila Function in the Absence of Cajal Bodies. Mol. Biol. Cell 2009, 20, 5250–5259. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Yang, J.; van Nues, R.; Watzinger, P.; Kötter, P.; Lafontaine, D.L.J.; Granneman, S.; Entian, K.-D. Specialized Box C/D SnoRNPs Act as Antisense Guides to Target RNA Base Acetylation. PLoS Genet. 2017, 13, e1006804. [Google Scholar] [CrossRef] [PubMed]
- Bazeley, P.S.; Shepelev, V.; Talebizadeh, Z.; Butler, M.G.; Fedorova, L.; Filatov, V.; Fedorov, A. SnoTARGET Shows That Human Orphan SnoRNA Targets Locate Close to Alternative Splice Junctions. Gene 2008, 408, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Kehr, S.; Bartschat, S.; Stadler, P.F.; Tafer, H. PLEXY: Efficient Target Prediction for Box C/D SnoRNAs. Bioinformatics 2011, 27, 279–280. [Google Scholar] [CrossRef] [Green Version]
- Sharma, E.; Sterne-Weiler, T.; O’Hanlon, D.; Blencowe, B.J. Global Mapping of Human RNA-RNA Interactions. Mol. Cell 2016, 62, 618–626. [Google Scholar] [CrossRef] [Green Version]
- Kishore, S.; Stamm, S. The SnoRNA HBII-52 Regulates Alternative Splicing of the Serotonin Receptor 2C. Science 2006, 311, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Vitali, P.; Basyuk, E.; Le Meur, E.; Bertrand, E.; Muscatelli, F.; Cavaillé, J.; Huttenhofer, A. ADAR2-Mediated Editing of RNA Substrates in the Nucleolus Is Inhibited by C/D Small Nucleolar RNAs. J. Cell Biol. 2005, 169, 745–753. [Google Scholar] [CrossRef]
- Doe, C.M.; Relkovic, D.; Garfield, A.S.; Dalley, J.W.; Theobald, D.E.H.; Humby, T.; Wilkinson, L.S.; Isles, A.R. Loss of the Imprinted SnoRNA Mbii-52 Leads to Increased 5htr2c Pre-RNA Editing and Altered 5HT2CR-Mediated Behaviour. Hum. Mol. Genet. 2009, 18, 2140–2148. [Google Scholar] [CrossRef]
- Brüning, L.; Hackert, P.; Martin, R.; Davila Gallesio, J.; Aquino, G.R.R.; Urlaub, H.; Sloan, K.E.; Bohnsack, M.T. RNA Helicases Mediate Structural Transitions and Compositional Changes in Pre-Ribosomal Complexes. Nat. Commun. 2018, 9, 5383. [Google Scholar] [CrossRef]
- Sardana, R.; Liu, X.; Granneman, S.; Zhu, J.; Gill, M.; Papoulas, O.; Marcotte, E.M.; Tollervey, D.; Correll, C.C.; Johnson, A.W. The DEAH-Box Helicase Dhr1 Dissociates U3 from the Pre-RRNA to Promote Formation of the Central Pseudoknot. PLoS Biol. 2015, 13, e1002083. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, P.; Hackert, P.; Memet, I.; Sloan, K.E.; Bohnsack, M.T. The Human RNA Helicase DHX37 Is Required for Release of the U3 SnoRNP from Pre-Ribosomal Particles. RNA Biol. 2019, 16, 54–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aquino, G.R.R.; Krogh, N.; Hackert, P.; Martin, R.; Gallesio, J.D.; van Nues, R.W.; Schneider, C.; Watkins, N.J.; Nielsen, H.; Bohnsack, K.E.; et al. RNA Helicase-Mediated Regulation of SnoRNP Dynamics on Pre-Ribosomes and RRNA 2′-O-Methylation. Nucleic Acids Res. 2021, 49. [Google Scholar] [CrossRef]
- Leeds, N.B.; Small, E.C.; Hiley, S.L.; Hughes, T.R.; Staley, J.P. The Splicing Factor Prp43p, a DEAH Box ATPase, Functions in Ribosome Biogenesis. Mol. Cell. Biol. 2006, 26, 513–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasler, D.; Meduri, R.; Bąk, M.; Lehmann, G.; Heizinger, L.; Wang, X.; Li, Z.-T.; Sement, F.M.; Bruckmann, A.; Dock-Bregeon, A.-C.; et al. The Alazami Syndrome-Associated Protein LARP7 Guides U6 Small Nuclear RNA Modification and Contributes to Splicing Robustness. Mol. Cell 2020, 77, 1014–1031.e13. [Google Scholar] [CrossRef]
- Huang, C.; Shi, J.; Guo, Y.; Huang, W.; Huang, S.; Ming, S.; Wu, X.; Zhang, R.; Ding, J.; Zhao, W.; et al. A SnoRNA Modulates MRNA 3′ End Processing and Regulates the Expression of a Subset of MRNAs. Nucleic Acids Res. 2017, 45, 8647–8660. [Google Scholar] [CrossRef] [Green Version]
- Siprashvili, Z.; Webster, D.E.; Johnston, D.; Shenoy, R.M.; Ungewickell, A.J.; Bhaduri, A.; Flockhart, R.; Zarnegar, B.J.; Che, Y.; Meschi, F.; et al. The Noncoding RNAs SNORD50A and SNORD50B Bind K-Ras and Are Recurrently Deleted in Human Cancer. Nat. Genet. 2016, 48, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Che, Y.; Siprashvili, Z.; Kovalski, J.R.; Jiang, T.; Wozniak, G.; Elcavage, L.; Khavari, P.A. KRAS Regulation by Small Non-Coding RNAs and SNARE Proteins. Nat. Commun. 2019, 10, 5118. [Google Scholar] [CrossRef]
- Su, X.; Feng, C.; Wang, S.; Shi, L.; Gu, Q.; Zhang, H.; Lan, X.; Zhao, Y.; Qiang, W.; Ji, M.; et al. The Noncoding RNAs SNORD50A and SNORD50B-Mediated TRIM21-GMPS Interaction Promotes the Growth of P53 Wild-Type Breast Cancers by Degrading P53. Cell Death Differ. 2021. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-S.; Camacho, C.V.; Nagari, A.; Malladi, V.S.; Challa, S.; Kraus, W.L. Activation of PARP-1 by SnoRNAs Controls Ribosome Biogenesis and Cell Growth via the RNA Helicase DDX21. Mol. Cell 2019, 75, 1270–1285.e14. [Google Scholar] [CrossRef]
- D’Souza, M.N.; Gowda, N.K.C.; Tiwari, V.; Babu, R.O.; Anand, P.; Dastidar, S.G.; Singh, R.; Selvaraja, B.; Pal, R.; Ramesh, A.; et al. FMRP Interacts with C/D Box SnoRNA in the Nucleus and Regulates Ribosomal RNA Methylation. iScience 2018, 9, 399–411. [Google Scholar] [CrossRef] [Green Version]
- Chen, E.; Sharma, M.R.; Shi, X.; Agrawal, R.K.; Joseph, S. Fragile X Mental Retardation Protein Regulates Translation by Binding Directly to the Ribosome. Mol. Cell 2014, 54, 407–417. [Google Scholar] [CrossRef] [Green Version]
- Simsek, D.; Tiu, G.C.; Flynn, R.A.; Byeon, G.W.; Leppek, K.; Xu, A.F.; Chang, H.Y.; Barna, M. The Mammalian Ribo-Interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell 2017, 169, 1051–1065.e18. [Google Scholar] [CrossRef] [Green Version]
- Brandis, K.A.; Gale, S.; Jinn, S.; Langmade, S.J.; Dudley-Rucker, N.; Jiang, H.; Sidhu, R.; Ren, A.; Goldberg, A.; Schaffer, J.E.; et al. Box C/D Small Nucleolar RNA (SnoRNA) U60 Regulates Intracellular Cholesterol Trafficking. J. Biol. Chem. 2013, 288, 35703–35713. [Google Scholar] [CrossRef] [Green Version]
- Michel, C.I.; Holley, C.L.; Scruggs, B.S.; Sidhu, R.; Brookheart, R.T.; Listenberger, L.L.; Behlke, M.A.; Ory, D.S.; Schaffer, J.E. Small Nucleolar RNAs U32a, U33 and U35a Are Critical Mediators of Metabolic Stress. Cell Metab. 2011, 14, 33–44. [Google Scholar] [CrossRef] [Green Version]
- Li, M.W.; Sletten, A.C.; Lee, J.; Pyles, K.D.; Matkovich, S.J.; Ory, D.S.; Schaffer, J.E. Nuclear Export Factor 3 Regulates Localization of Small Nucleolar RNAs. J. Biol. Chem. 2017, 292, 20228–20239. [Google Scholar] [CrossRef] [Green Version]
- Huo, C.-Y.; Chang, M.-L.; Cheng, H.; Ma, T.-T.; Fu, Y.; Wang, Y.; Wang, Y.-Y.; Kan, Y.-C.; Li, D.-D. Small Nucleolar RNA of Silkworm Can Translocate from the Nucleolus to the Cytoplasm under Abiotic Stress. Cell Biol. Int. 2021, 45. [Google Scholar] [CrossRef] [PubMed]
- Rimer, J.M.; Lee, J.; Holley, C.L.; Crowder, R.J.; Chen, D.L.; Hanson, P.I.; Ory, D.S.; Schaffer, J.E. Long-Range Function of Secreted Small Nucleolar RNAs That Direct 2′-O-Methylation. J. Biol. Chem. 2018, 293, 13284–13296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanford, J.R.; Wang, X.; Mort, M.; Vanduyn, N.; Cooper, D.N.; Mooney, S.D.; Edenberg, H.J.; Liu, Y. Splicing Factor SFRS1 Recognizes a Functionally Diverse Landscape of RNA Transcripts. Genome Res. 2009, 19, 381–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Änkö, M.-L.; Müller-McNicoll, M.; Brandl, H.; Curk, T.; Gorup, C.; Henry, I.; Ule, J.; Neugebauer, K.M. The RNA-Binding Landscapes of Two SR Proteins Reveal Unique Functions and Binding to Diverse RNA Classes. Genome Biol. 2012, 13, R17. [Google Scholar] [CrossRef] [Green Version]
- Sperling, R. Small Non-Coding RNA within the Endogenous Spliceosome and Alternative Splicing Regulation. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194406. [Google Scholar] [CrossRef]
- Schubert, T.; Pusch, M.C.; Diermeier, S.; Benes, V.; Kremmer, E.; Imhof, A.; Längst, G. Df31 Protein and SnoRNAs Maintain Accessible Higher-Order Structures of Chromatin. Mol. Cell 2012, 48, 434–444. [Google Scholar] [CrossRef] [Green Version]
- Dudnakova, T.; Dunn-Davies, H.; Peters, R.; Tollervey, D. Mapping Targets for Small Nucleolar RNAs in Yeast. Wellcome Open Res. 2018, 3. [Google Scholar] [CrossRef]
- Labialle, S.; Cavaillé, J. Do Repeated Arrays of Regulatory Small-RNA Genes Elicit Genomic Imprinting? Concurrent Emergence of Large Clusters of Small Non-Coding RNAs and Genomic Imprinting at Four Evolutionarily Distinct Eutherian Chromosomal Loci. Bioessays 2011, 33, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, T.; del Gaudio, D.; German, J.R.; Shinawi, M.; Peters, S.U.; Person, R.E.; Garnica, A.; Cheung, S.W.; Beaudet, A.L. Prader-Willi Phenotype Caused by Paternal Deficiency for the HBII-85 C/D Box Small Nucleolar RNA Cluster. Nat. Genet. 2008, 40, 719–721. [Google Scholar] [CrossRef] [Green Version]
- de Smith, A.J.; Purmann, C.; Walters, R.G.; Ellis, R.J.; Holder, S.E.; Van Haelst, M.M.; Brady, A.F.; Fairbrother, U.L.; Dattani, M.; Keogh, J.M.; et al. A Deletion of the HBII-85 Class of Small Nucleolar RNAs (SnoRNAs) Is Associated with Hyperphagia, Obesity and Hypogonadism. Hum. Mol. Genet. 2009, 18, 3257–3265. [Google Scholar] [CrossRef]
- Duker, A.L.; Ballif, B.C.; Bawle, E.V.; Person, R.E.; Mahadevan, S.; Alliman, S.; Thompson, R.; Traylor, R.; Bejjani, B.A.; Shaffer, L.G.; et al. Paternally Inherited Microdeletion at 15q11.2 Confirms a Significant Role for the SNORD116 C/D Box SnoRNA Cluster in Prader-Willi Syndrome. Eur. J. Hum. Genet. 2010, 18, 1196–1201. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Potter, K.J.; Burnett, L.C.; Orsso, C.E.; Inman, M.; Ryman, D.C.; Haqq, A.M. Prader-Willi-Like Phenotype Caused by an Atypical 15q11.2 Microdeletion. Genes 2020, 11, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skryabin, B.V.; Gubar, L.V.; Seeger, B.; Pfeiffer, J.; Handel, S.; Robeck, T.; Karpova, E.; Rozhdestvensky, T.S.; Brosius, J. Deletion of the MBII-85 SnoRNA Gene Cluster in Mice Results in Postnatal Growth Retardation. PLoS Genet. 2007, 3, e235. [Google Scholar] [CrossRef]
- Ding, F.; Li, H.H.; Zhang, S.; Solomon, N.M.; Camper, S.A.; Cohen, P.; Francke, U. SnoRNA Snord116 (Pwcr1/MBII-85) Deletion Causes Growth Deficiency and Hyperphagia in Mice. PLoS ONE 2008, 3, e1709. [Google Scholar] [CrossRef] [Green Version]
- Polex-Wolf, J.; Lam, B.Y.; Larder, R.; Tadross, J.; Rimmington, D.; Bosch, F.; Cenzano, V.J.; Ayuso, E.; Ma, M.K.; Rainbow, K.; et al. Hypothalamic Loss of Snord116 Recapitulates the Hyperphagia of Prader-Willi Syndrome. J. Clin. Investig. 2018, 128, 960–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adhikari, A.; Copping, N.A.; Onaga, B.; Pride, M.C.; Coulson, R.L.; Yang, M.; Yasui, D.H.; LaSalle, J.M.; Silverman, J.L. Cognitive Deficits in the Snord116 Deletion Mouse Model for Prader-Willi Syndrome. Neurobiol. Learn. Mem. 2019, 165, 106874. [Google Scholar] [CrossRef]
- Lui, L.M.; Uzilov, A.V.; Bernick, D.L.; Corredor, A.; Lowe, T.M.; Dennis, P.P. Methylation Guide RNA Evolution in Archaea: Structure, Function and Genomic Organization of 110 C/D Box SRNA Families across Six Pyrobaculum Species. Nucleic Acids Res. 2018, 46, 5678–5691. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baldini, L.; Charpentier, B.; Labialle, S. Emerging Data on the Diversity of Molecular Mechanisms Involving C/D snoRNAs. Non-Coding RNA 2021, 7, 30. https://doi.org/10.3390/ncrna7020030
Baldini L, Charpentier B, Labialle S. Emerging Data on the Diversity of Molecular Mechanisms Involving C/D snoRNAs. Non-Coding RNA. 2021; 7(2):30. https://doi.org/10.3390/ncrna7020030
Chicago/Turabian StyleBaldini, Laeya, Bruno Charpentier, and Stéphane Labialle. 2021. "Emerging Data on the Diversity of Molecular Mechanisms Involving C/D snoRNAs" Non-Coding RNA 7, no. 2: 30. https://doi.org/10.3390/ncrna7020030
APA StyleBaldini, L., Charpentier, B., & Labialle, S. (2021). Emerging Data on the Diversity of Molecular Mechanisms Involving C/D snoRNAs. Non-Coding RNA, 7(2), 30. https://doi.org/10.3390/ncrna7020030






